Chromosome-Level Assembly of Male Opsariichthys bidens Genome Provides Insights into the Regulation of the GnRH Signaling Pathway and Genome Evolution
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
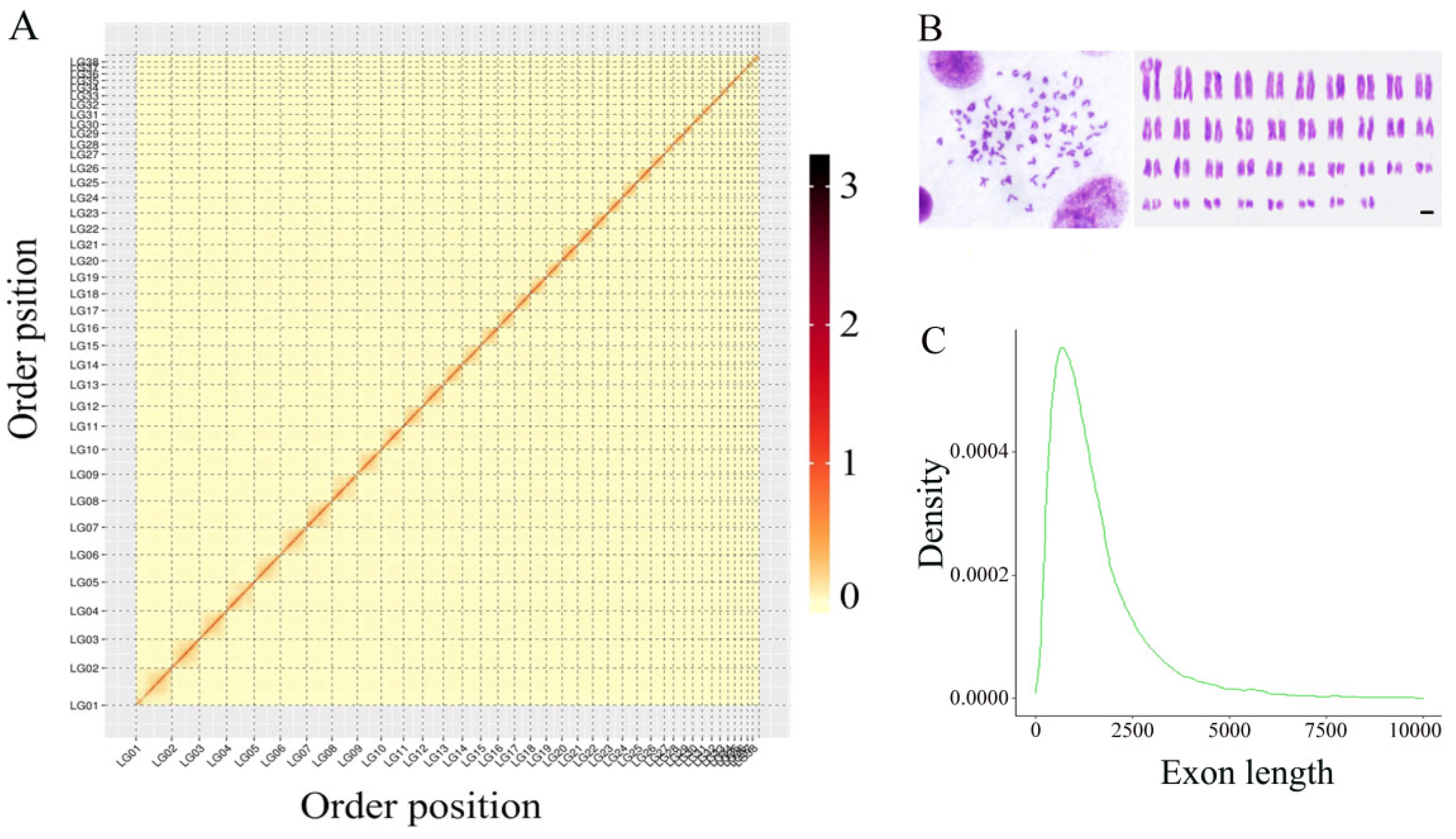
2.1. Sample Collection, Sequencing, and Karyotype Analysis
2.2. Genome Assembly
2.3. Genome Annotation
2.4. Genome Evolution Analysis
2.5. Comparison Analysis of Chromosomes
3. Results
3.1. Genome Assembly and Integrity Assessment
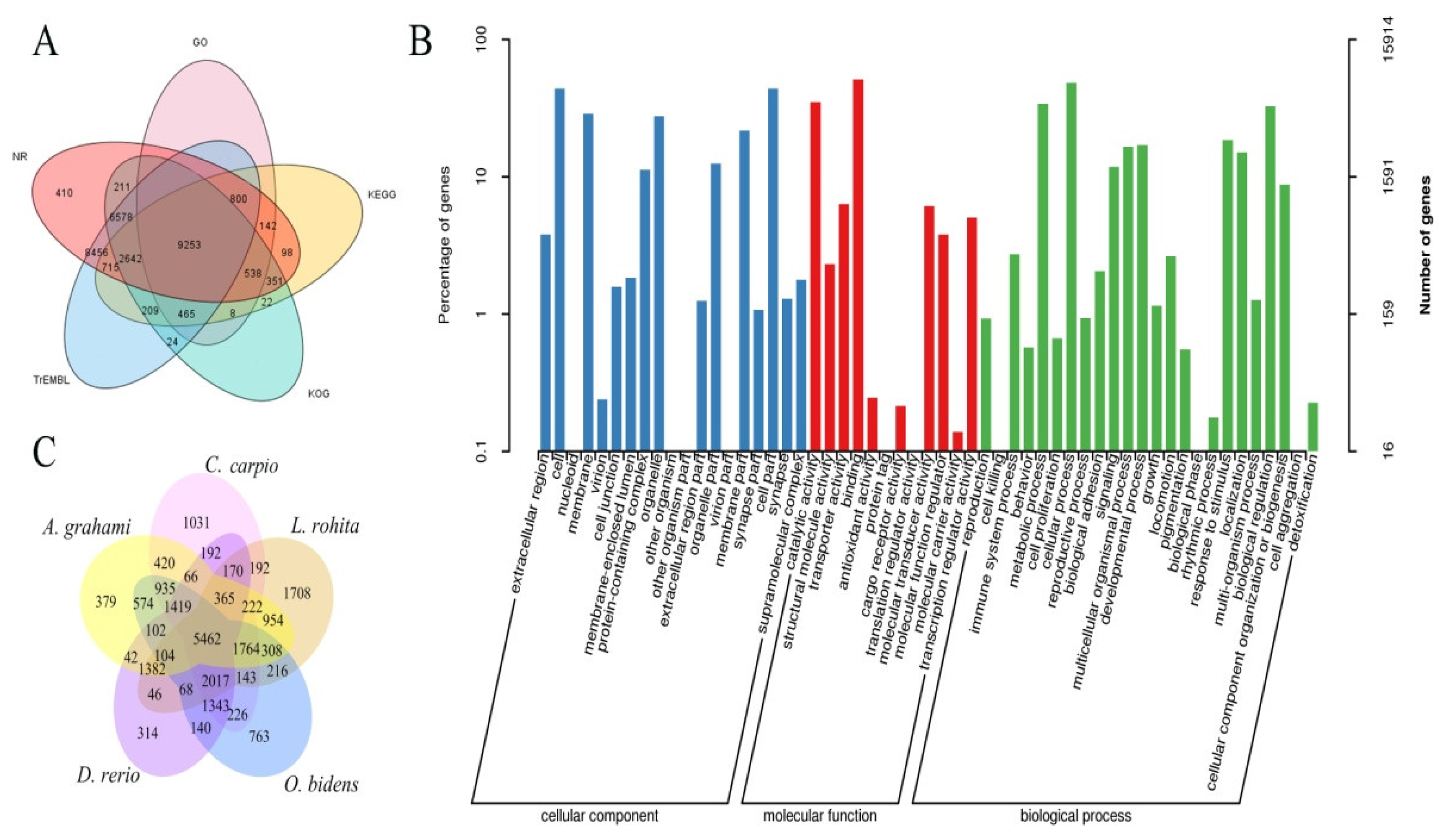
3.2. Genome Annotation
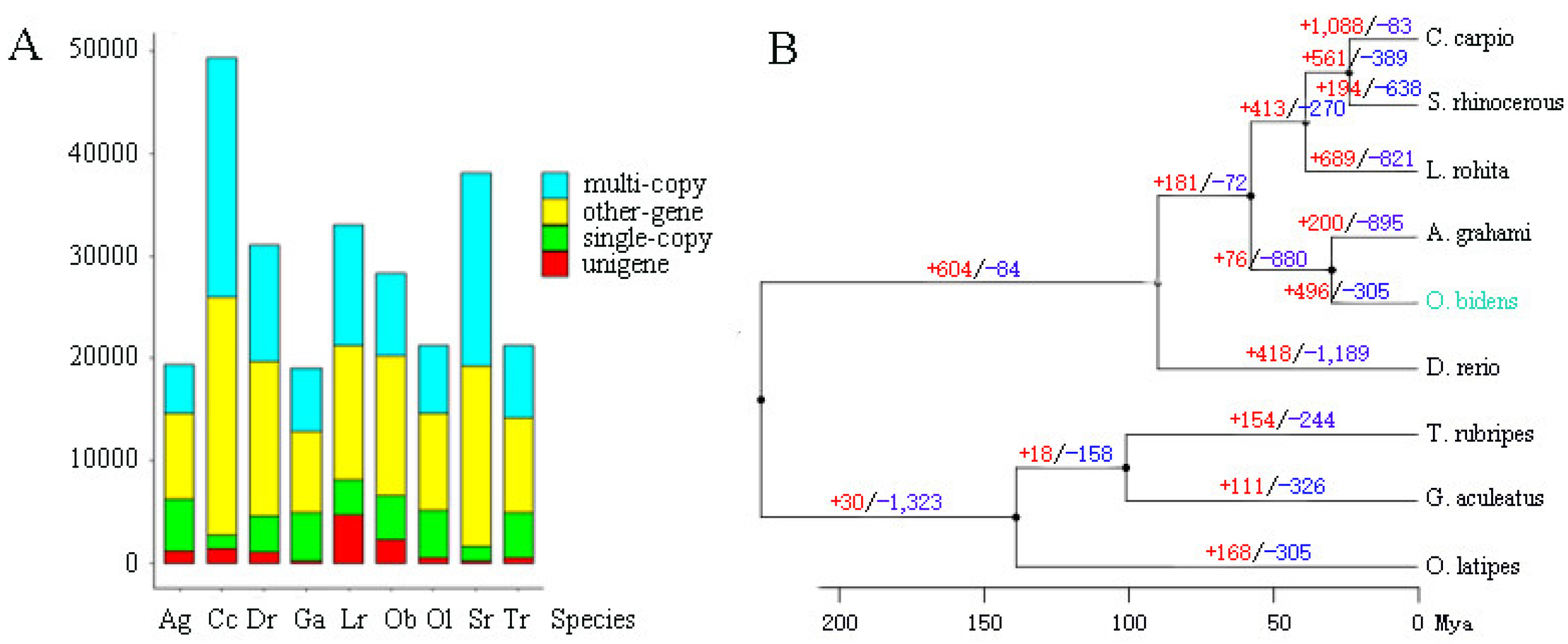
3.3. Genome Phylogeny, Expansion, and Contraction of Gene Families
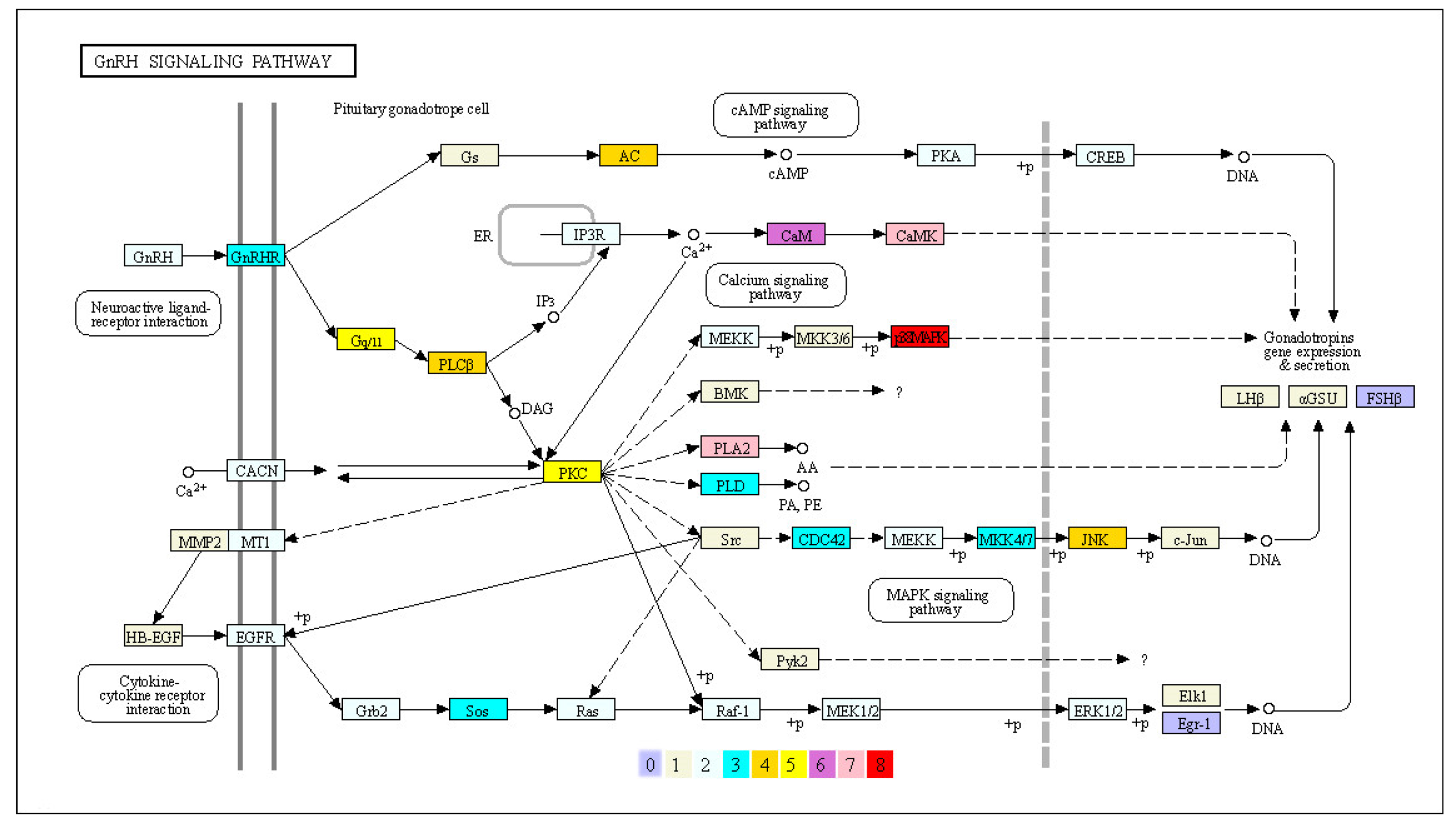
3.4. Expanded Genes Involved in the GnRH Signaling Pathway
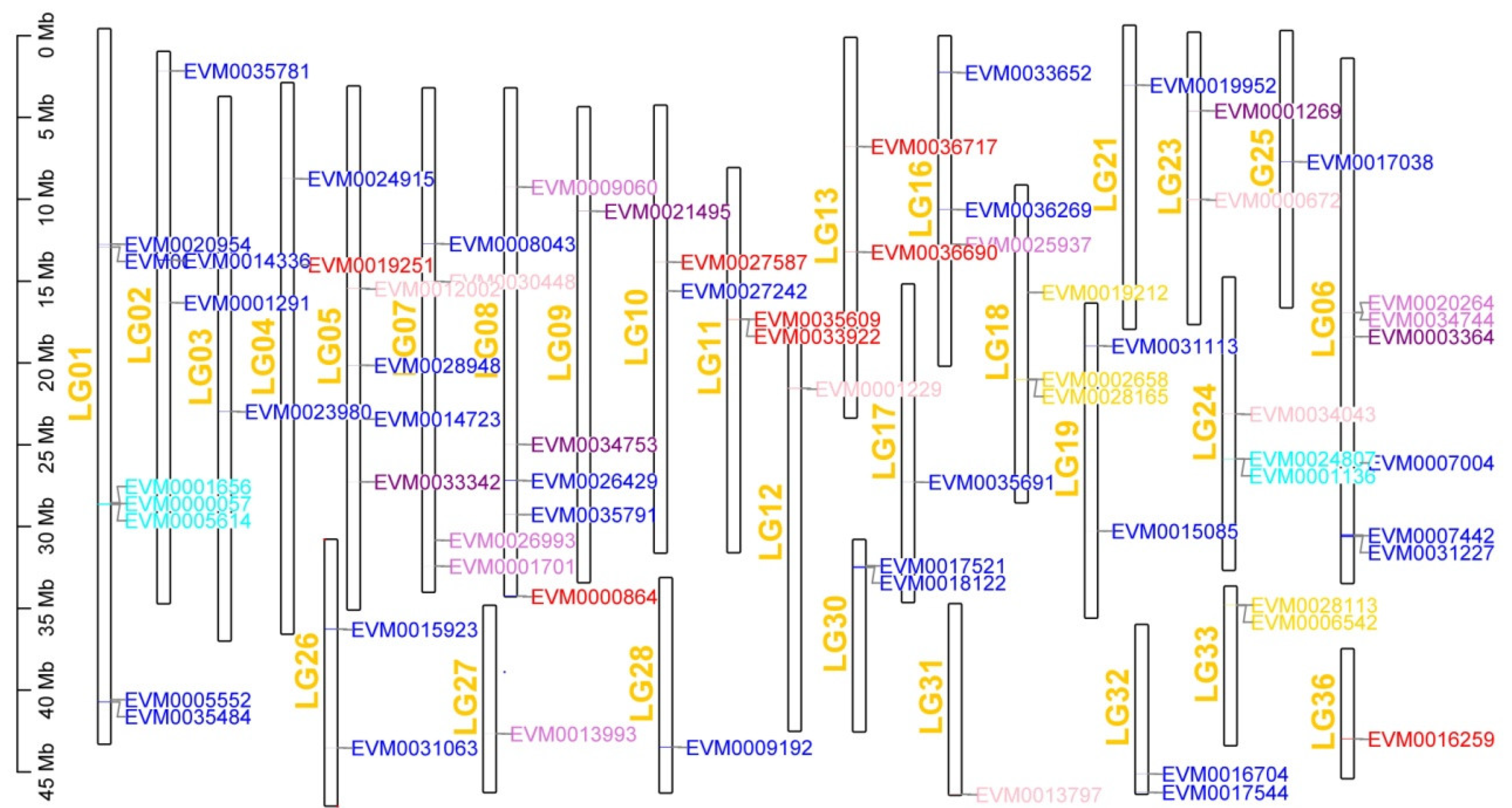
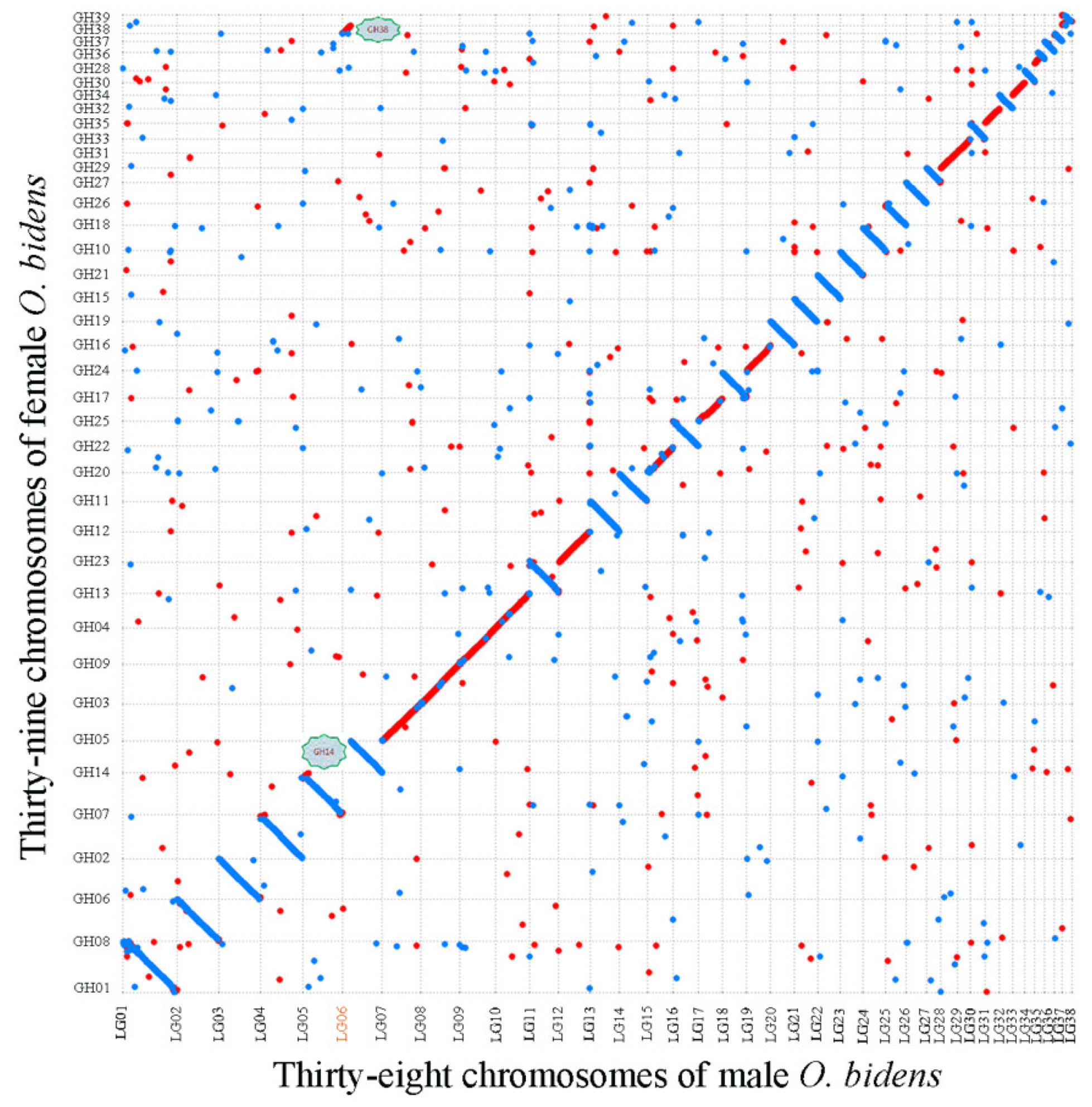
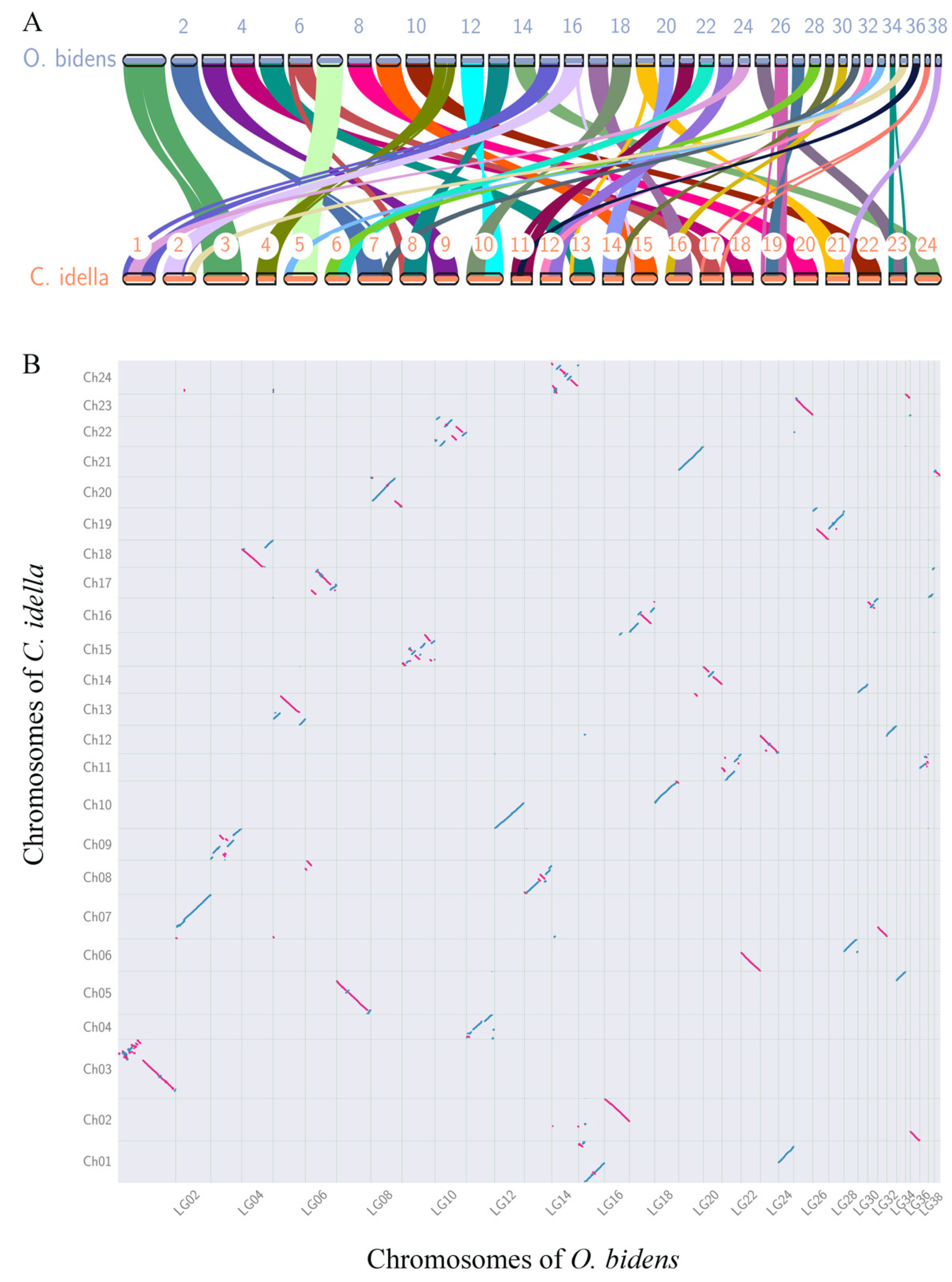
3.5. Chromosome Breakage and Reunion Verified by Synteny Comparison
4. Discussion
4.1. Quality of the Assembled Genome at the Chromosome Level
4.2. Male Sexual Dimorphism-Related GnRH Signaling Pathway
4.3. Mechanism Underlying O. bidens Chromosome Number Variety
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Fu, S.J.; Peng, Z.; Cao, Z.D.; Peng, J.L.; He, X.K.; Xu, D.; Zhang, A.J. Habitat-specific locomotor variation among Chinese hook snout carp (Opsariichthys bidens) along a river. PLoS ONE 2012, 7, e40791. [Google Scholar] [CrossRef]
- Du, G. Biological characteristics and artificial breeding technology of Opsariichthys bidens. North. Chin. Fish. 2021, 40, 50–52. [Google Scholar]
- Chen, W.; Schmidt, B.V.; He, S. The potential colonization histories of Opsariichthys bidens (Cyprinidae) in China using Bayesian binary MCMC analysis. Gene 2018, 676, 1–8. [Google Scholar] [CrossRef]
- Perdices, A.; Sayanda, D.; Coelho, M.M. Mitochondrial diversity of Opsariichthys bidens (Teleostei, Cyprinidae) in three Chinese drainages. Mol. Phylogenet. Evol. 2005, 37, 920–927. [Google Scholar] [CrossRef]
- Potau, N.; Ibanez, L.; Sentis, M.; Carrascosa, A. Sexual dimorphism in the maturation of the pituitary-gonadal axis, assessed by GnRH agonist challenge. Eur. J. Endocrinol. 1999, 141, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Tang, R.; Zhu, Y.; Gan, W.; Zhang, Y.; Yao, Z.; Ren, J.; Li, M. De novo transcriptome analysis of gonads reveals the sex-associated genes in Chinese hook snout carp Opsariichthys bidens. Aquac. Rep. 2022, 23, 101068. [Google Scholar] [CrossRef]
- Tang, R.; Xu, C.; Zhu, Y.; Yan, J.; Yao, Z.; Zhou, W.; Gui, L.; Li, M. Identification and expression analysis of sex biased miRNAs in chinese hook snout carp Opsariichthys bidens. Front. Genet. 2022, 13, 990683. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Kan, Y.; Zhong, Y.; Jawad, M.; Wei, W.; Gu, K.; Gui, L.; Li, M. Generation of a Normal Long-Term-Cultured Chinese Hook Snout Carp Spermatogonial Stem Cell Line Capable of Sperm Production In Vitro. Biology 2022, 11, 1069. [Google Scholar] [CrossRef]
- Lian, Q.; Fu, G.; Liu, S. Sexual dimorphism of morphological characteristics in Chinese hooksnout carp (Opsariichthys bidens). Mod. Agric. Technol. 2017, 22, 226–229. [Google Scholar]
- Xu, X.; Guan, W.; Niu, B.; Guo, D.; Xie, Q.P.; Zhan, W.; Yi, S.; Lou, B. Chromosome-Level Assembly of the Chinese Hooksnout Carp (Opsariichthys bidens) Genome Using PacBio Sequencing and Hi-C Technology. Front. Genet. 2021, 12, 788547. [Google Scholar] [CrossRef] [PubMed]
- Kirkpatrick, M. The Evolution of Genome Structure by Natural and Sexual Selection. J. Hered. 2017, 108, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Hua, H.; Luo, Z.; Zhang, Q.; Chen, M.; Gong, J.; Wei, X.; Huang, Z.; Huang, X.; Wang, Q. Whole-Genome Sequencing of 117 Chromosome Segment Substitution Lines for Genetic Analyses of Complex Traits in Rice. Rice 2022, 15, 5. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Zhang, X.; Wang, X.; Li, J.; Liu, G.; Kuang, Y.; Xu, J.; Zheng, X.; Ren, L.; Wang, G.; et al. Genome sequence and genetic diversity of the common carp, Cyprinus carpio. Nat. Genet. 2014, 46, 1212–1219. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhang, G.; Shao, C.; Huang, Q.; Liu, G.; Zhang, P.; Song, W.; An, N.; Chalopin, D.; Volff, J.N.; et al. Whole-genome sequence of a flatfish provides insights into ZW sex chromosome evolution and adaptation to a benthic lifestyle. Nat. Genet. 2014, 46, 253–260. [Google Scholar] [CrossRef]
- Wang, Y.; Lu, Y.; Zhang, Y.; Ning, Z.; Li, Y.; Zhao, Q.; Lu, H.; Huang, R.; Xia, X.; Feng, Q.; et al. The draft genome of the grass carp (Ctenopharyngodon idellus) provides insights into its evolution and vegetarian adaptation. Nat. Genet. 2015, 47, 625–631, Erratum in Nat. Genet. 2015, 47, 962. [Google Scholar] [CrossRef]
- Shao, C.; Bao, B.; Xie, Z.; Chen, X.; Li, B.; Jia, X.; Yao, Q.; Ortí, G.; Li, W.; Li, X.; et al. The genome and transcriptome of Japanese flounder provide insights into flatfish asymmetry. Nat. Genet. 2017, 49, 119–124. [Google Scholar] [CrossRef] [PubMed]
- Gong, G.; Dan, C.; Xiao, S.; Guo, W.; Huang, P.; Xiong, Y.; Wu, J.; He, Y.; Zhang, J.; Li, X.; et al. Chromosomal-level assembly of yellow catfish genome using third-generation DNA sequencing and Hi-C analysis. Gigascience 2018, 7, giy120. [Google Scholar] [CrossRef] [PubMed]
- Jian, J.; Yang, L.; Gan, X.; Wu, B.; Gao, L.; Zeng, H.; Wang, X.; Liang, Z.; Wang, Y.; Fang, L.; et al. Whole genome sequencing of silver carp (Hypophthalmichthys molitrix) and bighead carp (Hypophthalmichthys nobilis) provide novel insights into their evolution and speciation. Mol. Ecol. Resour. 2021, 21, 912–923. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, X.Y.; Xu, W.J.; Wang, K.; Wu, B.; Xu, M.; Chen, Y.; Miao, L.J.; Wang, Z.W.; Li, Z.; et al. Comparative genome anatomy reveals evolutionary insights into a unique amphitriploid fish. Nat. Ecol. Evol. 2022, 6, 1354–1366. [Google Scholar] [CrossRef]
- Gui, J.; Zhou, L.; Li, X. Rethinking fish biology and biotechnologies in the challenge era for burgeoning genome resources and strengthening food security. Water Biol. Secur. 2022, 1, 100002. [Google Scholar] [CrossRef]
- Chen, Y. Fauna Sinica: Osteichthyes Cypriniformes (II) [M]; Science Press: Beijing, China, 1998; pp. 46–49. [Google Scholar]
- Li, Y.; Li, K. Karyotypes of Opsariichthys bidens and Misgurnus anguillicaudatus (PISCES), with a consideration of the relationship between polymorphism of chromosome number and taxonomy of fish. J. Wuhan Univ. (Nat. Sci. Ed.) 1987, 1, 107–132. [Google Scholar]
- Xiao, Y.; Xiao, Z.; Ma, D.; Liu, J.; Li, J. Genome sequence of the barred knifejaw Oplegnathus fasciatus (Temminck & Schlegel, 1844): The first chromosome-level draft genome in the family Oplegnathidae. Gigascience 2019, 8, giz013. [Google Scholar] [PubMed]
- Ueno, K.; Takai, A. Multiple sex chromosome system of X1X1X2X2/X1X2)Y type in lutjanid fish, Lutjanus quinquelineatus (Perciformes). Genetica 2008, 132, 35–41. [Google Scholar] [CrossRef]
- Ranallo-Benavidez, T.R.; Jaron, K.S.; Schatz, M.C. GenomeScope 2.0 and Smudgeplot for reference-free profiling of polyploid genomes. Nat. Commun. 2020, 11, 1432. [Google Scholar] [CrossRef]
- Rao, S.S.; Huntley, M.H.; Durand, N.C.; Stamenova, E.K.; Bochkov, I.D.; Robinson, J.T.; Sanborn, A.L.; Machol, I.; Omer, A.D.; Lander, E.S.; et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 2014, 159, 1665–1680. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Wang, X.; Guo, H.; Zhang, X.; Zhang, M.; Tang, W. Chromosome-level genome assembly of the endangered humphead wrasse Cheilinus undulatus: Insight into the expansion of opsin genes in fishes. Mol. Ecol. Resour. 2021, 21, 2388–2406. [Google Scholar] [CrossRef]
- Wang, J.; Qin, Q.; Chen, S.; Liu, S.; Duan, W.; Liu, J.; Zhang, C.; Luo, K.; Xiao, J.; Liu, Y. Formation and biological characterization of three new types of improved crucian carp. Sci. China C Life Sci. 2008, 51, 544–551. [Google Scholar] [CrossRef] [PubMed]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017, 27, 722–736. [Google Scholar] [CrossRef] [PubMed]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Parra, G.; Bradnam, K.; Korf, I. CEGMA: A pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics 2007, 23, 1061–1067. [Google Scholar] [CrossRef] [PubMed]
- Simao, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef] [PubMed]
- Servant, N.; Varoquaux, N.; Lajoie, B.R.; Viara, E.; Chen, C.J.; Vert, J.P.; Heard, E.; Dekker, J.; Barillot, E. HiC-Pro: An optimized and flexible pipeline for Hi-C data processing. Genome Biol. 2015, 16, 259. [Google Scholar] [CrossRef] [PubMed]
- Burton, J.N.; Adey, A.; Patwardhan, R.P.; Qiu, R.; Kitzman, J.O.; Shendure, J. Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions. Nat. Biotechnol. 2013, 31, 1119–1125. [Google Scholar] [CrossRef]
- Xu, Z.; Wang, H. LTR_FINDER: An efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res. 2007, 35, W265–W268. [Google Scholar] [CrossRef]
- Price, A.L.; Jones, N.C.; Pevzner, P.A. De novo identification of repeat families in large genomes. Bioinformatics 2005, 21 (Suppl. S1), i351–i358. [Google Scholar] [CrossRef]
- Hoede, C.; Arnoux, S.; Moisset, M.; Chaumier, T.; Inizan, O.; Jamilloux, V.; Quesneville, H. PASTEC: An automatic transposable element classification tool. PLoS ONE 2014, 9, e91929. [Google Scholar] [CrossRef]
- Bao, W.; Kojima, K.K.; Kohany, O. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob. DNA 2015, 6, 11. [Google Scholar] [CrossRef]
- Tarailo-Graovac, M.; Chen, N. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr. Protoc. Bioinform. 2009, 4, 10. [Google Scholar] [CrossRef]
- Stanke, M.; Schoffmann, O.; Morgenstern, B.; Waack, S. Gene prediction in eukaryotes with a generalized hidden Markov model that uses hints from external sources. BMC Bioinform. 2006, 7, 62. [Google Scholar] [CrossRef]
- Keilwagen, J.; Hartung, F.; Grau, J. GeMoMa: Homology-Based Gene Prediction Utilizing Intron Position Conservation and RNA-seq Data. Gene Predict. 2019, 1962, 161–177. [Google Scholar]
- Tang, S.; Lomsadze, A.; Borodovsky, M. Identification of protein coding regions in RNA transcripts. Nucleic Acids Res. 2015, 43, e78. [Google Scholar] [CrossRef] [PubMed]
- Haas, B.J.; Salzberg, S.L.; Zhu, W.; Pertea, M.; Allen, J.E.; Orvis, J.; White, O.; Buell, C.R.; Wortman, J.R. Automated eukaryotic gene structure annotation using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome Biol. 2008, 9, R7. [Google Scholar] [CrossRef]
- Griffiths-Jones, S.; Moxon, S.; Marshall, M.; Khanna, A.; Eddy, S.R.; Bateman, A. Rfam: Annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 2005, 33, D121–D124. [Google Scholar] [CrossRef]
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Boeckmann, B.; Bairoch, A.; Apweiler, R.; Blatter, M.C.; Estreicher, A.; Gasteiger, E.; Martin, M.J.; Michoud, K.; O’Donovan, C.; Phan, I.; et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003, 31, 365–370. [Google Scholar] [CrossRef]
- Birney, E.; Clamp, M.; Durbin, R. GeneWise and Genomewise. Genome Res. 2004, 14, 988–995. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Stoeckert, C.J., Jr.; Roos, D.S. OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome Res. 2003, 13, 2178–2189. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z. PAML: A program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 1997, 13, 555–556. [Google Scholar] [CrossRef]
- De Bie, T.; Cristianini, N.; Demuth, J.P.; Hahn, M.W. CAFE: A computational tool for the study of gene family evolution. Bioinformatics 2006, 22, 1269–1271. [Google Scholar] [CrossRef]
- Hunter, S.; Apweiler, R.; Attwood, T.K.; Bairoch, A.; Bateman, A.; Binns, D.; Bork, P.; Das, U.; Daugherty, L.; Duquenne, L.; et al. InterPro: The integrative protein signature database. Nucleic Acids Res. 2009, 37, D211–D215. [Google Scholar] [CrossRef]
- Marcais, G.; Delcher, A.L.; Phillippy, A.M.; Coston, R.; Salzberg, S.L.; Zimin, A. MUMmer4: A fast and versatile genome alignment system. PLoS Comput. Biol. 2018, 14, e1005944. [Google Scholar] [CrossRef] [PubMed]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [PubMed]
- Nattestad, M.; Schatz, M.C. Assemblytics: A web analytics tool for the detection of variants from an assembly. Bioinformatics 2016, 32, 3021–3023. [Google Scholar] [CrossRef]
- Wu, C.S.; Ma, Z.Y.; Zheng, G.D.; Zou, S.M.; Zhang, X.J.; Zhang, Y.A. Chromosome-level genome assembly of grass carp (Ctenopharyngodon idella) provides insights into its genome evolution. BMC Genom. 2022, 23, 271. [Google Scholar] [CrossRef]
- Tang, H.; Krishnakumar, V.; Li, J. Jcvi: Jcvi Utility Libraries, Version 0.5.7. Zenodo. 2015. Available online: https://zenodo.org/record/31631#.Y0d0aExByUk (accessed on 18 August 2022).
- Zheng, W.; Huang, L.; Huang, J.; Wang, X.; Chen, X.; Zhao, J.; Guo, J.; Zhuang, H.; Qiu, C.; Liu, J.; et al. High genome heterozygosity and endemic genetic recombination in the wheat stripe rust fungus. Nat. Commun. 2013, 4, 2673. [Google Scholar] [CrossRef]
- Fu, B.; Zhou, Y.; Liu, H.; Yu, X.; Tong, J. Updated Genome Assembly of Bighead Carp (Hypophthalmichthys nobilis) and Its Differences between Male and Female on Genomic, Transcriptomic, and Methylation Level. Front. Genet. 2021, 12, 728177. [Google Scholar] [CrossRef]
- Howe, K.; Clark, M.D.; Torroja, C.F.; Torrance, J.; Berthelot, C.; Muffato, M.; Collins, J.E.; Humphray, S.; McLaren, K.; Matthews, L.; et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature 2013, 496, 498–503. [Google Scholar] [CrossRef]
- Mascali, F.C.; Posner, V.M.; Marano, E.A.R.; del Pazo, F.; Hermida, M.; Sanchez, S.; Mazzoni, T.S.; Martinez, P.; Rubiolo, J.A.; Villanova, G.V. Development and validation of sex-specific markers in Piaractus mesopotamicus. Aquaculture 2022, 558, 738374. [Google Scholar] [CrossRef]
- Okubo, K.; Nagahama, Y. Structural and functional evolution of gonadotropin-releasing hormone in vertebrates. Acta Physiol. 2008, 193, 3–15. [Google Scholar] [CrossRef] [PubMed]
- Fernald, R.D.; White, R.B. Gonadotropin-releasing hormone genes: Phylogeny, structure, and functions. Front. Neuroendocrinol. 1999, 20, 224–240. [Google Scholar] [CrossRef] [PubMed]
- Bosch, E.; Alviggi, C.; Lispi, M.; Conforti, A.; Hanyaloglu, A.C.; Chuderland, D.; Simoni, M.; Raine-Fenning, N.; Crepieux, P.; Kol, S.; et al. Reduced FSH and LH action: Implications for medically assisted reproduction. Hum. Reprod. 2021, 36, 1469–1480. [Google Scholar] [CrossRef]
- Wettschureck, N.; Offermanns, S. Mammalian G proteins and their cell type specific functions. Physiol. Rev. 2005, 85, 1159–1204. [Google Scholar] [CrossRef]
- Krsmanovic, L.Z.; Mores, N.; Navarro, C.E.; Arora, K.K.; Catt, K.J. An agonist-induced switch in G protein coupling of the gonadotropin-releasing hormone receptor regulates pulsatile neuropeptide secretion. Proc. Natl. Acad. Sci. USA 2003, 100, 2969–2974. [Google Scholar] [CrossRef] [PubMed]
- Ruf, F.; Fink, M.Y.; Sealfon, S.C. Structure of the GnRH receptor-stimulated signaling network: Insights from genomics. Front. Neuroendocrinol. 2003, 24, 181–199. [Google Scholar] [CrossRef]
- Du, Y.X.; Ma, K.Y.; Qiu, G.F. Discovery of the genes in putative GnRH signaling pathway with focus on characterization of GnRH- like receptor transcripts in the brain and ovary of the oriental river prawn Macrobrachium nipponense. Aquaculture 2015, 442, 1–11. [Google Scholar] [CrossRef]
- Illario, M.; Cavallo, A.L.; Bayer, K.U.; Di Matola, T.; Fenzi, G.; Rossi, G.; Vitale, M. Calcium/calmodulin-dependent protein kinase II binds to Raf-1 and modulates integrin-stimulated ERK activation. J. Biol. Chem. 2003, 278, 45101–45108. [Google Scholar] [CrossRef]
- Song, C.; Chang, X.J.; Bean, K.M.; Proia, M.S.; Knopf, J.L.; Kriz, R.W. Molecular characterization of cytosolic phospholipase A2-beta. J. Biol. Chem. 1999, 274, 17063–17067. [Google Scholar] [CrossRef] [PubMed]
- Melamed, P.; Savulescu, D.; Lim, S.; Wijeweera, A.; Luo, Z.; Luo, M.; Pnueli, L. Gonadotrophin-releasing hormone signalling downstream of calmodulin. J. Neuroendocrinol. 2012, 24, 1463–1475. [Google Scholar] [CrossRef] [PubMed]
- Fung, R.S.K.; Bai, J.; Yuen, K.W.Y.; Wong, A.O.L. Activin/follistatin system in grass carp pituitary cells: Regulation by local release of growth hormone and luteinizing hormone and its functional role in growth hormone synthesis and secretion. PLoS ONE 2017, 12, e0179789. [Google Scholar] [CrossRef]
- Chen, D.; Yang, W.; Han, S.; Yang, H.; Cen, X.; Liu, J.; Zhang, L.; Zhang, W. A Type IIb, but Not Type IIa, GnRH Receptor Mediates GnRH-Induced Release of Growth Hormone in the Ricefield Eel. Front. Endocrinol. 2018, 9, 721. [Google Scholar] [CrossRef] [PubMed]
- Fontaine, R.; Royan, M.R.; von Krogh, K.; Weltzien, F.A.; Baker, D.M. Direct and Indirect Effects of Sex Steroids on Gonadotrope Cell Plasticity in the Teleost Fish Pituitary. Front. Endocrinol. 2020, 11, 605068. [Google Scholar] [CrossRef] [PubMed]
- Klausen, C.; Chang, J.P.; Habibi, H.R. The effect of gonadotropin-releasing hormone on growth hormone and gonadotropin subunit gene expression in the pituitary of goldfish, Carassius auratus. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2001, 129, 511–516. [Google Scholar] [CrossRef]
- Zhou, H.; Jiang, Y.; Ko, W.K.; Li, W.; Wong, A.O. Paracrine regulation of growth hormone gene expression by gonadotrophin release in grass carp pituitary cells: Functional implications, molecular mechanisms and signal transduction. J. Mol. Endocrinol. 2005, 34, 415–432. [Google Scholar] [CrossRef]
- Bhandari, R.K.; Taniyama, S.; Kitahashi, T.; Ando, H.; Yamauchi, K.; Zohar, Y.; Ueda, H.; Urano, A. Seasonal changes of responses to gonadotropin-releasing hormone analog in expression of growth hormone/prolactin/somatolactin genes in the pituitary of masu salmon. Gen. Comp. Endocrinol. 2003, 130, 55–63. [Google Scholar] [CrossRef]
- Meyer, A.; Van de Peer, Y. From 2R to 3R: Evidence for a fish-specific genome duplication (FSGD). Bioessays 2005, 27, 937–945. [Google Scholar] [CrossRef]
- Jaillon, O.; Aury, J.M.; Brunet, F.; Petit, J.L.; Stange-Thomann, N.; Mauceli, E.; Bouneau, L.; Fischer, C.; Ozouf-Costaz, C.; Bernot, A.; et al. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature 2004, 431, 946–957. [Google Scholar] [CrossRef]
- David, L.; Blum, S.; Feldman, M.W.; Lavi, U.; Hillel, J. Recent duplication of the common carp (Cyprinus carpio L.) genome as revealed by analyses of microsatellite loci. Mol. Biol. Evol. 2003, 20, 1425–1434. [Google Scholar] [CrossRef]
- Wang, J.T.; Li, J.T.; Zhang, X.F.; Sun, X.W. Transcriptome analysis reveals the time of the fourth round of genome duplication in common carp (Cyprinus carpio). BMC Genom. 2012, 13, 96. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Xiao, Z.; Ma, D.; Zhao, C.; Liu, L.; Wu, H.; Nie, W.; Xiao, S.; Liu, J.; Li, J.; et al. Chromosome-Level Genome Reveals the Origin of Neo-Y Chromosome in the Male Barred Knifejaw Oplegnathus fasciatus. iScience 2020, 23, 101039. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Sember, A.; Zhu, Q.; Oliveira, E.A.; Liehr, T.; Al-Rikabi, A.B.H.; Xiao, Z.; Song, H.; Cioffi, M.B. Deciphering the Origin and Evolution of the X1X2Y System in Two Closely-Related Oplegnathus Species (Oplegnathidae and Centrarchiformes). Int. J. Mol. Sci. 2019, 20, 3571. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, K.; Hong, Y.; Gui, J.; Zhou, T. Research on the karyotypes of Chinese Cyprinidae fishes VΙΙ. Analysis of the karyotypes and systematic relationship of seven Leuciscinae species. J. Genet. 1985, 5, 367–372. [Google Scholar]

| Types | Number | Length (bp) | Rate (%) |
|---|---|---|---|
| Class I | 1,139,895 | 232,332,806 | 23.34 |
| DIRS | 105,253 | 29,919,018 | 3.01 |
| LARD | 655,447 | 119,102,747 | 11.99 |
| LINE | 223,903 | 52,367,782 | 5.27 |
| Copia | 7429 | 2,540,946 | 0.26 |
| Gypsy | 61,709 | 19,444,768 | 1.96 |
| PLE | 24,473 | 8,156,447 | 0.82 |
| SINE | 1725 | 316,380 | 0.03 |
| Unknown | 47,754 | 15,326,877 | 1.54 |
| Class II | 785,886 | 137,824,754 | 12.59 |
| Crypton | 11,798 | 1,634,260 | 0.16 |
| Helitron | 35,267 | 9,129,144 | 0.92 |
| Maverick | 4578 | 567,260 | 0.06 |
| TIR | 601,449 | 113,701,347 | 11.45 |
| Unknown | 132,794 | 17,751,657 | 1.79 |
| SSR | 7405 | 5,817,413 | 0.59 |
| Unknown | 145,781 | 33,606,673 | 3.38 |
| Total | 2,078,967 | 357,309,255 | 43.23 |
| ncRNA | Number in of Loci | Average Length (bp) | Family |
|---|---|---|---|
| tRNA | 3280 | 74.37 | 25 |
| rRNA | 5616 | 149.31 | 4 |
| miRNA | 450 | 87.52 | 88 |
| snRNA | 449 | 151.74 | 7 |
| snoRNA | 339 | 185.61 | 4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, D.; Gui, L.; Zhu, Y.; Xu, C.; Zhou, W.; Li, M. Chromosome-Level Assembly of Male Opsariichthys bidens Genome Provides Insights into the Regulation of the GnRH Signaling Pathway and Genome Evolution. Biology 2022, 11, 1500. https://doi.org/10.3390/biology11101500
Liu D, Gui L, Zhu Y, Xu C, Zhou W, Li M. Chromosome-Level Assembly of Male Opsariichthys bidens Genome Provides Insights into the Regulation of the GnRH Signaling Pathway and Genome Evolution. Biology. 2022; 11(10):1500. https://doi.org/10.3390/biology11101500
Chicago/Turabian StyleLiu, Dong, Lang Gui, Yefei Zhu, Cong Xu, Wenzong Zhou, and Mingyou Li. 2022. "Chromosome-Level Assembly of Male Opsariichthys bidens Genome Provides Insights into the Regulation of the GnRH Signaling Pathway and Genome Evolution" Biology 11, no. 10: 1500. https://doi.org/10.3390/biology11101500
APA StyleLiu, D., Gui, L., Zhu, Y., Xu, C., Zhou, W., & Li, M. (2022). Chromosome-Level Assembly of Male Opsariichthys bidens Genome Provides Insights into the Regulation of the GnRH Signaling Pathway and Genome Evolution. Biology, 11(10), 1500. https://doi.org/10.3390/biology11101500






