Simple Summary
The methanogenesis pathway via methanogens dates back to the Hadean or Archaean Earth. These methanogens are considered to have a thermophilic origin and are presently ubiquitously distributed across anaerobic environments. The class Methanobacteria comprises methanogens that are found extensively in geothermal environments, such as hot springs and hydrothermal vents, and their evolutionary history and how they adapted to different temperatures remain unclear. In this study, we isolated a novel species of the class Methanobacteria from a natural hot spring in Tengchong, China. This species can produce methane, utilizing hydrogen and carbon dioxide, at 65 °C. In addition, we found that members of the class Methanobacteria originated in a geothermal niche and then evolved to adapt to ambient temperatures; during this process, thermal adaptation genes were lost and a wide range of metabolic genes were acquired. This research on methanogen evolution will help us understand how life originated in geothermal environments and then spread extensively across present-day Earth.
Abstract
Methanogens can produce methane in anaerobic environments via the methanogenesis pathway, and are regarded as one of the most ancient life forms on Earth. They are ubiquitously distributed across distinct ecosystems and are considered to have a thermophilic origin. In this study, we isolated, pure cultured, and completely sequenced a single methanogen strain DL9LZB001, from a hot spring at Tengchong in Southwest China. DL9LZB001 is a thermophilic and hydrogenotrophic methanogen with an optimum growth temperature of 65 °C. It is a putative novel species, which has been named Methanothermobacter tengchongensis—a Class I methanogen belonging to the class Methanobacteria. Comparative genomic and ancestral analyses indicate that the class Methanobacteria originated in a hyperthermal environment and then evolved to adapt to ambient temperatures. This study extends the understanding of methanogens living in geothermal niches, as well as the origin and evolutionary history of these organisms in ecosystems with different temperatures.
1. Introduction
Methanogens are considered one of the most ancient life forms on Earth, with an evolutionary history dating back more than 3.4 billion years [1,2]. The methanogenesis pathway is considered one of the most ancient metabolic pathways and may even be shared by the last universal common ancestor (LUCA) [3]. The methanogenesis pathway can be classified into four major types, based on metabolic substrate utilization, i.e., hydrogenotrophic (H2/CO2 and formate), methylotrophic (methanol, methylamine, methanethiol), acetoclastic (acetate), and other complex carbon sources such as coal [4,5]. It has been estimated that about one billion tons of methane are produced via these methanogenesis pathways and enter the atmosphere every year, accounting for ~1.6% of annual global carbon fixation [6]. Methane is the second most potent greenhouse gas on Earth and significantly influences global climate change [7]. Therefore, methanogens are crucial for the stability of Earth’s ecosystems [6], and increasing attention has recently been focused on methanogens.
Previous studies have shown that the methanogenesis pathway may have existed in the common ancestor of the Euryarchaeota and TACK superphyla, including Ca. Verstraetearchaeota [8], Ca. Bathyarchaeota [9], Ca. Nezhaarchaeota and Thaumarchaeota [10,11], close to the last common archaea ancestor [12], which has been postulated to be a thermophilic methanogen [8,9,10,11,13]. However, none of the TACK methanogens have been pure cultured, and their actual optimum growth temperatures have not been verified. Pure cultured methanogen strains to date have only been from the phylum Euryarchaeota, and can be categorized into three major groups: (1) the Class I methanogens, which include three classes (Methanobacteria, Methanococci, and Methanopyri); (2) Class II methanogens, referred to as the class Methanomicrobia; (3) Class III methanogens, including the order Methanomassiliicoccales, Ca. Methanofastidiosales and Ca. Nuwarchaeales [12,14,15]. Most Class II and III methanogens are found in temperate environments, while many of the Class I methanogens are thermophilic and can be found in hydrothermal vents or hot springs [12,14,15]. Within the Euryarchaeota group, the class Methanobacteria—a Class I methanogen—comprises thermophilic and non-thermophilic members [16,17,18,19,20]; however, the evolutionary history of this ancient methanogen branch remains elusive.
The isolated strains in the class Methanobacteria have diverse temperature adaptation ranges and are divided into five genera: Methanothermus, Methanothermobacter, Methanobrevibacter, Methanosphaera, and Methanobacterium [16,17,18,19,20]. The deep-branching genus Methanothermus can grow on H2 and CO2, and its optimum growth temperature range is 80–88 °C [20,21]. In contrast, the optimum growth temperature for Methanothermobacter is 55–70 °C, with H2 or formate and CO2 as metabolic substrates [17,22]. However, the genera Methanobrevibacter, Methanosphaera, and Methanobacterium have optimum growth temperature ranges of 28–40 °C [18,23,24,25]. It remains unclear whether the class Methanobacteria has a thermophilic origin or how their descendants adapted to temperate environments. In this study, we isolated and pure cultured a single Methanobacteria strain DL9LZB001, of the genus Methanothermobacter, from a hot spring in Tengchong, China. DL9LZB001 is postulated as a novel species and has been named Methanothermobacter tengchongensis. We sequenced and analyzed its genome to deepen our understanding of the evolutionary history of the class Methanobacteria.
2. Materials and Methods
2.1. Sampling, Enrichment, Isolation, and Pure Culturing
Our samples were collected from a hot spring in Tengchong, China (98.82350° N, 25.04233° W) in January 2020. The temperature of the sampling site was 65.7 °C, and the pH was 6.66. Sediments and upper water were collected and stored in anaerobic bottles with nitrogen as the headspace.
During the primary enrichment, we mixed each set of sediments collected with its corresponding upper water sample in an anaerobic tank. We transferred 100 mL of the mixture into 125 mL sterile anaerobic bottles with butyl rubber stoppers and then amended it with various substrates, such as methylamine, sodium formate, and methanol. We pressurized H2 (100%, 10 psi) into these bottles and maintained them at 65 °C for approximately one year. Then, we inoculated the primary enriched product into a modified MB medium [26], and each group was enriched with the corresponding previously amended substrates. The modified MB medium contained the following, per liter: 900 mL distilled water, 1.00 g NH4Cl, 0.30 g K2HPO4·3H2O, 0.30 g KH2PO4, 0.20 g KCl, 0.50 g MgCl2·6H2O, 2.00 g NaCl, 0.05 g CaCl2, 1.00 mL resazurin solution (0.5 g·L−1), and 1.00 mL trace element solution [26,27]. After the medium was autoclaved and then cooled, we transferred a 100 mL mixture solution (containing 1.00 mL vitamin mixture solution [27], 4.00 g bicarbonate, and 0.5 g Na2S·9H2O) into the medium via a 0.22 μm filter in the anaerobic tank. The pH value was regulated to ~7. We then inoculated approximately 5 mL of enriched product into a 95 mL modified MB medium in 125 mL bottles in an anaerobic tank, maintained at 65 °C for 30 days before the next round of enrichment.
The strains were isolated using a Hungate rolling tube [28] by adding 2% gellan gum (w/v), 1.00 g·L−1 yeast extract, 2.00 g·L−1 tryptone, 10 mM methylamine, and 10 mM sodium formate into the modified MB medium described earlier; H2 (100%, 10 psi) was then pressurized into the tube. Subsequently, all the colonies—with different colors, sizes, and shapes—from the middle of the tube were collected and transferred into a 5 mL modified MB medium with additional methylamine, sodium formate, yeast extract, and tryptone, in 20 mL bottles filled with H2/CO2 (80%/20%, 10 psi). We repeated this process three times and used antibiotics for purification, including ampicillin sodium, kanamycin sulfate, streptomycin sulfate, and vancomycin hydrochloride at the same final concentration of 200 mg·L−1. Finally, we serially cultured the strains in a modified MB medium with H2, CO2, and bicarbonate as the only energy and carbon sources. The cell concentrations were monitored spectrophotometrically at an optical density of 600 nm (OD600), with 1 mL double distilled water as the blank. The pH range for growth in the modified MB medium was determined to be 4.0–10.0, by using different pH buffers: (1) 0.1 M citrate and 0.1 M sodium citrate for pH 4 and pH 5; (2) 0.2 M Na2HPO4 and 0.2 M NaH2PO4 for pH 6, pH 7, and pH 8; (3) 0.1 M NaHCO3 and 0.1 M Na2CO3 for pH 9 and pH 10 [29]. The salinity range for cell growth was determined at NaCl concentrations of 2, 10, 20, 30, and 40 g·L−1, at 0.2%, 1%, 2%, 3%, and 4% (w/v), respectively.
2.2. Headspace Gas Analysis
The methanogenesis of the enrichments and cultures was monitored using a gas chromatograph (GC, Agilent, California, USA) and a flame ionization detector (FID). The injection temperature, column temperature, and FID temperature were 60 °C, 80 °C, and 300 °C, respectively. Before monitoring the samples, we tested pure methane (99.9%) at concentrations of 0%, 20%, 40%, 60%, 80%, and 100% to establish a standard curve between various concentrations of methane and the peak areas of the GC results. Subsequently, the methane concentrations in the headspace of the samples were calculated using the results for the corresponding peak area.
2.3. Microscopy Observation
The coenzyme F420 contains a fluorescent component [30], and the oxidation states of the coenzyme F420 can absorb 420 nm ultraviolet light and excite 470 nm blue-green fluorescence [30]. Isolated cells from pure cultures were observed under a fluorescence microscope (BX63 Olympus, Tokyo, Japan) with 405 nm ultraviolet light as the light source. For observation under electron microscopes, we centrifuged the cell suspension at 6000× g for one minute and washed it three times with 0.5 mL sterile water. We then fixed the cells on a silicon pellet illuminated by a white light source and observed them under a scanning electron microscope (VEGA 3, TESCAN, Brno-Kohoutovice, Czech Republic). Alternatively, the cells were fixed on copper grids and observed under a transmission electron microscope (Tecnai G2, Thermo Fisher Scientific, Waltham, MA, USA).
2.4. ToF-SIMS Analysis
Cell morphology and composition were imaged using time-of-flight secondary ion mass spectrometry (ToF-SIMS) analysis. ToF-SIMS can directly distinguish between different cell-surface components because of its high mass resolution and spatial resolution. For this study, we used ION-ToF ToF-SIMS 5 at the Shanghai Jiao Tong University Instrument Analysis Centre, and the pressure of the analysis chamber was maintained below 1.1 × 10−9 mbar. The fast imaging mode with a pulsed 30 keV Bi3+ (0.16~0.18 pA pulsed current) ion beam was applied for high lateral resolution mapping (<100 nm) analysis, which showed the 2D or 3D distribution of elements and molecules in cells; the typical analysis area was 50 × 50 μm2, the lateral resolution as low as 60 nm, and the sensitivity as high as ppm–ppb. Through spectrometry analysis, the specific peaks of target cells, i.e., C2SO2+, PO3−, and PO2−, were identified. The distribution of C2SO2+, PO3−, and PO2− acquired by the SIMS mapping method only instructed the distribution of target cells, and even some non-target cells existing in samples, as cells’ chemical composition could be differentiated at the same position. Therefore, ToF-SIMS analysis can be seen as a combination between electron microscopy and mass spectrum analysis, and is presented as a supplementary certification of the TEM/SEM results.
2.5. DNA Extraction, Sequencing, and Analysis
DNA was extracted from the pure culture using a Bacterial Genome DNA Rapid Extraction Kit (Huiling Biotechnology Co., Ltd., Shanghai, China), following the manufacturer’s instructions. The exact concentration of the DNA was measured using a Qubit 4 Fluorometer (Thermo Fisher Scientific, Waltham, MA, USA), and DNA purity and integrity were detected via agarose gel electrophoresis. Genome sequencing was completed using PacBio sequencing technology and Illumina PE150 sequencing for high-accuracy assembly. The genome was assembled using SMRT Link software (version 5.0.1) [31], and the protein-coding genes were predicted using GeneMarkS software (Version 4.17) [32]. The tRNA was predicted by the software tRNAscan-SE (version 1.3.1) [33], the rRNA genes were predicted using RNAmmer software (version 1.2) [34], and small RNA was identified against the Rfam database [35] using CMsearch (version 1.1rc4) [36]. The genomic island was predicted using IslandPath-DIOMB (version 0.2) [37], and the CRISPR (clustered regularly interspaced short palindromic repeat) sequences were predicted using CRISPRdigger software (version 1.0) [38]. A display of the entire genomic map was generated using Circos (version 0.69) [39], along with the prediction results of the protein-coding genes.
2.6. Phylogenetic Analyses
The genome of our isolated strain and 233 reference genomes of representative archaea from the four superphyla, i.e., Euryarchaeota, TACK, Asgard, and DPANN, were employed to construct a phylogenomic tree based on the concatenated alignment of 37 marker genes [10,40]. The 233 reference genomes were downloaded from the NCBI prokaryote genome database [10]. Each of the 37 marker protein sequences translated from reference genomes was aligned using the rapid multiple sequence alignment program MAFFT (version 7.313), filtered using trimAI software (version 1.4. rev2) [10,41,42], and then concatenated into a single alignment. The phylogenetic tree was then constructed using IQ-Tree software (version 1.6.6) [43], with the model LG + C60 + F + G and a bootstrap value of 1000 [10]. We also downloaded from the NCBI database the completely sequenced genomes of all the isolated strains from the classes Thermococci, Methanobacteria, Methanococci, and Methanopyri, and collected in situ environmental and pure culture information on these genomes. A phylogenetic tree for evolutionary analysis was constructed using only the completely sequenced genomes of Class I methanogens, employing the same method.
2.7. Comparative Genomic and Ancestral Analyses
The amino acid identities (AAI) of the isolated strains of the classes Thermococci, Methanobacteria, Methanococci, and Methanopyri were calculated using the CompareM (version 0.0.23, https://github.com/dparks1134/CompareM (accessed on 12 May 2022)) software toolkit, and the average nucleotide identities (ANI) were calculated using fastANI software (version 1.33) [44]. The protein sequences of all the completely sequenced genomes were predicted using Prodigal gene prediction software (version 2.6.3) [45]. We then identified all the clusters of orthologous groups (COG) of these strains using Orthofinder software (version 2.5.4) [46], and annotated these COG using BlastKOALA (https://www.kegg.jp/blastkoala/ (accessed on 24 June 2022)). Combined with the phylogenetic tree of the isolated strains, the gene sets of the ancestor nodes were predicted using a posterior probability algorithm in Count software (version 9.1106 RC1) [47]. Finally, by comparing the gene sets of different ancestor nodes, we identified the lost and the gained gene sets of each predicted ancestor node.
3. Results and Discussion
3.1. Enrichment and Isolation of a Thermophilic Methanogen from a Hot Spring
The sediment and upper water samples were collected from a hot spring in Tengchong, China in January 2020 (Figure 1A). This hot spring (pH: 6.66, temperature: 65.7 °C) is a natural environment with abundant vegetation (Figure 1B). Initial enrichment was made using the collected sediment and corresponding upper water sample combined with potential methanogenesis substrates (for details, see Section 2.1) for approximately one year (Figure 1C). A modified MB medium was used in the subsequent enrichments to gradually remove the sediments from the medium (Figure 1C) [26]. To eliminate bacteria from the enrichments, cultures with H2/CO2 as the headspace gas and bicarbonate as the only carbon source in a liquid medium were established using four different kinds of antibiotics, and rapid methane production was detected. After this round of enrichment, the Hungate rolling tube technique was performed, and one strain of methanogens from this enrichment was successfully isolated; the colony was white, approximately 1–2 mm in diameter, and convex (Figure 1D). After another two rounds of colony isolation via the Hungate rolling tube, and several instances of continuous culture, the strain was purified by checking the cell morphology under an optical microscope and an electron microscope. During continuous culture, the only carbon sources in the medium were CO2 and bicarbonate, without any organics, indicating that the strain DL9LZB001 is autotrophic archaea.

Figure 1.
Sampling, enrichment, and isolation of methanogens from hot spring sediments. (A) Tengchong is located in the Yunnan province in Southwest China. (B) The sampling site was a natural hot spring and the sediments were brownish. (C,D) We enriched methanogens with methanogenesis substrates, and isolated methanogens using Hungate rolling tubes.
3.2. Morphology and Physiological Characteristics of Strain DL9LZB001
Strain DL9LZB001 was directly observed under a fluorescence microscope (for details, see Section 2.3 and Figure 2A). The DL9LZB001 cells were straight slender rods 2.80–6.43 μm in length (average length = 4.72 ± 1.01 μm, n = 9), with an average diameter of 0.436 ± 0.052 μm (n = 9). The cells often occurred in pairs, similar to Methanothermobacter marburgensis Marburg (Figure 2A–C). M. marburgensis Marburg is a pure-cultured strain isolated from the sewage sludge of an artificial digestor (Table 1) and has the closest ANI match to strain DL9LZB001 [48]. We did not observe a flagellum (Figure 2B–D), indicating that the strain DL9LZB001 is nonmotile, similar to most strains in the genus Methanothermobacter [17]. We performed ToF-SIMS analysis and 3D surface reconstruction. C2SO+, PO2−, and CN− fragments (Figure 2E–G) were employed to represent the cell components.
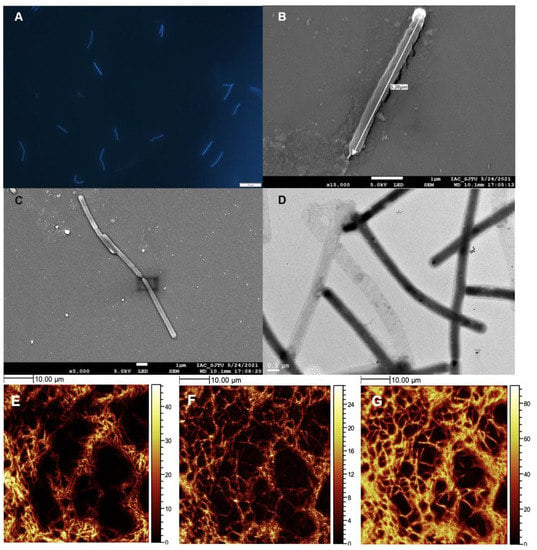
Figure 2.
Cells of Methanothermobacter tengchongensis DL9LZB001 under a microscope and in ToF-SIMS images. (A) Under a fluorescence microscope with 405 nm ultraviolet light as the light source. (B,C) Under a scanning electron microscope. The length of DL9LZB001 in these images is 5.26 μm, and the diameter is approximately 0.4 μm. (D) DL9LZB001 cells under a transmission electron microscope. (E–G) ToF-SIMS ion images of dried cultured cell aggregates showing their morphology and components. The lighter parts in the pictures are signals of (E) C2SO2+, (F) a combination of PO3−, PO2−, and PO−, and (G) CN−. The darker parts are sections without signal (without cells). The scale bar on the right displays the strength of the signal.

Table 1.
Descriptive and catabolic features of M. tengchongensis DL9LZB001 compared with other members of the genus Methanothermobacter.
The temperature range for growth was tested by measuring the turbidity (OD600) of the pure cultures at temperatures of 40, 50, 60, 65, 70, and 80 °C (Figure 3, n = 3). No growth was observed at 40 °C or 80 °C. The culture maintained at 65 °C grew better than those at 50, 60, and 70 °C during the exponential growth phase. Therefore, the optimum growth temperature for M. tengchongensis DL9LZB001 is approximately 65 °C, close to that of M. marburgensis Marburg (Table 1) and the environmental temperature of the sample location. The pH and salinity (NaCl%, w/v) ranges for growth at 65 °C for the strain DL9LZB001 are pH 6–8 and ≤1%, respectively (Table 1, n = 3); both values are less than those for M. marburgensis Marburg (Table 1).
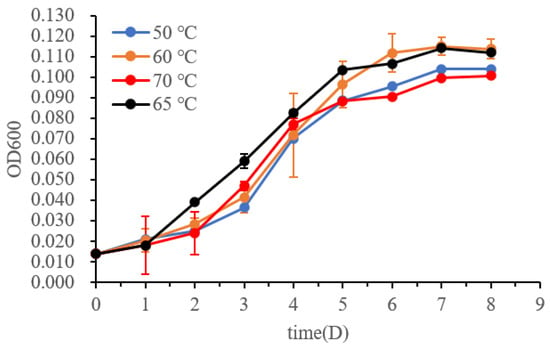
Figure 3.
Growth curves of M. tengchongensis DL9LZB001 at different temperatures.
3.3. General Genome Characteristics of the Strain DL9LZB001
Sequencing of the strain DL9LZB001 genome on the PacBio platforms generated 108,026 reads, accounting for 1,072,592,174 bases of the total sequence information. The final assembly and correction using Illumina PE150 produced a closed circular chromosome of DL9LZB001 with a size of 1,674,288 bases and a GC content of 48.39% (Figure 4). Gene prediction using GeneMarkS revealed 1802 protein-coding genes, accounting for 1,522,086 bases (90.91% of the chromosome size), and the number of tRNA genes was found to be 36 (Table 2). Using IslandPath-DIOMB we identified six genomic islands in the chromosome, accounting for 69,449 bases (4.15% of the chromosome size). No predicted prophage gene was identified in the chromosome; however, four CRISPR sequences were predicted by the CRISPRdigger software (Figure 4).
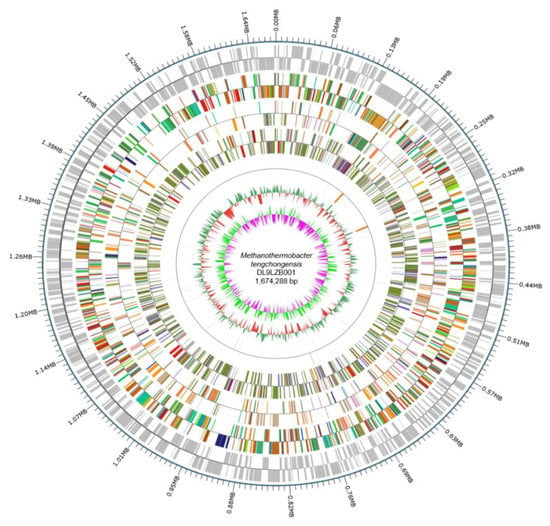
Figure 4.
The entire genome map of M. tengchongensis DL9LZB001. The outermost circle is the position of the genome sequence. From the outside to the inside: coding genes, gene function annotation results, ncRNA, genome GC contents, and genome GC skew values. We calculated the GC contents and GC skew values based on window bp (chromosome length/1000) and step bp (chromosome length/1000). The red inward part indicates that the GC content in that area is lower than the average of the entire genome, while the green outward part shows higher GC content. The heights of the peaks indicate the difference in value between the GC content of that area and the average GC content. The algorithm of the skew value is (G – C)/(G + C), and the pink inward part indicates that the content of G in that area is lower than that of C, while for the light green outward part the reverse applies. The heights of the peaks indicate the value difference.

Table 2.
General genome features of M. tengchongensis DL9LZB001.
To determine the phylogenetic position of the strain DL9LZB001, a phylogenetic tree was constructed based on 37 marker protein sequences with 233 representative archaea genomes from four superphyla: DPANN, Asgard, TACK, and Euryarchaeota (Supplementary Table S2) [10]. The constructed phylogenetic tree is rooted in the superphylum DPANN and displays only the phylogeny of the Class I methanogens. Among the Class I methanogens, the class Methanobacteria is clustered with the classes Methanopyri and Methanococci of the phylum Euryarchaeota (Figure 5). The strain DL9LZB001 is clustered with M. marburgensis Marburg and M. thermautotrophicus DeltaH within the genus Methanothermobacter (Figure 5). Genome comparison results indicate that the strain most closely related to DL9LZB001 is M. marburgensis Marburg; the ANI between M. marburgensis Marburg and strain DL9LZB001 is 94.13% with an AAI of 94.87% (Supplementary Tables S4 and S5), indicating that the strain DL9LZB001 may represent a novel species, named Methanothermobacter tengchongensis DL9LZB001.
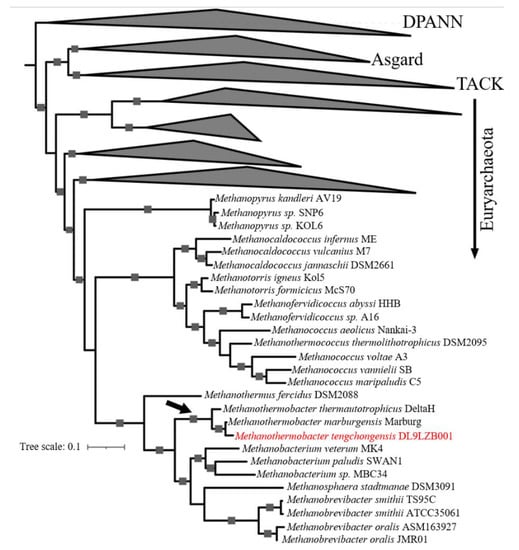
Figure 5.
Phylogenomic affiliation of M. tengchongensis DL9LZB001. We constructed the phylogenomic tree based on 37 marker-protein sequences, with 233 representative archaea genomes from four superphyla: (from top to bottom) DPANN, Asgard, TACK, and Euryarchaeota [10]. The tree is rooted in the superphylum DPANN. The position of the genus Methanothermobacter is labeled with a black arrow, and DL9LZB001 is highlighted in red. Bootstrap values larger than 80 are marked as squares on the tree branch.
The metabolic pathways of the strain DL9LZB001 were constructed based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) (Figure 6). DL9LZB001 uses CO2 or bicarbonate as the electron acceptor and H2 as the electron donor for the hydrogenotrophic methanogenesis pathway, which comprises seven independent enzymic reaction steps (Figure 6). Briefly, the CO2 is first reduced to formyl-methanofuran (formyl-MFR) by formylmethanofuran dehydrogenase (Fwd). The formyl group is then transferred to tetrahydromethanopterin (H4MPT). The formyl-H4MPT is dehydrated to methenyl-H4MPT, reduced to methylene-H4MPT, then reduced to methyl-H4MPT (Figure 6). Then, the methyl group is transferred to the coenzyme M (HS-CoM) by methyl-H4MPT methyltransferase (Mtr), a sodium ion transporter. The final and rate-limiting step of the methanogenesis pathway is the reduction of the methyl coenzyme M (methyl-S-CoM) by methyl-coenzyme M reductase (Mcr), with the coenzyme B (HS-CoB) as the electron donor. For recycling HS-CoM and HS-CoB, coenzyme M and coenzyme B heterodisulfide (CoM-S-S-CoB) are reduced by the heterodisulfide reductase (Hdr). During this process, three coenzyme F420 (reduced) molecules and one H2 molecule or four coenzyme F420 (reduced) molecules are consumed in total to generate one methane molecule (Figure 6). The reduction of coenzyme F420 (oxidized) is catalyzed by methyl-viologen-reducing hydrogenase (Mvh), with H2 as the electron donor. Therefore, four H2 molecules are consumed to generate one methane molecule. DL9LZB001 also contains formate dehydrogenase (Fdh), which can catalyze four formate molecules to four CO2 molecules and produce four coenzyme F420 (reduced) molecules for the subsequent generation of one methane molecule.
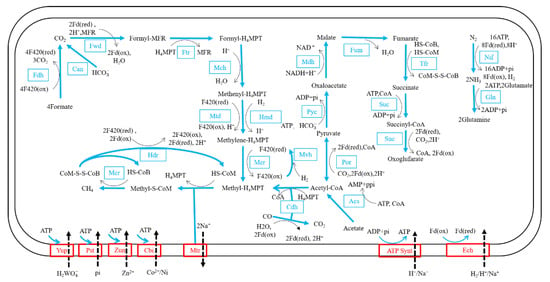
Figure 6.
Reconstructed metabolic pathways of M. tengchongensis DL9LZB001. We reconstructed the main pathways based on the KEGG annotation. All enzymes and reaction directions are marked in blue, except for enzymes belonging to transporters which are marked in red with dashed lines to show transporting directions.
Our annotation results indicate that DL9LZB001 may also use acetate for methanogenesis (Figure 6). The first step is acetate activation by acetyl-CoA synthetase (Acs) with ATP as the energy donor. The acetyl-CoA is then cleaved into methyl and carbonyl groups by acetyl-CoA decarbonylase (Cdh). However, we failed to detect methane production via pure culturing under 100% N2 of DL9LZB001 with 10 mM acetate and 5 g·L−1 bicarbonate as the only carbon source (65 °C, pH7, 0.2% NaCl). The known acetoclastic methanogens primarily belong to the order Methanosarcinales and the genus Methanothrix [52,53]; however, no acetate-utilizing ability has been reported in the genus Methanothermobacter. Therefore, the actual functions of Acs and Cdh require further research. The strain DL9LZB001 uses an incomplete tricarboxylic acid cycle for carbon fixation and may have the potential to transform nitrogen to ammonia (Figure 6). No intact sulfur pathway was discovered in this strain. We found only that it may have the potential to oxidize thiosulfate to sulfite using the enzyme thiosulfate sulfurtransferase, or to sulfate using S-sulfosulfanyl-L-cysteine sulfohydrolase. It also includes several ABC-type transporters for the uptake of tungstate, phosphate, zinc, cobalt, and nickel, with ATP as the energy driver (Figure 6).
3.4. The Evolutionary History of the Class Methanobacteria
For evolutionary analyses, we collected from the GTDB database all completely sequenced genomes of the isolated strains from the classes Thermococci, Methanobacteria, Methanococci, and Methanopyri (https://gtdb.ecogenomic.org/ (accessed on 26 February 2022), Supplementary Table S1), including the strain DL9LZB001. A phylogenetic tree was constructed, rooted in the class Thermococci (Figure 7). We also added to the phylogenetic tree the environmental information corresponding to each isolated strain (Figure 7). Most of the published pure cultured and completely sequenced strains of the genus Methanothermobacter were isolated from artificial environments (Figure 7), whereas DL9LZB001 was isolated from a natural hot spring (Figure 1). Adding this newly isolated strain from a natural environment to construct the evolutionary history of the class Methanobacteria may increase the habitat diversity on the ancestral node of the genus Methanothermobacter. All the COG of these genomes were identified using the Orthofinder software and were subsequently annotated. The gene sets of ancestor nodes were predicted by a posterior probability algorithm in the Count software, and the gene set of the common ancestor of the class Methanobacteria was constructed.
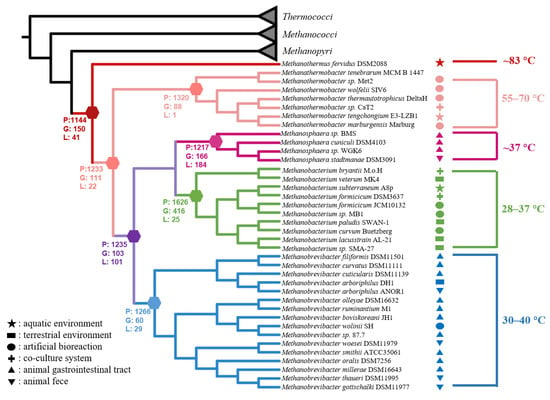
Figure 7.
Phylogenomic tree of the class Methanobacteria, based on 37 marker protein sequences with the completely sequenced genomes of isolated strains from Thermococci, Methanococci, Mathanopyri, and Methanobacteria. We rooted the tree in the class Thermococci1 and labeled the optimum growth temperature ranges with corresponding colors as the genus within the class Methanobacteria. The number of present genes (P), gained genes (G), and lost genes (L) on the speculated common ancestor nodes were also labeled. The genus Methanothermus (83 °C) is the deepest clade, followed by the genus Methanothermobacter (55–70 °C). Other clades are the genera Methanobrevibacter (30–40 °C), Methanosphaera (37 °C), and Methanobacterium (28–37 °C). We also labeled the in situ environments of strains with different geometries. The star denotes that the strains were isolated from natural aquatic environments, including hot springs or groundwater. The rectangle denotes that the strains were isolated from natural terrestrial environments, including permafrost, peat soil, and wet wood. The ellipse indicates that the strains were isolated from artificial bioreactions, including digesters and fermenters. The plus sign indicates that the strains were isolated from co-culture systems. The regular triangle indicates that the strains were isolated from gastrointestinal tracts of animals including mammals and termites. The del operator indicates that the strains were isolated from animal feces including those from mammals and geese.
Based on the KEGG annotation, the common ancestor of the class Methanobacteria possesses hydrogenotrophic pathways, and it can use formate for methanogenesis (Supplementary Table S3). However, none of the known nitrogen and sulfur metabolic pathways are complete, indicating that it may have alternative pathways for nitrogen and sulfur assimilation. The Methanobacteria ancestor also codes for most subunits of phosphate, tungstate, molybdate, zinc, cobalt, nickel, and biotin ABC-type transporters. Previous phylogenomic analysis suggests that the Class I methanogens including the class Methanobacteria may have originated before the Great Oxygenation Event (GOE), which is consistent with their strictly anaerobic features [54]. There have also been studies focused on the evolution of the genus Methanosphaera within the class Methanobacteria that confirm its monophyletic origin [55]. Nonetheless, to our knowledge, no research has systemically focused on the evolutionary history of the class Methanobacteria.
Based on the phylogenetic results and environmental information, we found a clear trend indicating that the optimum growth temperatures for different genera decreased gradually during the evolution of the class Methanobacteria. At the deep-branching position of the class Methanobacteria, the first diverging clade was the genus Methanothermus (83 °C), followed by the genus Methanothermobacter (55–70 °C), with other members of the class being the mesophilic genera (28–40 °C) including Methanobrevibacter, Methanosphaera, and Methanobacterium (Figure 7). Therefore, we hypothesize that the loss of genes related to thermal adaptation by the ancestor nodes led to mesophilic adaptation.
Compared with the deep-branching genus Methanothermus, other nodes lost genes related to the reverse gyrase gene (rgy) (Supplementary Table S3). By consuming ATP, Rgy can modify the topological state of DNA by introducing positive supercoils, which are very important for the hyperthermal adaptation of DNA and may also be involved in unwinding DNA strands under a positive supercoil state to allow subsequent transcription or replication [56]. We propose that the loss of rgy might be one of the factors that altered the living conditions of the common ancestor of the genus Methanothermobacter and other mesophilic methanogens, resulting in a shift in their temperature adaptation ability from hyperthermal (83 °C) to thermal or ambient adaptation (28–70 °C). By comparing the common ancestor of the thermal methanogens (50–70 °C) and mesophilic methanogens (28–40 °C), we found that the latter lost the genes coding for enzymes that may be related to thermal adaptation—such as the integral membrane protein that may help to stabilize the membrane under thermal conditions, and the enzyme that repairs DNA, prevents DNA lesions, or facilitates DNA transcription (Supplementary Table S3)—which may have helped the methanogens to adapt to thermal environments.
The three mesophilic genera Methanobrevibacter (30–40 °C), Methanosphaera (37 °C), and Methanobacterium (28–37 °C) cluster together and share a mesophilic common ancestor (Figure 7). However, the methanogenesis pathway of the genus Methanosphaera, which gains energy by reducing methanol to methane only when H2 is the electron donor, is the most specific in the entire class Methanobacteria. Our results show that the common ancestor of the genus Methanosphaera lost genes coding for the complex that catalyzes the reversible cleavage of acetyl-CoA and enzymes related to the hydrogenotrophic methanogenesis pathway, i.e., the sulfur-carrier protein required for the activity of Fdh, the subunit E of the Fwd, coenzyme F420-dependent NADP oxidoreductase, and even part of the Mtr complex related to energy conservation (Figure 6, Supplementary Table S3). However, it gained genes encoding for enzymes, such as methanol cobalamin methyltransferase, corrinoid iron-sulfur protein (which transfers methyl in the Wood-Ljungdahl pathway), and the subunit A of methanol-coenzyme M methyltransferase (MtaA). Mta catalyzes methyl group transfers from methanol to coenzyme M to form methyl coenzyme M [55]. We propose that gaining these genes occasioned the transformation of the genus Methanosphaera from hydrogenotrophic metabolism to hydrogen-dependent methanol metabolism.
Interestingly, strains of the genera Methanosphaera and Methanobrevibacter were all isolated from the gastrointestinal systems of mammals or termites or the feces of mammals or geese, except for Methanobrevibacter arboriphilus DH1, which was isolated from wet wood, and Methanobrevibacter smithii ATCC35061 from a primary sewage digester (Figure 7) [18,20,57]. However, all the strains of the genus Methanobacterium were isolated from natural systems (Figure 7) [19]. Compared with the genus Methanobacterium, which inhabits natural environments, the common ancestor of the host-associated genus Methanobrevibacter did not experience considerable gene gain or loss (Figure 7, Supplementary Table S3). The potential reason for this may be that both genera continue to depend on H2/CO2 utilization as the anaerobic intestine environments of the host may be similar to ancient natural anaerobic environments. However, the common ancestor of another host-associated genus, Methanosphaera, experienced significant gene gain and gene loss, and obtained energy from the hydrogen-dependent reduction of methanol to methane (Supplementary Table S3). Previous research indicates that the methanol concentration in gastrointestinal systems (10 μM in cockroaches, 23–72 μM in the rumen, and 70 μM in humans) is higher than in natural environments where methanol is generally around or below the micromolar level [58,59,60,61]. Therefore, this specific selection force may have driven the ancestor of the genus Methanosphaera to become better adapted to methanol-rich gastrointestinal systems. Therefore, the host-adapted genera Methanobrevibacter and Methanosphaera may have evolved independently to utilize different substrates in intestinal systems. It should be fascinating to study what produced this massive difference between these two closely related genera, which inhabit similar environments.
4. Conclusions
Methanogens are one of the most ancient life forms on Earth and have immensely impacted the global climate since their origin. They exist in an extensive range of all kinds of anaerobic ecosystems and are considered to have originated in geothermal environments. In this study, we isolated and pure cultured one methanogen, temporarily named Methanothermobacter tengchongensis DL9LZB001, from a hot spring at Tengchong and completely sequenced its genome. DL9LZB001 utilizes H2 and CO2 for methanogenesis, with an optimum growth temperature of 65 °C. Genome analysis demonstrates that it has a complete hydrogenotrophic methanogenesis pathway and a potential acetate metabolic pathway. Phylogenetic analysis indicates that the strain DL9LZB001 belongs to the class Methanobacteria of the Class I methanogens, and its closest relative is the strain M. marburgensis Marburg. Comparative genomic and ancestral analysis of isolated strains among the Class I methanogens indicates that the class Methanobacteria had a hyperthermal origin, from which it gradually evolved to be thermophilic and then finally to be mesophilic. The evolutionary history of the class Methanobacteria is an odyssey of leaving high-temperature niches, which have the greatest potential to be cradles of life. These ancient strains gradually lost their thermal adaptation and came to occupy niches with different temperatures.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/biology11101514/s1, Table S1: environmental information of isolated strains within the class Methanobacteria; Table S2: information of 37 marker-protein sequences; Table S3: common ancestor gene annotations within the class Methanobacteria; Table S4: AAI from strain DL9LZB001 to the isolated strains within the class Methanobacteria; Table S5: ANI from strain DL9LZB001 to the isolated strains within the class Methanobacteria.
Author Contributions
Conceptualization, Y.W. and F.W.; methodology, Z.L., J.D. and Y.W.; investigation, H.W., J.W., Y.C., L.L. and T.Y.; writing—original draft preparation, Z.L. and Y.W.; writing—review and editing, Y.W. and F.W. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Natural Science Foundation of China, grant number 92051116, 42272354, 91951209, 41921006, 41867067. Natural Science Foundation of Shanghai, grant number 20ZR1428000.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Genome sequence can be downloaded from eLMSG (https://www.biosino.org/elmsg/index) with code: https://www.biosino.org/elmsg/record/MSG083741?code=4644.
Acknowledgments
We thank Xue Ding, Nannan Zhang, Limin Sun, Zhe Xie, and Qian Luo for instructions on instrument usage, as well as Chen Li, Yujue Wang, and Ya Guo from SJTU HPC for useful discussion of software.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ueno, Y.; Yamada, K.; Yoshida, N.; Maruyama, S.; Isozaki, Y. Evidence from fluid inclusions for microbial methanogenesis in the early Archaean era. Nature 2006, 440, 516–519. [Google Scholar] [CrossRef] [PubMed]
- Battistuzzi, F.U.; Feijao, A.; Hedges, S.B. A genomic timescale of prokaryote evolution: Insights into the origin of methanogenesis, phototrophy, and the colonization of land. BMC Evol. Biol. 2004, 4, 44. [Google Scholar] [CrossRef] [PubMed]
- Borrel, G.; Adam, P.S.; Gribaldo, S. Methanogenesis and the Wood-Ljungdahl Pathway: An Ancient, Versatile, and Fragile Association. Genome Biol. Evol. 2016, 8, 1706–1711. [Google Scholar] [CrossRef]
- Liu, Y.; Whitman, W.B. Metabolic, phylogenetic, and ecological diversity of the methanogenic archaea. Ann. N. Y. Acad. Sci. 2008, 1125, 171–189. [Google Scholar] [CrossRef] [PubMed]
- Mayumi, D.; Mochimaru, H.; Tamaki, H.; Yamamoto, K.; Yoshioka, H.; Suzuki, Y.; Kamagata, Y.; Sakata, S. Methane production from coal by a single methanogen. Science 2016, 354, 222–225. [Google Scholar] [CrossRef] [PubMed]
- Thauer, R.K.; Kaster, A.K.; Seedorf, H.; Buckel, W.; Hedderich, R. Methanogenic archaea: Ecologically relevant differences in energy conservation. Nat. Rev. Microbiol. 2008, 6, 579–591. [Google Scholar] [CrossRef] [PubMed]
- Kiehl, J.T.; Trenberth, K.E. Earth’s annual global mean energy budget. Bull. Am. Meteorol. Soc. 1997, 78, 197–208. [Google Scholar] [CrossRef]
- Berghuis, B.A.; Yu, F.B.; Schulz, F.; Blainey, P.C.; Woyke, T.; Quake, S.R. Hydrogenotrophic methanogenesis in archaeal phylum Verstraetearchaeota reveals the shared ancestry of all methanogens. Proc. Natl. Acad. Sci. USA 2019, 116, 5037–5044. [Google Scholar] [CrossRef]
- Evans, P.N.; Parks, D.H.; Chadwick, G.L.; Robbins, S.J.; Orphan, V.J.; Golding, S.D.; Tyson, G.W. Methane metabolism in the archaeal phylum Bathyarchaeota revealed by genome-centric metagenomics. Science 2015, 350, 434–438. [Google Scholar] [CrossRef]
- Wang, Y.; Wegener, G.; Hou, J.; Wang, F.; Xiao, X. Expanding anaerobic alkane metabolism in the domain of Archaea. Nat. Microbiol. 2019, 4, 595–602. [Google Scholar] [CrossRef]
- Hua, Z.S.; Wang, Y.L.; Evans, P.N.; Qu, Y.N.; Goh, K.M.; Rao, Y.Z.; Qi, Y.L.; Li, Y.X.; Huang, M.J.; Jiao, J.Y.; et al. Insights into the ecological roles and evolution of methyl-coenzyme M reductase-containing hot spring Archaea. Nat. Commun. 2019, 10, 4574. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wegener, G.; Williams, T.A.; Xie, R.; Hou, J.; Wang, F.; Xiao, X. A methylotrophic origin of methanogenesis and early divergence of anaerobic multicarbon alkane metabolism. Sci. Adv. 2021, 7, eabj1453. [Google Scholar] [CrossRef] [PubMed]
- Boyd, J.A.; Jungbluth, S.P.; Leu, A.O.; Evans, P.N.; Woodcroft, B.J.; Chadwick, G.L.; Orphan, V.J.; Amend, J.P.; Rappé, M.S.; Tyson, G.W. Divergent methyl-coenzyme M reductase genes in a deep-subseafloor Archaeoglobi. BioRxiv 2018. [Google Scholar] [CrossRef] [PubMed]
- Adam, P.S.; Borrel, G.; Brochier-Armanet, C.; Gribaldo, S. The growing tree of Archaea: New perspectives on their diversity, evolution and ecology. ISME J. 2017, 11, 2407–2425. [Google Scholar] [CrossRef]
- Bapteste, E.; Brochier, C.; Boucher, Y. Higher-level classification of the Archaea: Evolution of methanogenesis and methanogens. Archaea 2005, 1, 353–363. [Google Scholar] [CrossRef]
- Stetter, K.O. Methanothermus. In Bergey’s Manual of Systematics of Archaea and Bacteria; Wiley: Hoboken, NJ, USA, 2015; pp. 1–4. [Google Scholar]
- Boone, D.R. Methanothermobacter. In Bergey’s Manual of Systematics of Archaea and Bacteria; Wiley: Hoboken, NJ, USA, 2015; pp. 1–8. [Google Scholar]
- Miller, T.L. Methanobrevibacter. In Bergey’s Manual of Systematics of Archaea and Bacteria; Wiley: Hoboken, NJ, USA, 2015; pp. 1–14. [Google Scholar]
- Boone, D.R. Methanobacterium. In Bergey’s Manual of Systematics of Archaea and Bacteria; Wiley: Hoboken, NJ, USA, 2015; pp. 1–8. [Google Scholar]
- Miller, T.L. Methanosphaera. In Bergey’s Manual of Systematics of Archaea and Bacteria; Wiley: Hoboken, NJ, USA, 2015; pp. 1–6. [Google Scholar]
- Validation of the publication of new names and new combinations previously effectively published outside the IJSB. List No. 63. Int. J. Syst. Bacteriol. 1997, 47, 1274. [CrossRef]
- Nakamura, K.; Takahashi, A.; Mori, C.; Tamaki, H.; Mochimaru, H.; Nakamura, K.; Takamizawa, K.; Kamagata, Y. Methanothermobacter tenebrarum sp. nov., a hydrogenotrophic, thermophilic methanogen isolated from gas-associated formation water of a natural gas field. Int. J. Syst. Evol. Microbiol. 2013, 63, 715–722. [Google Scholar] [CrossRef]
- Leadbetter, J.R.; Crosby, L.D.; Breznak, J.A. Methanobrevibacter filiformis sp. nov., A filamentous methanogen from termite hindguts. Arch. Microbiol. 1998, 169, 287–292. [Google Scholar] [CrossRef]
- Cadillo-Quiroz, H.; Brauer, S.L.; Goodson, N.; Yavitt, J.B.; Zinder, S.H. Methanobacterium paludis sp. nov. and a novel strain of Methanobacterium lacus isolated from northern peatlands. Int. J. Syst. Evol. Microbiol. 2014, 64, 1473–1480. [Google Scholar] [CrossRef]
- Krivushin, K.V.; Shcherbakova, V.A.; Petrovskaya, L.E.; Rivkina, E.M. Methanobacterium veterum sp. nov., from ancient Siberian permafrost. Int. J. Sys.t Evol. Microbiol. 2010, 60, 455–459. [Google Scholar] [CrossRef]
- Xue, Y.; Xu, Y.; Liu, Y.; Ma, Y.; Zhou, P. Thermoanaerobacter tengcongensis sp. nov., a novel anaerobic, saccharolytic, thermophilic bacterium isolated from a hot spring in Tengcong, China. Int. J. Syst. Evol. Microbiol. 2001, 51, 1335–1341. [Google Scholar] [CrossRef] [PubMed]
- Plugge, C.M. Anoxic Media Design, Preparation, and Considerations. Methods Enzymol. 2005, 397, 3–16. [Google Scholar] [PubMed]
- Gordon, D.F.; Stutman, M.; Loesche, W.J. Improved isolation of anaerobic bacteria from the gingival crevice area of man. Appl. Microbiol. 1971, 21, 1046–1050. [Google Scholar] [CrossRef]
- Yang, S.; Li, X.; Xiao, X.; Zhuang, G.; Zhang, Y. Sphingomonas profundi sp. nov., isolated from deep-sea sediment of the Mariana Trench. Int. J. Syst. Evol. Microbiol. 2020, 70, 3809–3815. [Google Scholar] [CrossRef] [PubMed]
- Vetriani, C. Archaea, Origin of. In Encyclopedia of Biodiversity, 2nd ed.; Levin, S.A., Ed.; Academic Press: Waltham, MA, USA, 2001; pp. 218–226. [Google Scholar]
- Ardui, S.; Ameur, A.; Vermeesch, J.R.; Hestand, M.S. Single molecule real-time (SMRT) sequencing comes of age: Applications and utilities for medical diagnostics. Nucleic Acids Res. 2018, 46, 2159–2168. [Google Scholar] [CrossRef] [PubMed]
- Besemer, J.; Lomsadze, A.; Borodovsky, M. GeneMarkS: A self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 2001, 29, 2607–2618. [Google Scholar] [CrossRef] [PubMed]
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 1997, 25, 955–964. [Google Scholar] [CrossRef]
- Lagesen, K.; Hallin, P.; Rodland, E.A.; Staerfeldt, H.H.; Rognes, T.; Ussery, D.W. RNAmmer: Consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007, 35, 3100–3108. [Google Scholar] [CrossRef]
- Gardner, P.P.; Daub, J.; Tate, J.G.; Nawrocki, E.P.; Kolbe, D.L.; Lindgreen, S.; Wilkinson, A.C.; Finn, R.D.; Griffiths-Jones, S.; Eddy, S.R.; et al. Rfam: Updates to the RNA families database. Nucleic Acids Res. 2009, 37, D136–D140. [Google Scholar] [CrossRef]
- Nawrocki, E.P.; Kolbe, D.L.; Eddy, S.R. Infernal 1.0: Inference of RNA alignments. Bioinformatics 2009, 25, 1335–1337. [Google Scholar] [CrossRef]
- Hsiao, W.; Wan, I.; Jones, S.J.; Brinkman, F.S. IslandPath: Aiding detection of genomic islands in prokaryotes. Bioinformatics 2003, 19, 418–420. [Google Scholar] [CrossRef]
- Grissa, I.; Vergnaud, G.; Pourcel, C. CRISPRFinder: A web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 2007, 35, W52–W57. [Google Scholar] [CrossRef] [PubMed]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [PubMed]
- Hug, L.A.; Baker, B.J.; Anantharaman, K.; Brown, C.T.; Probst, A.J.; Castelle, C.J.; Butterfield, C.N.; Hernsdorf, A.W.; Amano, Y.; Ise, K.; et al. A new view of the tree of life. Nat. Microbiol. 2016, 1, 16048. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Capella-Gutierrez, S.; Silla-Martinez, J.M.; Gabaldon, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Jain, C.; Rodriguez, R.L.; Phillippy, A.M.; Konstantinidis, K.T.; Aluru, S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 2018, 9, 5114. [Google Scholar] [CrossRef]
- Hyatt, D.; Chen, G.L.; Locascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef]
- Emms, D.M.; Kelly, S. OrthoFinder: Phylogenetic orthology inference for comparative genomics. BioRxiv 2018. [Google Scholar] [CrossRef]
- Csuros, M. Count: Evolutionary analysis of phylogenetic profiles with parsimony and likelihood. Bioinformatics 2010, 26, 1910–1912. [Google Scholar] [CrossRef] [PubMed]
- Wasserfallen, A.; Nolling, J.; Pfister, P.; Reeve, J.; Conway de Macario, E. Phylogenetic analysis of 18 thermophilic Methanobacterium isolates supports the proposals to create a new genus, Methanothermobacter gen. nov., and to reclassify several isolates in three species, Methanothermobacter thermautotrophicus comb. nov., Methanothermobacter wolfeii comb. nov., and Methanothermobacter marburgensis sp. nov. Int. J. Syst. Evol. Microbiol. 2000, 50, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Zeikus, J.G.; Wolfe, R.S. Methanobacterium thermoautotrophicus sp. n., an anaerobic, autotrophic, extreme thermophile. J. Bacteriol. 1972, 109, 707–715. [Google Scholar] [CrossRef] [PubMed]
- Kotelnikova, S.V.; Obraztsova, A.Y.; Gongadze, G.M.; Laurinavichius, K.S. Methanobacterium thermoflexum sp. nov. and Methanobacterium defluvii sp. nov., Thermophilic Rod-Shaped Methanogens Isolated from Anaerobic Digestor Sludge. Syst. Appl. Microbiol. 1993, 16, 427–435. [Google Scholar] [CrossRef]
- Winter, J.; Lerp, C.; Zabel, H.P.; Wildenauer, F.X.; König, H.; Schindler, F. Methanobacterium wolfei, sp. nov., a New Tungsten-Requiring, Thermophilic, Autotrophic Methanogen. Syst. Appl. Microbiol. 1984, 5, 457–466. [Google Scholar] [CrossRef]
- Oren, A. The Family Methanotrichaceae. In The Prokaryotes; Springer: Berlin/Heidelberg, Germany, 2014; pp. 297–306. [Google Scholar]
- Oren, A. The Family Methanosarcinaceae. In The Prokaryotes; Springer: Berlin/Heidelberg, Germany, 2014; pp. 259–281. [Google Scholar]
- Lyu, Z.; Lu, Y. Metabolic shift at the class level sheds light on adaptation of methanogens to oxidative environments. ISME J. 2018, 12, 411–423. [Google Scholar] [CrossRef]
- Hoedt, E.C.; Parks, D.H.; Volmer, J.G.; Rosewarne, C.P.; Denman, S.E.; McSweeney, C.S.; Muir, J.G.; Gibson, P.R.; Cuiv, P.O.; Hugenholtz, P.; et al. Culture- and metagenomics-enabled analyses of the Methanosphaera genus reveals their monophyletic origin and differentiation according to genome size. ISME J. 2018, 12, 2942–2953. [Google Scholar] [CrossRef]
- Lulchev, P.; Klostermeier, D. Reverse gyrase--recent advances and current mechanistic understanding of positive DNA supercoiling. Nucleic Acids Res. 2014, 42, 8200–8213. [Google Scholar] [CrossRef]
- Kandler, O. Zellwandstrukturen bei Methan-Bakterien. Naturwissenschaften 1979, 66, 95–105. [Google Scholar] [CrossRef]
- Borrel, G.; McCann, A.; Deane, J.; Neto, M.C.; Lynch, D.B.; Brugere, J.F.; O’Toole, P.W. Genomics and metagenomics of trimethylamine-utilizing Archaea in the human gut microbiome. ISME J. 2017, 11, 2059–2074. [Google Scholar] [CrossRef]
- Sprenger, W.W.; Hackstein, J.H.; Keltjens, J.T. The competitive success of Methanomicrococcus blatticola, a dominant methylotrophic methanogen in the cockroach hindgut, is supported by high substrate affinities and favorable thermodynamics. FEMS Microbiol. Ecol. 2007, 60, 266–275. [Google Scholar] [CrossRef] [PubMed]
- Mausz, M.A.; Chen, Y. Microbiology and Ecology of Methylated Amine Metabolism in Marine Ecosystems. Curr. Issues Mol. Biol. 2019, 33, 133–148. [Google Scholar] [CrossRef] [PubMed]
- Ametaj, B.N.; Zebeli, Q.; Saleem, F.; Psychogios, N.; Lewis, M.J.; Dunn, S.M.; Xia, J.G.; Wishart, D.S. Metabolomics reveals unhealthy alterations in rumen metabolism with increased proportion of cereal grain in the diet of dairy cows. Metabolomics 2010, 6, 583–594. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).