The Role of Long Noncoding RNAs on Male Infertility: A Systematic Review and In Silico Analysis
Abstract
Simple Summary
Abstract
1. Introduction
2. Methods
2.1. Search Strategy
2.2. Inclusion/Exclusion Criteria
2.3. Extraction and Analysis
2.4. In Silico Analyses: Identification of Dysregulated lncRNAs, Target Genes and Functional Enrichment Analysis
3. Results
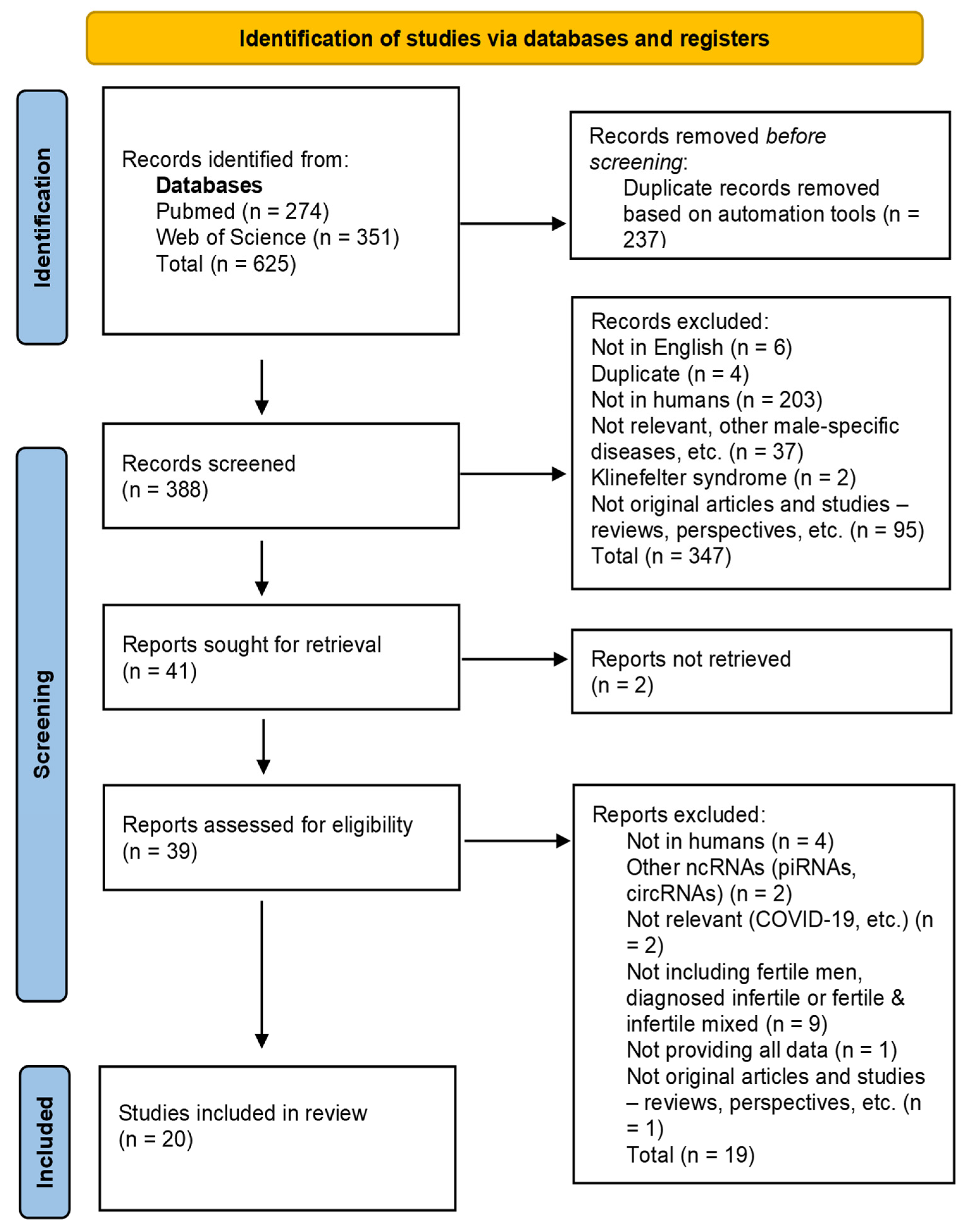
3.1. Study Selection
3.2. Study Characteristics (Included Studies)
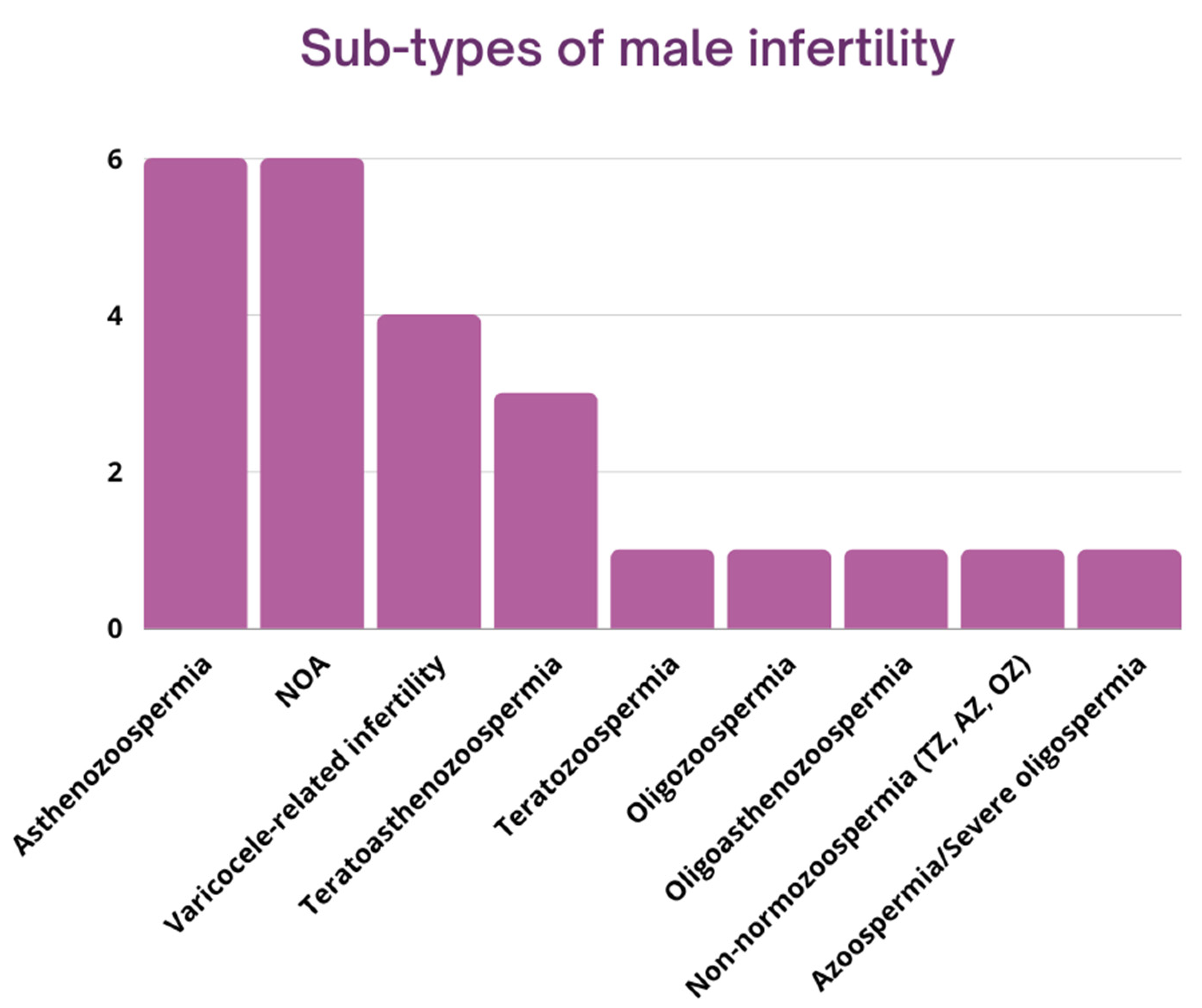
3.3. Subtypes of Male Infertility
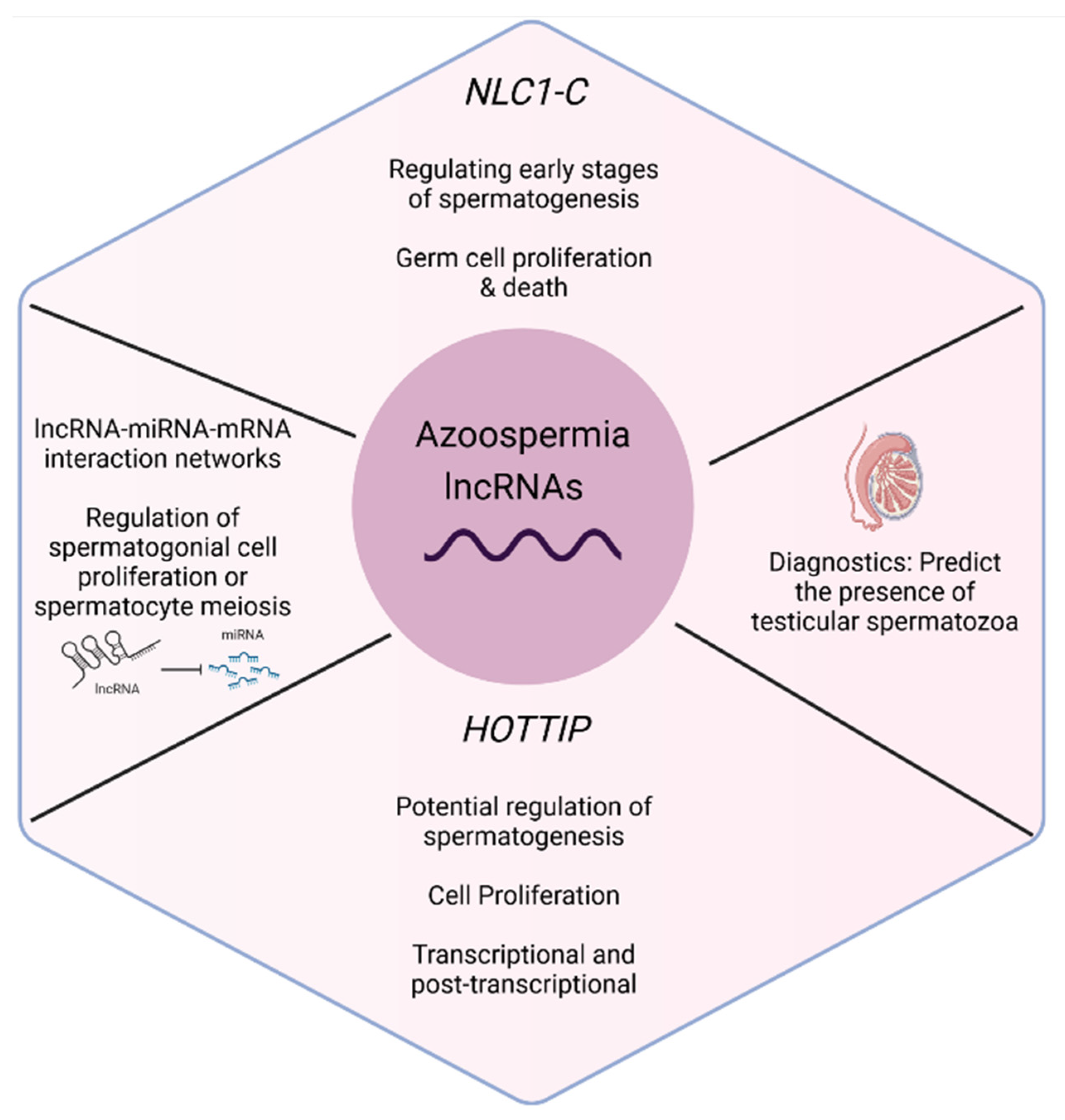
3.3.1. Azoospermia (n = 6)
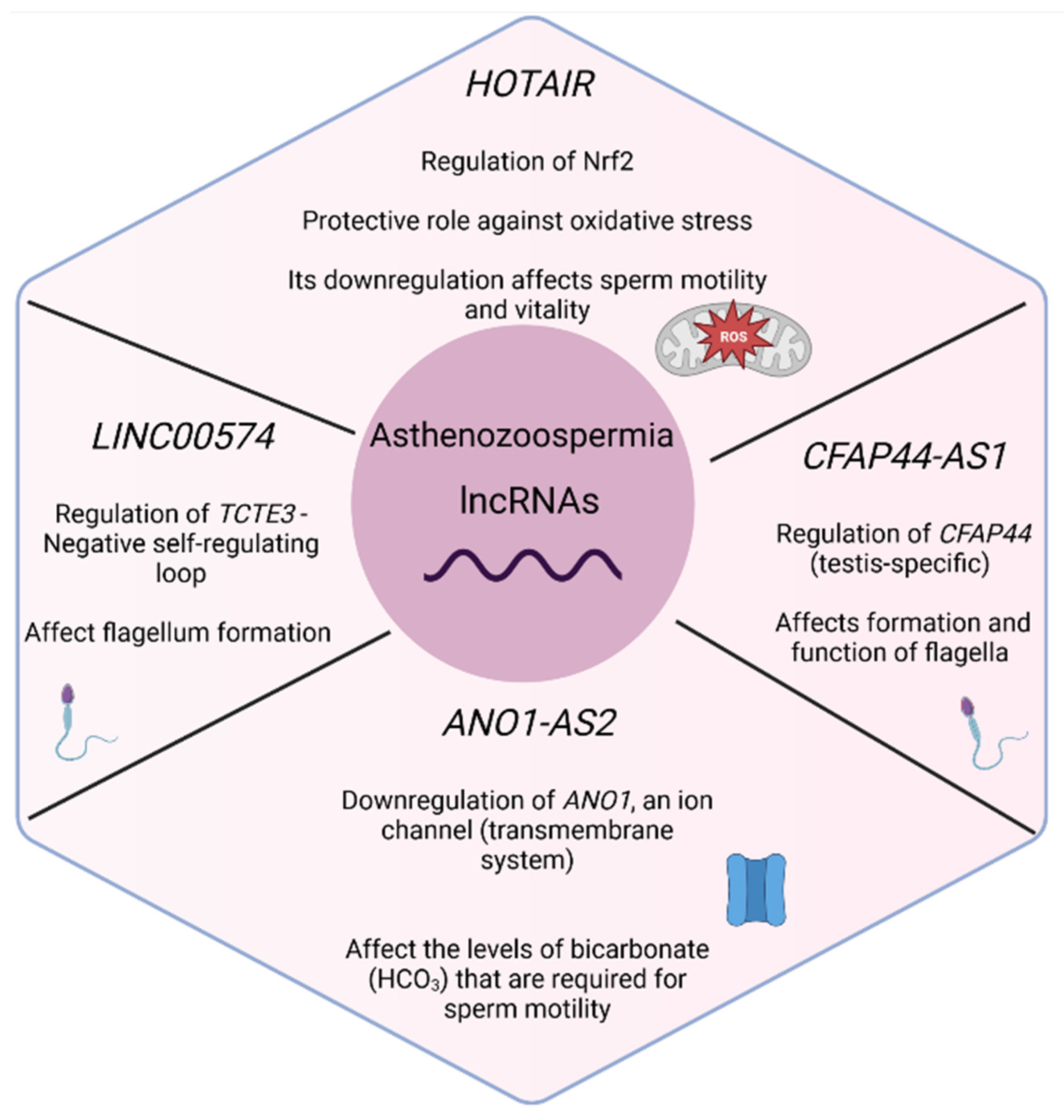
3.3.2. Asthenozoospermia (n = 6)
3.3.3. Teratozoospermia (n = 1)
3.3.4. Oligozoospermia (n = 1)
3.3.5. Oligoasthenozoospermia (n = 1)
3.3.6. Teratoasthenozoospermia (n = 3)
3.3.7. Varicocele-Related Male Infertility (n = 4)
3.4. Interactions between lncRNAs-miRNAs and Putative Target Genes
3.5. Exploring Variants on lncRNAs (n = 3)
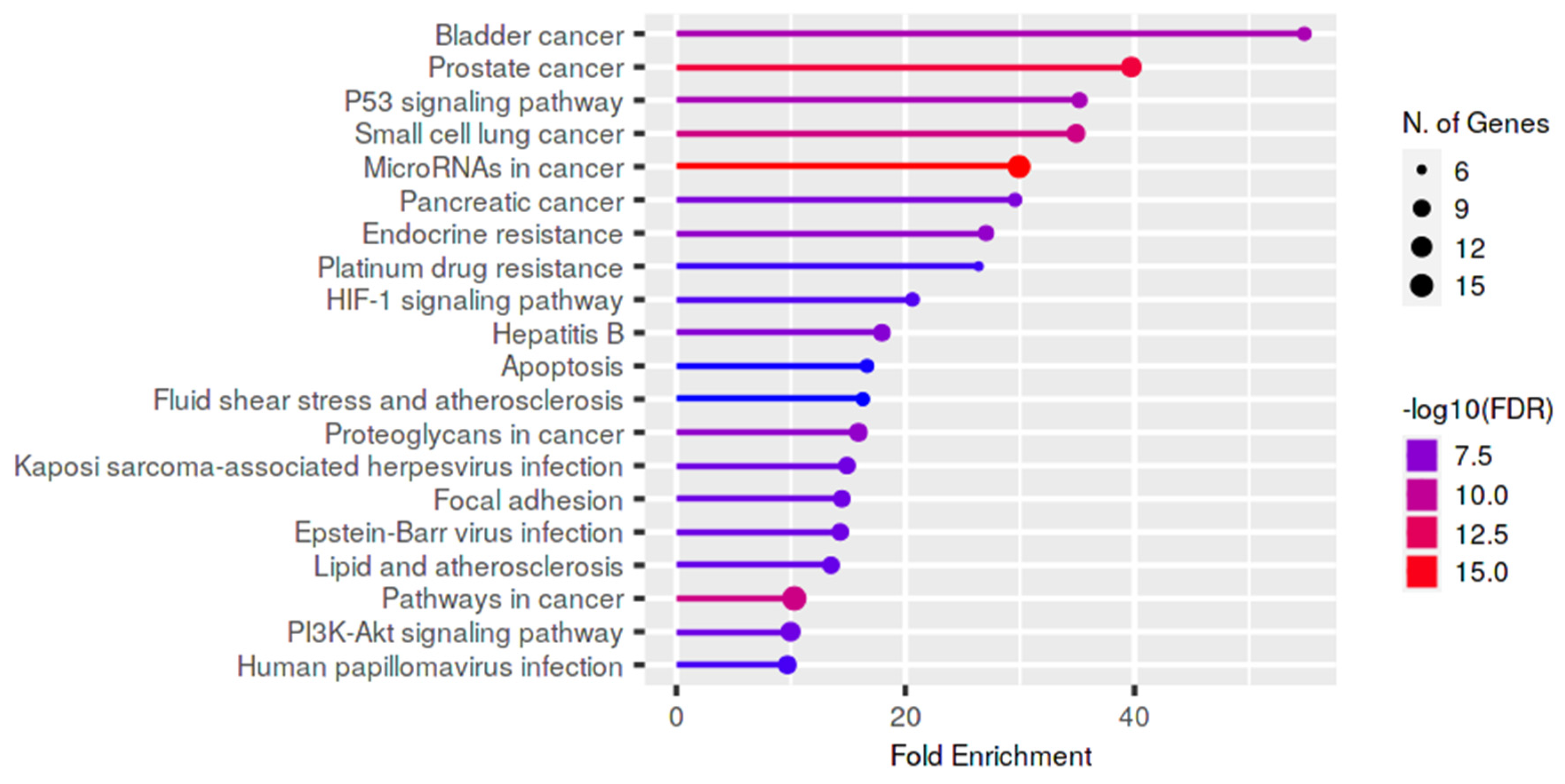
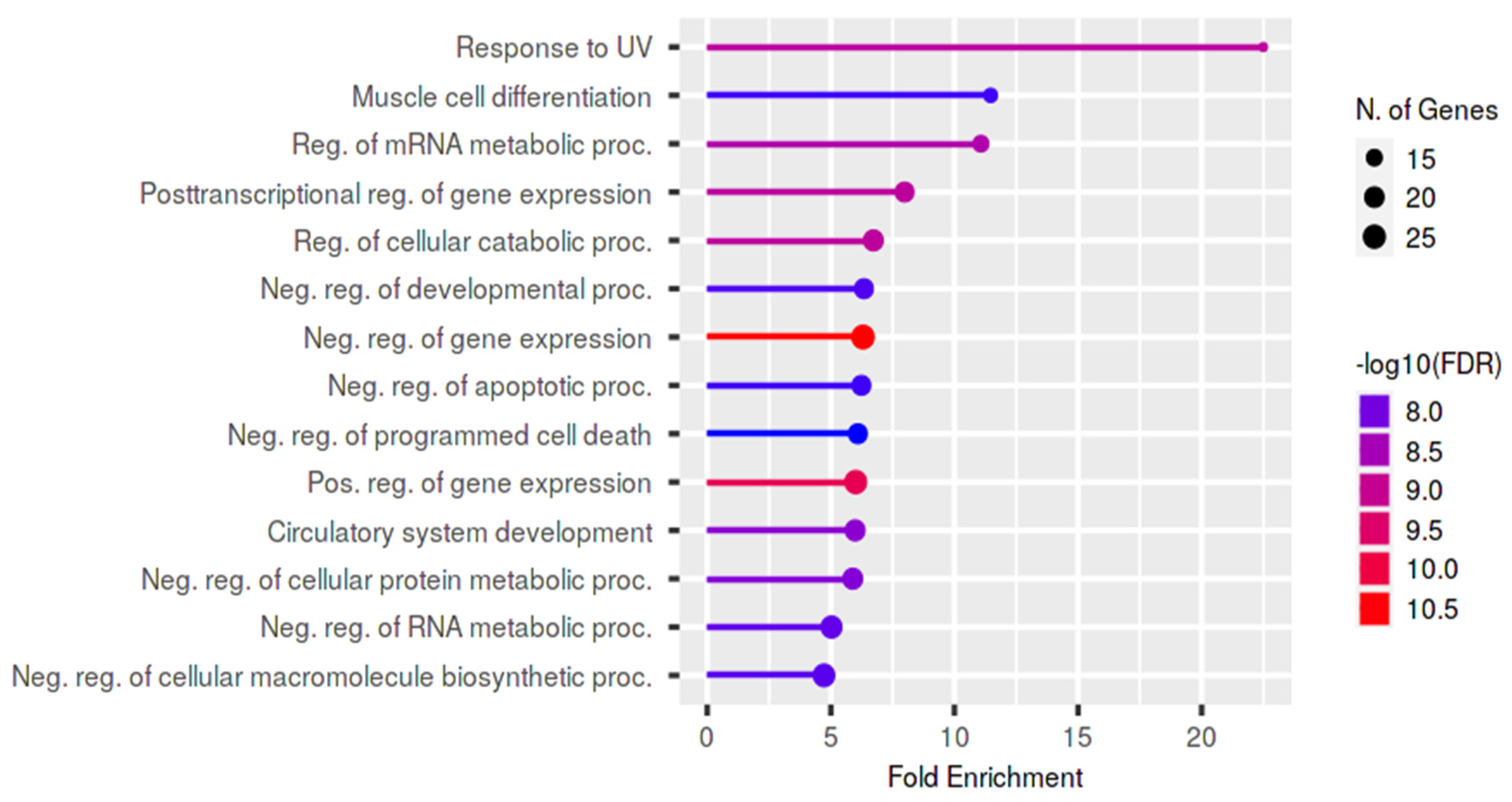
3.6. In Silico Analysis
4. Discussion
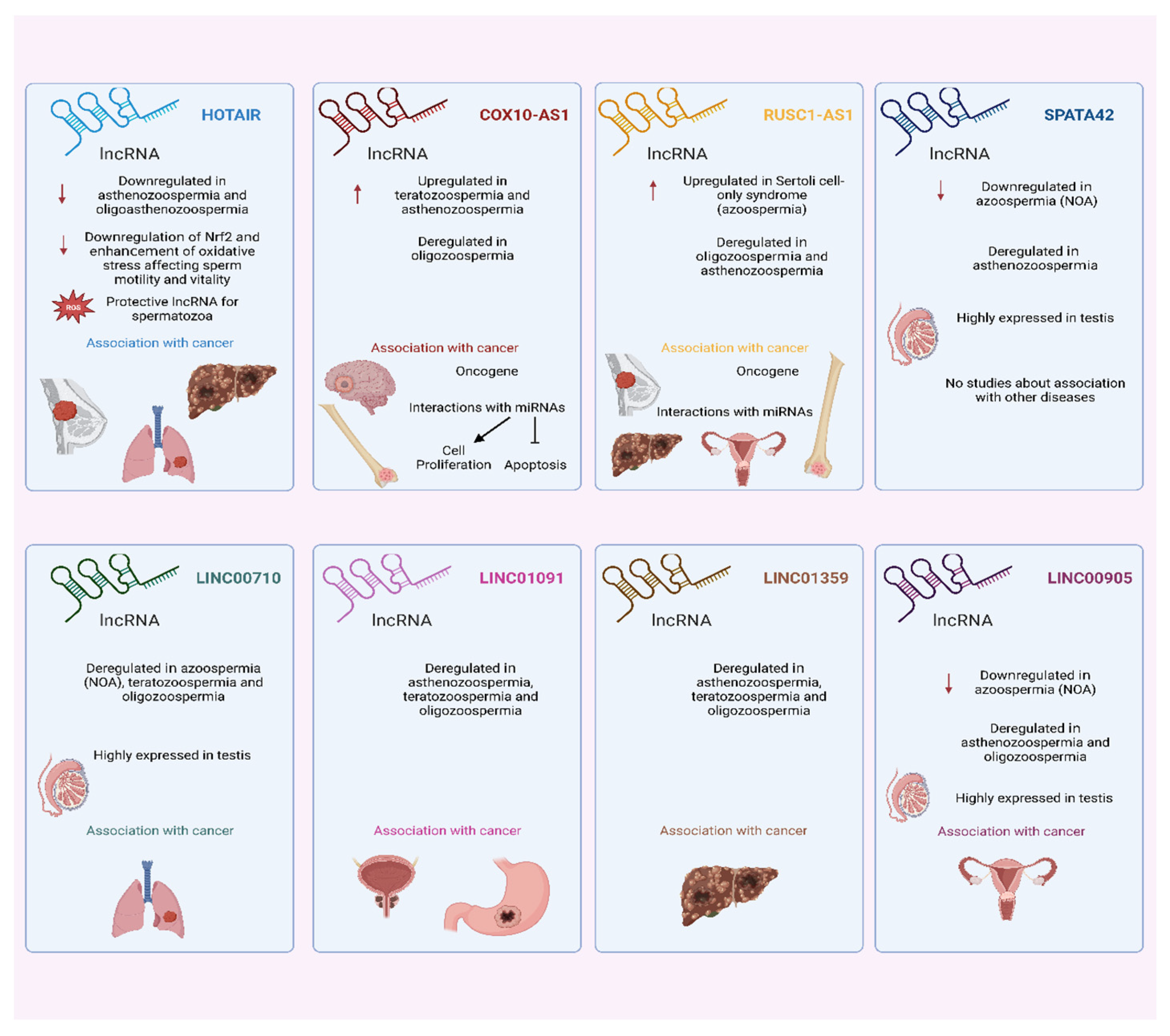
4.1. Main Findings
4.2. In Silico Analysis: Male Infertility and Cancer
4.3. Limitations
4.4. Strengths
4.5. Directions for Future Research
4.6. Challenges of Studying lncRNAs
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Vander Borght, M.; Wyns, C. Fertility and Infertility: Definition and Epidemiology. Clin. Biochem. 2018, 62, 2–10. [Google Scholar] [CrossRef] [PubMed]
- Nik Hazlina, N.H.; Norhayati, M.N.; Shaiful Bahari, I.; Nik Muhammad Arif, N.A. Worldwide Prevalence, Risk Factors and Psychological Impact of Infertility among Women: A Systematic Review and Meta-Analysis. BMJ Open 2022, 12, e057132. [Google Scholar] [CrossRef] [PubMed]
- Pandruvada, S.; Royfman, R.; Shah, T.A.; Sindhwani, P.; Dupree, J.M.; Schon, S.; Avidor-Reiss, T. Lack of Trusted Diagnostic Tools for Undetermined Male Infertility. J. Assist. Reprod. Genet. 2021, 38, 265–276. [Google Scholar] [CrossRef]
- Agarwal, A.; Baskaran, S.; Parekh, N.; Cho, C.L.; Henkel, R.; Vij, S.; Arafa, M.; Panner Selvam, M.K.; Shah, R. Male Infertility. Lancet 2021, 397, 319–333. [Google Scholar] [CrossRef]
- Sudhakar, D.V.S.; Shah, R.; Gajbhiye, R.K. Genetics of Male Infertility – Present and Future: A Narrative Review. J. Hum. Reprod. Sci. 2021, 14, 217–227. [Google Scholar] [CrossRef] [PubMed]
- Hangauer, M.J.; Vaughn, I.W.; McManus, M.T. Pervasive Transcription of the Human Genome Produces Thousands of Previously Unidentified Long Intergenic Noncoding RNAs. PLoS Genet. 2013, 9, 1003569. [Google Scholar] [CrossRef] [PubMed]
- Palazzo, A.F.; Lee, E.S. Non-Coding RNA: What Is Functional and What Is Junk? Front. Genet. 2015, 5, 2. [Google Scholar] [CrossRef]
- Quinn, J.J.; Chang, H.Y. Unique Features of Long Non-Coding RNA Biogenesis and Function. Nat. Rev. Genet. 2016, 17, 47–62. [Google Scholar] [CrossRef]
- Wu, R.; Su, Y.; Wu, H.; Dai, Y.; Zhao, M.; Lu, Q. Characters, Functions and Clinical Perspectives of Long Non-Coding RNAs. Mol. Genet. Genomics 2016, 291, 1013–1033. [Google Scholar] [CrossRef] [PubMed]
- Statello, L.; Guo, C.J.; Chen, L.L.; Huarte, M. Gene Regulation by Long Non-Coding RNAs and Its Biological Functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef] [PubMed]
- Joshi, M.; Rajender, S. Long Non-Coding RNAs (LncRNAs) in Spermatogenesis and Male Infertility. Reprod. Biol. Endocrinol. 2020, 18, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Dieter, C.; Lemos, N.E.; de Faria Corrêa, N.R.; Assmann, T.S.; Crispim, D. The Impact of LncRNAs in Diabetes Mellitus: A Systematic Review and In Silico Analyses. Front. Endocrinol. 2021, 12, 602597. [Google Scholar] [CrossRef] [PubMed]
- Uchida, S.; Dimmeler, S. Long Noncoding RNAs in Cardiovascular Diseases. Circ. Res. 2015, 116, 737–750. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.; Zuo, X.; Deng, H.; Liu, X.; Liu, L.; Ji, A. Roles of Long Noncoding RNAs in Brain Development, Functional Diversification and Neurodegenerative Diseases. Brain Res. Bull. 2013, 97, 69–80. [Google Scholar] [CrossRef]
- Mukherjee, A.; Koli, S.; Reddy, K.V.R. Regulatory Non-Coding Transcripts in Spermatogenesis: Shedding Light on ‘Dark Matter’. Andrology 2014, 2, 360–369. [Google Scholar] [CrossRef]
- Johnsson, P.; Lipovich, L.; Grandér, D.; Morris, K.V. Evolutionary Conservation of Long Noncoding RNAs; Sequence, Structure, Function. Biochim. Biophys. Acta 2014, 1840, 1063–1071. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 Statement: An Updated Guideline for Reporting Systematic Reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef]
- Agarwal, A.; Leisegang, K.; Majzoub, A.; Henkel, R.; Finelli, R.; Selvam, M.K.P.; Tadros, N.; Parekh, N.; Ko, E.Y.; Cho, C.L.; et al. Utility of Antioxidants in the Treatment of Male Infertility: Clinical Guidelines Based on a Systematic Review and Analysis of Evidence. World J. Mens. Health 2021, 39, 1–58. [Google Scholar] [CrossRef]
- Ross, C.; Morriss, A.; Khairy, M.; Khalaf, Y.; Braude, P.; Coomarasamy, A.; El-Toukhy, T. A Systematic Review of the Effect of Oral Antioxidants on Male Infertility. Reprod. Biomed. Online 2010, 20, 711–723. [Google Scholar] [CrossRef]
- Quan, J.; Pan, X.; Zhao, L.; Li, Z.; Dai, K.; Yan, F.; Liu, S.; Ma, H.; Lai, Y. LncRNA as a Diagnostic and Prognostic Biomarker in Bladder Cancer: A Systematic Review and Meta-Analysis. Onco. Targets. Ther. 2018, 11, 6415–6424. [Google Scholar] [CrossRef]
- Tian, T.; Wang, M.; Lin, S.; Guo, Y.; Dai, Z.; Liu, K.; Yang, P.; Dai, C.; Zhu, Y.; Zheng, Y.; et al. The Impact of LncRNA Dysregulation on Clinicopathology and Survival of Breast Cancer: A Systematic Review and Meta-Analysis. Mol. Ther.-Nucleic Acids 2018, 12, 359–369. [Google Scholar] [CrossRef] [PubMed]
- Li, J.H.; Liu, S.; Zhou, H.; Qu, L.H.; Yang, J.H. StarBase v2.0: Decoding MiRNA-CeRNA, MiRNA-NcRNA and Protein-RNA Interaction Networks from Large-Scale CLIP-Seq Data. Nucleic Acids Res. 2014, 42, D92–D97. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the Unification of Biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Consortium, T.G.O. The Gene Ontology Resource: Enriching a GOld Mine. Nucleic Acids Res. 2021, 49, D325–D334. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a Reference Resource for Gene and Protein Annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef]
- Lü, M.; Tian, H.; Cao, Y.X.; He, X.; Chen, L.; Song, X.; Ping, P.; Huang, H.; Sun, F. Downregulation of MiR-320a/383-Sponge-like Long Non-Coding RNA NLC1-C (Narcolepsy Candidate-Region 1 Genes) Is Associated with Male Infertility and Promotes Testicular Embryonal Carcinoma Cell Proliferation. Cell Death Dis. 2015, 6, E1960. [Google Scholar] [CrossRef]
- Xie, Y.; Yao, J.; Zhang, X.; Chen, J.; Gao, Y.; Zhang, C.; Chen, H.; Wang, Z.; Zhao, Z.; Chen, W.; et al. A Panel of Extracellular Vesicle Long Noncoding RNAs in Seminal Plasma for Predicting Testicular Spermatozoa in Nonobstructive Azoospermia Patients. Hum. Reprod. 2020, 35, 2413–2427. [Google Scholar] [CrossRef]
- Li, M.; Li, J.; Zhang, C.; Hou, S.; Weng, B. MIR210HG Is Aberrantly Expressed in the Seminal Plasma of Varicocele Patients and Associated with Varicocele-Related Dyszoospermia. Andrologia 2022, 54, e14277. [Google Scholar] [CrossRef] [PubMed]
- Bo, H.; Liu, Z.; Zhu, F.; Zhou, D.; Tan, Y.; Zhu, W.; Fan, L. Long Noncoding RNAs Expression Profile and Long Noncoding RNA-Mediated Competing Endogenous RNA Network in Nonobstructive Azoospermia Patients. Epigenomics 2020, 12, 673–684. [Google Scholar] [CrossRef]
- Saberiyan, M.; Mirfakhraie, R.; Moghni, M.; Teimori, H. Study of Linc00574 Regulatory Effect on the TCTE3 Expression in Sperm Motility. Reprod. Sci. 2021, 28, 159–165. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, P.; Song, D.; Xiong, S.; Zhang, H.; Fu, J.; Gao, F.; Chen, H.; Zeng, X. Expression Profiles and Characteristics of Human LncRNA in Normal and Asthenozoospermia Sperm. Biol. Reprod. 2019, 100, 982–993. [Google Scholar] [CrossRef] [PubMed]
- Eggers, S.; DeBoer, K.D.; Van Den Bergen, J.; Gordon, L.; White, S.J.; Jamsai, D.; McLachlan, R.I.; Sinclair, A.H.; O’Bryan, M.K. Copy Number Variation Associated with Meiotic Arrest in Idiopathic Male Infertility. Fertil. Steril. 2015, 103, 214–219. [Google Scholar] [CrossRef] [PubMed]
- Sabetian, S.; Zarei, M.; Jahromi, B.N.; Morowvat, M.H.; Tabei, S.M.B.; Cava, C. Exploring the Dysregulated MRNAs-MiRNAs-LncRNAs Interactions Associated to Idiopathic Non-Obstructive Azoospermia. J. Biomol. Struct. Dyn. 2022, 40, 5956–5964. [Google Scholar] [CrossRef]
- Sanei-Ataabadi, N.; Mowla, S.J.; Nasr-Esfahani, M.H. Transcript Isoforms of SLC7A11-AS1 Are Associated With Varicocele-Related Male Infertility. Front. Genet. 2020, 11, 1015. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Liu, Z.; Li, X.; Zhang, P.; Wang, J.; Zhu, D.; Chen, X.; Ye, L. Low Long Non-Coding RNA HOTAIR Expression Is Associated with down-Regulation of Nrf2 in the Spermatozoa of Patients with Asthenozoospermia or Oligoasthenozoospermia. Int. J. Clin. Exp. Pathol. 2015, 8, 14198–14205. [Google Scholar]
- Kamel, A.; Saberiyan, M.; Mirfakhraie, R.; Teimori, H. Reduced Expression of CFAP44 and CFAP44-AS1 May Affect Sperm Motility and Morphology. Andrologia 2022, 54, e14447. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.C.; Zhang, Y.; Yu, K.; Li, Y.; Yu, H.; Zhou, S.J.; Wang, Y.P.; Deng, S.L.; Tian, L. LncRNAs Induce Oxidative Stress and Spermatogenesis by Regulating Endoplasmic Reticulum Genes and Pathways. Aging 2021, 13, 13764–13787. [Google Scholar] [CrossRef] [PubMed]
- Ataabadi, N.S.; Mowla, S.J.; Aboutalebi, F.; Dormiani, K.; Kiani-Esfahani, A.; Tavalaee, M.; Nasr-Esfahani, M.H. Hypoxia-Related Long Noncoding RNAs Are Associated with Varicocele-Related Male Infertility. PLoS ONE 2020, 15, e0232357. [Google Scholar] [CrossRef]
- Su, Y.; Zhou, L.L.; Zhang, Y.Q.; Ni, L.Y. Long Noncoding RNA HOTTIP Is Associated with Male Infertility and Promotes Testicular Embryonal Carcinoma Cell Proliferation. Mol. Genet. Genom. Med. 2019, 7, e870. [Google Scholar] [CrossRef]
- Zhao, J.; Li, H.; Deng, H.; Zhu, L.; Zhou, B.; Yang, M.Q.; Liu, Q.R.; Luo, G.Q.; Yang, Y.; Ma, W.M. LncRNA Gadd7, Increased in Varicocele Patients, Suppresses Cell Proliferation and Promotes Cell Apoptosis. Oncotarget 2017, 9, 5105–5110. [Google Scholar] [CrossRef]
- Lu, H.; Xu, D.; Wang, P.; Sun, W.; Xue, X.; Hu, Y.; Xie, C.; Ma, Y. RNA-Sequencing and Bioinformatics Analysis of Long Noncoding RNAs and MRNAs in the Asthenozoospermia. Biosci. Rep. 2020, 40, BSR20194041. [Google Scholar] [CrossRef]
- Kyrgiafini, M.A.; Markantoni, M.; Sarafidou, T.; Chatziparasidou, A.; Christoforidis, N.; Mamuris, Z. Genome-Wide Association Study Identifies Candidate Markers Related to LincRNAs Associated with Male Infertility in the Greek Population. J. Assist. Reprod. Genet. 2020, 37, 2869–2881. [Google Scholar] [CrossRef] [PubMed]
- Saberiyan, M.; Mirfakhraie, R.; Gholami, D.; Dehdehi, L.; Teimori, H. Investigating the Regulatory Function of the ANO1-AS2 on the ANO1 Gene in Infertile Men with Asthenozoospermia and Terato-Asthenozoospermia. Exp. Mol. Pathol. 2020, 117, 104528. [Google Scholar] [CrossRef]
- Cerván-Martín, M.; Bossini-Castillo, L.; Rivera-Egea, R.; Garrido, N.; Luján, S.; Romeu, G.; Santos-Ribeiro, S.; Castilla, J.A.; del Carmen Gonzalvo, M.; Clavero, A.; et al. Effect and in Silico Characterization of Genetic Variants Associated with Severe Spermatogenic Disorders in a Large Iberian Cohort. Andrology 2021, 9, 1151–1165. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Wang, B. Identification of Male Infertility-Related Long Non-Coding RNAs and Their Functions Based on a Competing Endogenous RNA Network. J. Int. Med. Res. 2020, 48, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Wosnitzer, M.; Goldstein, M.; Hardy, M.P. Spermatogenesis Review of Azoospermia. Rev. Azoospermia 2014, 4, e28218. [Google Scholar] [CrossRef]
- Ghieh, F.; Mitchell, V.; Mandon-Pepin, B.; Vialard, F. Genetic Defects in Human Azoospermia. Basic Clin. Androl. 2019, 29, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Ghanami Gashti, N.; Sadighi Gilani, M.A.; Abbasi, M. Sertoli Cell-Only Syndrome: Etiology and Clinical Management. J. Assist. Reprod. Genet. 2021, 38, 559–572. [Google Scholar] [CrossRef] [PubMed]
- Yuen, W.; Golin, A.P.; Flannigan, R.; Schlegel, P.N. Histology and Sperm Retrieval among Men with Y Chromosome Microdeletions. Transl. Androl. Urol. 2021, 10, 1442–1456. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.S.; Lu, C.W.; Lin, T.Y.; Lin, P.Y.; Lin, Y.M. Causes and Clinical Features of Infertile Men With Nonobstructive Azoospermia and Histopathologic Diagnosis of Hypospermatogenesis. Urology 2017, 105, 62–68. [Google Scholar] [CrossRef]
- Nistal, M.; Paniagua, R. Testicular Biopsy. Contemporary Interpretation. Urol. Clin. N. Am. 1999, 26, 555–593. [Google Scholar] [CrossRef]
- Shahrokhi, S.Z.; Salehi, P.; Alyasin, A.; Taghiyar, S.; Deemeh, M.R. Asthenozoospermia: Cellular and Molecular Contributing Factors and Treatment Strategies. Andrologia 2020, 52, e13463. [Google Scholar] [CrossRef] [PubMed]
- Pedemonte, N.; Galietta, L.J.V. Structure and Function of TMEM16 Proteins (Anoctamins). Physiol. Rev. 2014, 94, 419–459. [Google Scholar] [CrossRef] [PubMed]
- Benedetto, R.; Ousingsawat, J.; Wanitchakool, P.; Zhang, Y.; Holtzman, M.J.; Amaral, M.; Rock, J.R.; Schreiber, R.; Kunzelmann, K. Epithelial Chloride Transport by CFTR Requires TMEM16A. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Wandernoth, P.M.; Mannowetz, N.; Szczyrba, J.; Grannemann, L.; Wolf, A.; Becker, H.M.; Sly, W.S.; Wennemuth, G. Normal Fertility Requires the Expression of Carbonic Anhydrases II and IV in Sperm. J. Biol. Chem. 2015, 290, 29202–29216. [Google Scholar] [CrossRef]
- Tang, S.; Wang, X.; Li, W.; Yang, X.; Li, Z.; Liu, W.; Li, C.; Zhu, Z.; Wang, L.; Wang, J.; et al. Biallelic Mutations in CFAP43 and CFAP44 Cause Male Infertility with Multiple Morphological Abnormalities of the Sperm Flagella. Am. J. Hum. Genet. 2017, 100, 854–864. [Google Scholar] [CrossRef]
- Coutton, C.; Escoffier, J.; Martinez, G.; Arnoult, C.; Ray, P.F. Teratozoospermia: Spotlight on the Main Genetic Actors in the Human. Hum. Reprod. Update 2014, 21, 455–485. [Google Scholar] [CrossRef]
- Cavarocchi, E.; Whitfield, M.; Saez, F.; Touré, A. Sperm Ion Transporters and Channels in Human Asthenozoospermia: Genetic Etiology, Lessons from Animal Models, and Clinical Perspectives. Int. J. Mol. Sci. 2022, 23, 3926. [Google Scholar] [CrossRef]
- Jensen, C.F.S.; Østergren, P.; Dupree, J.M.; Ohl, D.A.; Sønksen, J.; Fode, M. Varicocele and Male Infertility. Nat. Rev. Urol. 2017, 14, 523–533. [Google Scholar] [CrossRef]
- Wang, Y.J.; Zhang, R.Q.; Lin, Y.J.; Zhang, R.G.; Zhang, W.L. Relationship between Varicocele and Sperm DNA Damage and the Effect of Varicocele Repair: A Meta-Analysis. Reprod. Biomed. Online 2012, 25, 307–314. [Google Scholar] [CrossRef]
- Fang, Y.; Su, Y.; Xu, J.; Hu, Z.; Zhao, K.; Liu, C.; Zhang, H. Varicocele-Mediated Male Infertility: From the Perspective of Testicular Immunity and Inflammation. Front. Immunol. 2021, 12, 729539. [Google Scholar] [CrossRef] [PubMed]
- Brookheart, R.T.; Michel, C.I.; Listenberger, L.L.; Ory, D.S.; Schaffer, J.E. The Non-Coding RNA Gadd7 Is a Regulator of Lipid-Induced Oxidative and Endoplasmic Reticulum Stress. J. Biol. Chem. 2009, 284, 7446–7454. [Google Scholar] [CrossRef] [PubMed]
- Lewerenz, J.; Hewett, S.J.; Huang, Y.; Lambros, M.; Gout, P.W.; Kalivas, P.W.; Massie, A.; Smolders, I.; Methner, A.; Pergande, M.; et al. The Cystine/Glutamate Antiporter System x(c)(-) in Health and Disease: From Molecular Mechanisms to Novel Therapeutic Opportunities. Antioxid. Redox Signal. 2013, 18, 522–555. [Google Scholar] [CrossRef]
- Kotaja, N. MicroRNAs and Spermatogenesis. Fertil. Steril. 2014, 101, 1552–1562. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [PubMed]
- Salas-Huetos, A.; Blanco, J.; Vidal, F.; Mercader, J.M.; Garrido, N.; Anton, E. New Insights into the Expression Profile and Function of Micro-Ribonucleic Acid in Human Spermatozoa. Fertil. Steril. 2014, 102, 213–222. [Google Scholar] [CrossRef] [PubMed]
- Khawar, M.B.; Mehmood, R.; Roohi, N. MicroRNAs: Recent Insights towards Their Role in Male Infertility and Reproductive Cancers. Bosn. J. Basic Med. Sci. 2019, 19, 31. [Google Scholar] [CrossRef]
- Gou, L.T.; Dai, P.; Liu, M.F. Small Noncoding RNAs and Male Infertility. Wiley Interdiscip. Rev. RNA 2014, 5, 733–745. [Google Scholar] [CrossRef]
- Pratt, S.L.; Calcatera, S.M. Expression of MicroRNA in Male Reproductive Tissues and Their Role in Male Fertility. Reprod. Fertil. Dev. 2016, 29, 24–31. [Google Scholar] [CrossRef]
- Maatouk, D.M.; Loveland, K.L.; McManus, M.T.; Moore, K.; Harfe, B.D. Dicer1 Is Required for Differentiation of the Mouse Male Germline. Biol. Reprod. 2008, 79, 696–703. [Google Scholar] [CrossRef]
- Romero, Y.; Meikar, O.; Papaioannou, M.D.; Conne, B.; Grey, C.; Weier, M.; Pralong, F.; de Massy, B.; Kaessmann, H.; Vassalli, J.D.; et al. Dicer1 Depletion in Male Germ Cells Leads to Infertility Due to Cumulative Meiotic and Spermiogenic Defects. PLoS ONE 2011, 6, e25241. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, C.; Romero, Y.; Warnefors, M.; Bilican, A.; Borel, C.; Smith, L.B.; Kotaja, N.; Kaessmann, H.; Nef, S. Germ Cell-Specific Targeting of DICER or DGCR8 Reveals a Novel Role for Endo-SiRNAs in the Progression of Mammalian Spermatogenesis and Male Fertility. PLoS ONE 2014, 9, e107023. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Gao, C.; Lin, X.; Ning, Y.; He, W.; Zheng, C.; Zhang, D.; Yan, L.; Jiang, B.; Zhao, Y.; et al. The MicroRNA MiR-202 Prevents Precocious Spermatogonial Differentiation and Meiotic Initiation during Mouse Spermatogenesis. Development 2021, 148, dev199799. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Li, X.; Guo, J.; Zhang, P.; Zeng, W. The Roles of MicroRNAs in Regulation of Mammalian Spermatogenesis. J. Anim. Sci. Biotechnol. 2017, 8, 35. [Google Scholar] [CrossRef] [PubMed]
- Vashisht, A.; Gahlay, G.K. Using MiRNAs as Diagnostic Biomarkers for Male Infertility: Opportunities and Challenges. Mol. Hum. Reprod. 2020, 26, 199–214. [Google Scholar] [CrossRef] [PubMed]
- Salas-Huetos, A.; James, E.R.; Aston, K.I.; Carrell, D.T.; Jenkins, T.G.; Yeste, M. The Role of MiRNAs in Male Human Reproduction: A Systematic Review. Andrology 2020, 8, 7–26. [Google Scholar] [CrossRef]
- Yoon, J.H.; Abdelmohsen, K.; Gorospe, M. Functional Interactions among MicroRNAs and Long Noncoding RNAs. Semin. Cell Dev. Biol. 2014, 34, 9–14. [Google Scholar] [CrossRef]
- Paraskevopoulou, M.D.; Hatzigeorgiou, A.G. Analyzing MiRNA-LncRNA Interactions. Methods Mol. Biol. 2016, 1402, 271–286. [Google Scholar] [CrossRef]
- Zhao, B.; Xu, H.; Ai, X.; Adalat, Y.; Tong, Y.; Zhang, J.; Yang, S. Expression Profiles of Long Noncoding RNAs in Lung Adenocarcinoma. Onco. Targets. Ther. 2018, 11, 5383–5390. [Google Scholar] [CrossRef]
- López-Jiménez, E.; Andrés-León, E. The Implications of Ncrnas in the Development of Human Diseases. Non-Coding RNA 2021, 7, 17. [Google Scholar] [CrossRef]
- Heidary, Z.; Zaki-Dizaji, M.; Saliminejad, K.; Khorramkhorshid, H.R. Expression Analysis of the CRISP2, CATSPER1, PATE1 and SEMG1 in the Sperm of Men with Idiopathic Asthenozoospermia. J. Reprod. Infertil. 2019, 20, 70–75. [Google Scholar] [PubMed]
- Lappalainen, T.; MacArthur, D.G. From Variant to Function in Human Disease Genetics. Science 2021, 373, 1464–1468. [Google Scholar] [CrossRef] [PubMed]
- Uitterlinden, A.G. An Introduction to Genome-Wide Association Studies: GWAS for Dummies. Semin. Reprod. Med. 2016, 34, 196–204. [Google Scholar] [CrossRef] [PubMed]
- Zou, H.; Wu, L.X.; Tan, L.; Shang, F.F.; Zhou, H.H. Significance of Single-Nucleotide Variants in Long Intergenic Non-Protein Coding RNAs. Front. Cell Dev. Biol. 2020, 8, 347. [Google Scholar] [CrossRef]
- Aich, M.; Chakraborty, D. Role of LncRNAs in Stem Cell Maintenance and Differentiation. Curr. Top. Dev. Biol. 2020, 138, 73–112. [Google Scholar] [CrossRef]
- Jarroux, J.; Morillon, A.; Pinskaya, M. History, Discovery, and Classification of LncRNAs. Adv. Exp. Med. Biol. 2017, 1008, 1–46. [Google Scholar] [CrossRef]
- Yarani, R.; Mirza, A.H.; Kaur, S.; Pociot, F. The Emerging Role of LncRNAs in Inflammatory Bowel Disease. Exp. Mol. Med. 2018, 50, 1–14. [Google Scholar] [CrossRef]
- Vasudeva, K.; Dutta, A.; Munshi, A. Role of LncRNAs in the Development of Ischemic Stroke and Their Therapeutic Potential. Mol. Neurobiol. 2021, 58, 3712–3728. [Google Scholar] [CrossRef]
- Yuan, C.; Ning, Y.; Pan, Y. Emerging Roles of HOTAIR in Human Cancer. J. Cell. Biochem. 2020, 121, 3235–3247. [Google Scholar] [CrossRef]
- Zhao, W.; Geng, D.; Li, S.; Chen, Z.; Sun, M. LncRNA HOTAIR Influences Cell Growth, Migration, Invasion, and Apoptosis via the MiR-20a-5p/HMGA2 Axis in Breast Cancer. Cancer Med. 2018, 7, 842–855. [Google Scholar] [CrossRef]
- Loewen, G.; Jayawickramarajah, J.; Zhuo, Y.; Shan, B. Functions of LncRNA HOTAIR in Lung Cancer. J. Hematol. Oncol. 2014, 7, 90. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Peng, X.; Li, Y.; Zhang, X.; Ma, Y.; Wu, C.; Fan, Q.; Wei, S.; Li, H.; Liu, J. Long Non-Coding RNA HOTAIR Promotes Exosome Secretion by Regulating RAB35 and SNAP23 in Hepatocellular Carcinoma. Mol. Cancer 2019, 18, 78. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Li, X.; Wu, H.; Tang, Y.; Li, X.; Shi, Y. The COX10-AS1/MiR-641/E2F6 Feedback Loop Is Involved in the Progression of Glioma. Front. Oncol. 2021, 11, 648152. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Jiang, X.; Liang, A.; Zhu, R.; Yang, Y.; Zhong, L.; Wan, D. COX10-AS1 Facilitates Cell Proliferation and Inhibits Cell Apoptosis in Glioblastoma Cells at Post-Transcription Level. Neurochem. Res. 2020, 45, 2196–2203. [Google Scholar] [CrossRef]
- Rothzerg, E.; Ho, X.D.; Xu, J.; Wood, D.; Märtson, A.; Kõks, S. Upregulation of 15 Antisense Long Non-Coding RNAs in Osteosarcoma. Genes 2021, 12, 1132. [Google Scholar] [CrossRef]
- Hu, C.C.; Liang, Y.W.; Hu, J.L.; Liu, L.F.; Liang, J.W.; Wang, R. LncRNA RUSC1-AS1 Promotes the Proliferation of Breast Cancer Cells by Epigenetic Silence of KLF2 and CDKN1A. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 6602–6611. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.A.; Cheng, L.; Zhang, Y.; Peng, L.; Yang, H.G. LncRNA RUSC1-AS1 Promotes the Proliferation of Hepatocellular Carcinoma Cells through Modulating NOTCH Signaling. Neoplasma 2020, 67, 1204–1213. [Google Scholar] [CrossRef]
- Tong, C.J.; Deng, Q.C.; Ou, D.J.; Long, X.; Liu, H.; Huang, K. LncRNA RUSC1-AS1 Promotes Osteosarcoma Progression through Regulating the MiR-340-5p and PI3K/AKT Pathway. Aging 2021, 13, 20116–20130. [Google Scholar] [CrossRef]
- Wang, Q.; Zhang, C.; Cao, S.; Zhao, H.; Jiang, R.; Li, Y. Tumor-Derived Exosomes Orchestrate the MicroRNA-128-3p/ELF4/CDX2 Axis to Facilitate the Growth and Metastasis of Gastric Cancer via Delivery of LINC01091. Cell Biol. Toxicol. 2022. [Google Scholar] [CrossRef]
- Nikas, J.B.; Nikas, E.G. Genome-Wide DNA Methylation Model for the Diagnosis of Prostate Cancer. ACS Omega 2019, 4, 14902–14912. [Google Scholar] [CrossRef]
- Tan, C.; Cao, J.; Chen, L.; Xi, X.; Wang, S.; Zhu, Y.; Yang, L.; Ma, L.; Wang, D.; Yin, J.; et al. Noncoding RNAs Serve as Diagnosis and Prognosis Biomarkers for Hepatocellular Carcinoma. Clin. Chem. 2019, 65, 905–915. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, X.; Zhu, H.; Liu, Y.; Cao, J.; Li, D.; Ding, B.; Yan, W.; Jin, H.; Wang, S. Identification of Potential Prognostic Long Non-Coding RNA Biomarkers for Predicting Recurrence in Patients with Cervical Cancer. Cancer Manag. Res. 2020, 12, 719–730. [Google Scholar] [CrossRef] [PubMed]
- Eisenberg, M.L.; Li, S.; Brooks, J.D.; Cullen, M.R.; Baker, L.C. Increased Risk of Cancer in Infertile Men: Analysis of U.S. Claims Data. J. Urol. 2015, 193, 1596–1601. [Google Scholar] [CrossRef]
- Eisenberg, M.L.; Betts, P.; Herder, D.; Lamb, D.J.; Lipshultz, L.I. Increased Risk of Cancer among Azoospermic Men. Fertil. Steril. 2013, 100, 681–685. [Google Scholar] [CrossRef]
- Anderson, R.E.; Hanson, H.A.; Lowrance, W.T.; Redshaw, J.; Oottamasathien, S.; Schaeffer, A.; Johnstone, E.; Aston, K.I.; Carrell, D.T.; Cartwright, P.; et al. Childhood Cancer Risk in the Siblings and Cousins of Men with Poor Semen Quality. J. Urol. 2017, 197, 898–905. [Google Scholar] [CrossRef] [PubMed]
- Asadi, A.; Ghahremani, R.; Abdolmaleki, A.; Rajaei, F. Role of Sperm Apoptosis and Oxidative Stress in Male Infertility: A Narrative Review. Int. J. Reprod. Biomed. 2021, 19, 493–504. [Google Scholar] [CrossRef] [PubMed]
- Shukla, K.K.; Mahdi, A.A.; Singh, R. Apoptosis, Spermatogenesis and Male Infertility. Front. Biosci.-Elit. 2012, 4, 746–754. [Google Scholar] [CrossRef]
- Qiu, Q.; Yu, X.; Yao, C.; Hao, Y.; Fan, L.; Li, C.; Xu, P.; An, G.; Li, Z.; He, Z. FOXP3 Pathogenic Variants Cause Male Infertility through Affecting the Proliferation and Apoptosis of Human Spermatogonial Stem Cells. Aging 2019, 11, 12581–12599. [Google Scholar] [CrossRef]
- Inoue, S.; Tomasini, R.; Rufini, A.; Elia, A.J.; Agostini, M.; Amelio, I.; Cescon, D.; Dinsdale, D.; Zhou, L.; Harris, I.S.; et al. TAp73 Is Required for Spermatogenesis and the Maintenance of Male Fertility. Proc. Natl. Acad. Sci. USA 2014, 111, 1843–1848. [Google Scholar] [CrossRef]
- Levine, A.J.; Tomasini, R.; McKeon, F.D.; Mak, T.W.; Melino, G. The P53 Family: Guardians of Maternal Reproduction. Nat. Rev. Mol. Cell Biol. 2011, 12, 259–265. [Google Scholar] [CrossRef]
- Nagirnaja, L.; Aston, K.I.; Conrad, D.F. Genetic Intersection of Male Infertility and Cancer. Fertil. Steril. 2018, 109, 20–26. [Google Scholar] [CrossRef] [PubMed]
- Capogrosso, P.; Ventimiglia, E.; Boeri, L.; Cazzaniga, W.; Chierigo, F.; Montorsi, F.; Salonia, A. Male Infertility as a Proxy of the Overall Male Health Status. Minerva Urol. Nefrol. 2018, 70, 286–299. [Google Scholar] [CrossRef]
- Aznaourova, M.; Schmerer, N.; Schmeck, B.; Schulte, L.N. Disease-Causing Mutations and Rearrangements in Long Non-Coding RNA Gene Loci. Front. Genet. 2020, 11, 1485. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, S.; Lied, A.; Kulkarni, V.; Rucevic, M.; Martin, M.P.; Walker-Sperling, V.; Anderson, S.K.; Ewy, R.; Singh, S.; Nguyen, H.; et al. CCR5AS LncRNA Variation Differentially Regulates CCR5, Influencing HIV Disease Outcome. Nat. Immunol. 2019, 20, 824–834. [Google Scholar] [CrossRef] [PubMed]
- Wilson, C.; Kanhere, A. The Missing Link Between Cancer-Associated Variants and LncRNAs. Trends Genet. 2021, 37, 410–413. [Google Scholar] [CrossRef]
- Olazagoitia-Garmendia, A.; Sebastian-delaCruz, M.; Castellanos-Rubio, A. Involvement of LncRNAs in Celiac Disease Pathogenesis. Int. Rev. Cell Mol. Biol. 2021, 358, 241–264. [Google Scholar] [CrossRef]
- Han, Z.; Xue, W.; Tao, L.; Lou, Y.; Qiu, Y.; Zhu, F. Genome-Wide Identification and Analysis of the EQTL LncRNAs in Multiple Sclerosis Based on RNA-Seq Data. Brief. Bioinform. 2020, 21, 1023–1037. [Google Scholar] [CrossRef]
- Lee, P.H.; Lee, C.; Li, X.; Wee, B.; Dwivedi, T.; Daly, M. Principles and Methods of In-Silico Prioritization of Non-Coding Regulatory Variants. Hum. Genet. 2018, 137, 15–30. [Google Scholar] [CrossRef]
- Halvorsen, M.; Martin, J.S.; Broadaway, S.; Laederach, A. Disease-Associated Mutations That Alter the RNA Structural Ensemble. PLoS Genet. 2010, 6, e1001074. [Google Scholar] [CrossRef]
- Lappalainen, T.; Sammeth, M.; Friedländer, M.R.; ’T Hoen, P.A.C.; Monlong, J.; Rivas, M.A.; Gonzàlez-Porta, M.; Kurbatova, N.; Griebel, T.; Ferreira, P.G.; et al. Transcriptome and Genome Sequencing Uncovers Functional Variation in Humans. Nature 2013, 501, 506–511. [Google Scholar] [CrossRef]
- GTEx Consortium; Ardlie, K.G.; DeLuca, D.S.; Segrè, A.V.; Sullivan, T.J.; Young, T.R.; Gelfand, E.T.; Trowbridge, C.A.; Maller, J.B.; Tukiainen, T.; et al. The Genotype-Tissue Expression (GTEx) Pilot Analysis: Multitissue Gene Regulation in Humans. Science 2015, 348, 648–660. [Google Scholar] [CrossRef] [PubMed]
- Necsulea, A.; Soumillon, M.; Warnefors, M.; Liechti, A.; Daish, T.; Zeller, U.; Baker, J.C.; Grützner, F.; Kaessmann, H. The Evolution of LncRNA Repertoires and Expression Patterns in Tetrapods. Nature 2014, 505, 635–640. [Google Scholar] [CrossRef] [PubMed]
- Winge, S.B.; Dalgaard, M.D.; Belling, K.G.; Jensen, J.M.; Nielsen, J.E.; Aksglaede, L.; Schierup, M.H.; Brunak, S.; Skakkebæk, N.E.; Juul, A.; et al. Transcriptome Analysis of the Adult Human Klinefelter Testis and Cellularity-Matched Controls Reveals Disturbed Differentiation of Sertoli- and Leydig Cells. Cell Death Dis. 2018, 9, 1–14. [Google Scholar] [CrossRef]
- Salemi, M.; Cannarella, R.; Condorelli, R.A.; Cimino, L.; Ridolfo, F.; Giurato, G.; Romano, C.; La Vignera, S.; Calogero, A.E. Evidence for Long Noncoding RNA GAS5 Up-Regulationin Patients with Klinefelter Syndrome. BMC Med. Genet. 2019, 20, 4. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Teng, H.; Yao, F.; Yap, S.; Sun, Y.; Ma, L. Challenges and Strategies in Ascribing Functions to Long Noncoding RNAs. Cancers 2020, 12, 1458. [Google Scholar] [CrossRef] [PubMed]
- Freedman, J.E.; Miano, J.M. Challenges and Opportunities in Linking Long Noncoding RNAs to Cardiovascular, Lung, and Blood Diseases. Arterioscler. Thromb. Vasc. Biol. 2017, 37, 21. [Google Scholar] [CrossRef] [PubMed]
- Kopp, F.; Mendell, J.T. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 2018, 172, 393–407. [Google Scholar] [CrossRef]
- Dhanoa, J.K.; Sethi, R.S.; Verma, R.; Arora, J.S.; Mukhopadhyay, C.S. Long Non-Coding RNA: Its Evolutionary Relics and Biological Implications in Mammals: A Review. J. Anim. Sci. Technol. 2018, 60, 1–10. [Google Scholar] [CrossRef]


| Inclusion Criteria | Exclusion Criteria |
|---|---|
| Human participants | Animal and plant studies |
| Participants diagnosed with male infertility (seminogram) | Studies examining only the pregnancy outcome (no diagnosis of infertility) |
| Studies in English language | Reviews, perspectives, meta-analyses and other studies with non-original data |
| Studies not providing sufficient details about data | |
| No appropriate control group for comparison between fertile and infertile men or mixed sample of fertile and infertile males | |
| Studies on other types of noncoding RNAs (circRNAs, piRNAs, etc.) | |
| Studies not in English language | |
| Studies about Klinefelter syndrome |
| LncRNAs | Reference | Methodology | Samples | Tissue | Change of Expression |
|---|---|---|---|---|---|
| HOTAIR | Zhang et al. (2015) [35] | qPCR | 45 oligoasthenozoospermic patients, 45 healthy controls | Semen | Downregulated in patients |
| LncRNAs | Reference | Methodology | Samples | Tissue | Change of Expression |
|---|---|---|---|---|---|
| ANO1-AS2 (LINC02584) | Saberiyan et al. (2020) [43] | qPCR | 35 patients with TAZ, 34 people with normozoospermia (NZ, control) | Semen | Upregulated in patients |
| CFAP44-AS1 | Kamel et al. (2022) [36] | qPCR | 35 TAZ patients, 35 normozoospermic men | Semen | Downregulated in patients |
| LncRNAs | Reference | Methodology | Samples | Tissue | Change of expression |
|---|---|---|---|---|---|
| MIR210HG | Ataabadi et al. (2020) [38] | RNA sequencing, qPCR | 25 infertile men with varicocele, 17 fertile men as controls | Semen | Upregulation in patients |
| Li et al. (2022) [28] | qPCR | 188 VC patients, 92 healthy men | Seminal plasma | Upregulation in patients | |
| MLLT4-AS1 | Ataabadi et al. (2020) [38] | RNA sequencing, qPCR | 25 infertile men with varicocele, 17 fertile men as controls | Semen | Upregulation in patients |
| GADD7 | Zhao et al. (2017) [40] | qPCR | 56 patients with varicocele, 28 healthy controls | Semen | Upregulated in patients |
| SLC7A11-AS1 (isoform 6) | Sanei-Ataabadi, Mowla and Nasr-Esfahani, (2020) [34] | qPCR | 25 individuals with varicocele, 17 healthy donors with normal semen parameters | Semen | Upregulated in patients |
| Reference | Sub-Type of Male Infertility | lncRNA | miRNAs | Target Genes | Process Affected |
|---|---|---|---|---|---|
| Lü et al. (2015) [81] | NOA (MA, Hypo) | NLC1-C | miR-320a and miR-383 | - | Spermatogenesis, cell apoptosis and proliferation |
| Su et al. (2019) [39] | NOA (Hypo) | HOTTIP (ceRNA) | miR-128-3p | HOXA13 | Cell proliferation |
| Bo et al. (2020) [29] | NOA | LINC00467 | miR-500-3p | TDRD6, LRGUK | Spermatogenesis (Male gamete generation) |
| LncRNAs | References | Sub-Types of Male Infertility | Regulation |
|---|---|---|---|
| SPATA42 | Xie et al. (2020) [27] | NOA | Downregulated |
| Bo et al. (2020) [29] | NOA | Downregulated | |
| Lu et al. (2020) [41] | Asthenozoospermia | - | |
| LINC00301 | Xie et al. (2020) [27] | NOA | Downregulated |
| Sabetian et al. (2022) [33] | NOA | Downregulated | |
| ZNF503-AS1 | Xie et al. (2020) [27] | NOA | - |
| Lu et al. (2020) [41] | Asthenozoospermia | - | |
| LINC00863 | Xie et al. (2020) [27] | NOA | - |
| Lu et al. (2020) [41] | Asthenozoospermia | - | |
| PTOV1-AS2 | Xie et al. (2020) [27] | NOA | - |
| Sun et al. (2021) [37] | Oligozoospermia | - | |
| LINC00710 | Xie et al. (2020) [27] | NOA | - |
| Sun et al. (2021) [37] | Oligozoospermia | - | |
| Zhou and Wang, (2020) [45] | Teratozoospermia | - | |
| THUMPD3-AS1 | Xie et al. (2020) [27] | NOA | - |
| Lu et al. (2020) [41] | Asthenozoospermia | - | |
| LINC00847 | Xie et al. (2020) [27] | NOA | - |
| Sun et al. (2021) [37] | Oligozoospermia | - | |
| LINC00467 | Bo et al. (2020) [29] | NOA | Downregulated |
| Zhou and Wang, (2020) [45] | Teratozoospermia | Downregulated | |
| LINC00173 | Bo et al. (2020) [29] | NOA | Upregulated |
| Lu et al. (2020) [41] | Asthenozoospermia | - | |
| RUSC1-AS1 | Bo et al. (2020) [29] | NOA | Upregulated |
| Lu et al. (2020) [41] | Asthenozoospermia | - | |
| Sun et al. (2021) [37] | Oligozoospermia | - | |
| FAM230B | Sabetian et al. (2022) [33] | NOA | Downregulated |
| Lu et al. (2020) [41] | Asthenozoospermia | - | |
| LINC00905 | Sabetian et al. (2022) [33] | NOA | Downregulated |
| Lu et al. (2020) [41] | Asthenozoospermia | - | |
| Sun et al. (2021) [37] | Oligozoospermia | - | |
| MORC2-AS1 | Sabetian et al. (2022) [33] | NOA | Downregulated |
| Sun et al. (2021) [37] | Oligozoospermia | - | |
| MIR210HG | Ataabadi et al. (2020) [38] | Varicocele-related infertility | Upregulated |
| Li et al. (2022) [28] | Varicocele-related infertility | Upregulated | |
| LINC01039 | Lu et al. (2020) [41] | Asthenozoospermia | - |
| Sun et al. (2021) [37] | Oligozoospermia | - | |
| GLYCTK-AS1 | Lu et al. (2020) [41] | Asthenozoospermia | - |
| Sun et al. (2021) [37] | Oligozoospermia | - | |
| COX10-AS1 | Lu et al. (2020) [41] | Asthenozoospermia | Upregulated |
| Sun et al. (2021) [37] | Oligozoospermia | - | |
| Zhou and Wang (2020) [45] | Teratozoospermia | Upregulated | |
| LINC00894 | Lu et al. (2020) [41] | Asthenozoospermia | - |
| Sun et al. (2021) [37] | Oligozoospermia | - | |
| TRIM52-AS1 | Lu et al. (2020) [41] | Asthenozoospermia | - |
| Sun et al. (2021) [37] | Oligozoospermia | - | |
| HOTAIR | Lu et al. (2020) [41] | Asthenozoospermia | - |
| Zhang et al. (2015) [35] | Asthenozoospermia | Downregulated | |
| Zhang et al. (2015) [35] | Oligoasthenozoospermia | Downregulated | |
| CBR3-AS1 | Lu et al. (2020) [41] | Asthenozoospermia | - |
| Sun et al. (2021) [37] | Oligozoospermia | - | |
| LINC01091 | Lu et al. (2020) [41] | Asthenozoospermia | - |
| Sun et al. (2021) [37] | Oligozoospermia | - | |
| Zhou and Wang (2020) [45] | Teratozoospermia | - | |
| ZBED5-AS1 | Lu et al. (2020) [41] | Asthenozoospermia | - |
| Sun et al. (2021) [37] | Oligozoospermia | - | |
| VIPR1-AS1 | Lu et al. (2020) [41] | Asthenozoospermia | - |
| Sun et al. (2021) [37] | Oligozoospermia | - | |
| MYLK-AS1 | Lu et al. (2020) [41] | Asthenozoospermia | - |
| Sun et al. (2021) [37] | Oligozoospermia | - | |
| FARSA-AS1 | Lu et al. (2020) [41] | Asthenozoospermia | - |
| Sun et al. (2021) [37] | Oligozoospermia | - | |
| PPP3CB-AS1 | Lu et al. (2020) [41] | Asthenozoospermia | - |
| Sun et al. (2021) [37] | Oligozoospermia | - | |
| LINC01270 | Lu et al. (2020) [41] | Asthenozoospermia | - |
| Sun et al. (2021) [37] | Oligozoospermia | - | |
| SRRM2-AS1 | Lu et al. (2020) [41] | Asthenozoospermia | - |
| Sun et al. (2021) [37] | Oligozoospermia | - | |
| LINC01359 | Lu et al. (2020) [41] | Asthenozoospermia | - |
| Sun et al. (2021) [37] | Oligozoospermia | - | |
| Zhou and Wang (2020) [45] | Teratozoospermia | - | |
| UGDH-AS1 | Lu et al. (2020) [41] | Asthenozoospermia | - |
| Sun et al. (2021) [37] | Oligozoospermia | - | |
| MIR4435-2HG | Lu et al. (2020) [41] | Asthenozoospermia | - |
| Sun et al. (2021) [37] | Oligozoospermia | - | |
| ANO1-AS2 (LINC02584) | Saberiyan et al. (2020) [43] | Asthenozoospermia | Upregulated |
| Saberiyan et al. (2020) [43] | Teratoasthenozoospermia | Upregulated | |
| CFAP44-AS1 | Kamel et al. (2022) [36] | Asthenozoospermia | Downregulated |
| Kamel et al. (2022) [36] | Teratoasthenozoospermia | Downregulated | |
| RNF157-AS1 | Sun et al. (2021) [37] | Oligozoospermia | - |
| Zhou and Wang (2020) [45] | Teratozoospermia | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kyrgiafini, M.-A.; Sarafidou, T.; Mamuris, Z. The Role of Long Noncoding RNAs on Male Infertility: A Systematic Review and In Silico Analysis. Biology 2022, 11, 1510. https://doi.org/10.3390/biology11101510
Kyrgiafini M-A, Sarafidou T, Mamuris Z. The Role of Long Noncoding RNAs on Male Infertility: A Systematic Review and In Silico Analysis. Biology. 2022; 11(10):1510. https://doi.org/10.3390/biology11101510
Chicago/Turabian StyleKyrgiafini, Maria-Anna, Theologia Sarafidou, and Zissis Mamuris. 2022. "The Role of Long Noncoding RNAs on Male Infertility: A Systematic Review and In Silico Analysis" Biology 11, no. 10: 1510. https://doi.org/10.3390/biology11101510
APA StyleKyrgiafini, M.-A., Sarafidou, T., & Mamuris, Z. (2022). The Role of Long Noncoding RNAs on Male Infertility: A Systematic Review and In Silico Analysis. Biology, 11(10), 1510. https://doi.org/10.3390/biology11101510





