Genome-Wide Identification, Expression Profiling, and Characterization of Cyclin-like Genes Reveal Their Role in the Fertility of the Diamondback Moth
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Rearing of Plutella xylostella
2.2. Identification of Putative Cyclin-like Genes
2.3. Gene Structure and Motif Analysis
2.4. Genomic Location and Phylogenetic Analysis
2.5. Physiochemical Properties of Cyclin Genes
2.6. Cis-Regulatory Elements Analysis
2.7. Prediction of miRNAs Targeting the Cyclin Genes of P. xylostella
2.8. Stage- and Tissue-Specific Expression Profiling of Cyclin Genes
2.9. RNA Extraction, cDNA Preparation and RT-qPCR Analysis
2.10. PCR Amplification and Cloning of PxCyc B1 Gene
2.11. Double-Stranded RNA Synthesis, Microinjections, and RNAi Analysis
2.12. Statistical Analysis
3. Results
3.1. Identificationsand Subcellular Localization of Cyclin Genes in P. xylostella
3.2. Gene Structure and Conserved Domain
3.3. Genomic Location and Phylogenetic Analysis
3.4. Putative Cis-Regulatory Elements of Cyclin Genes
3.5. Identification of miRNA Targeting the Cyclin Genes
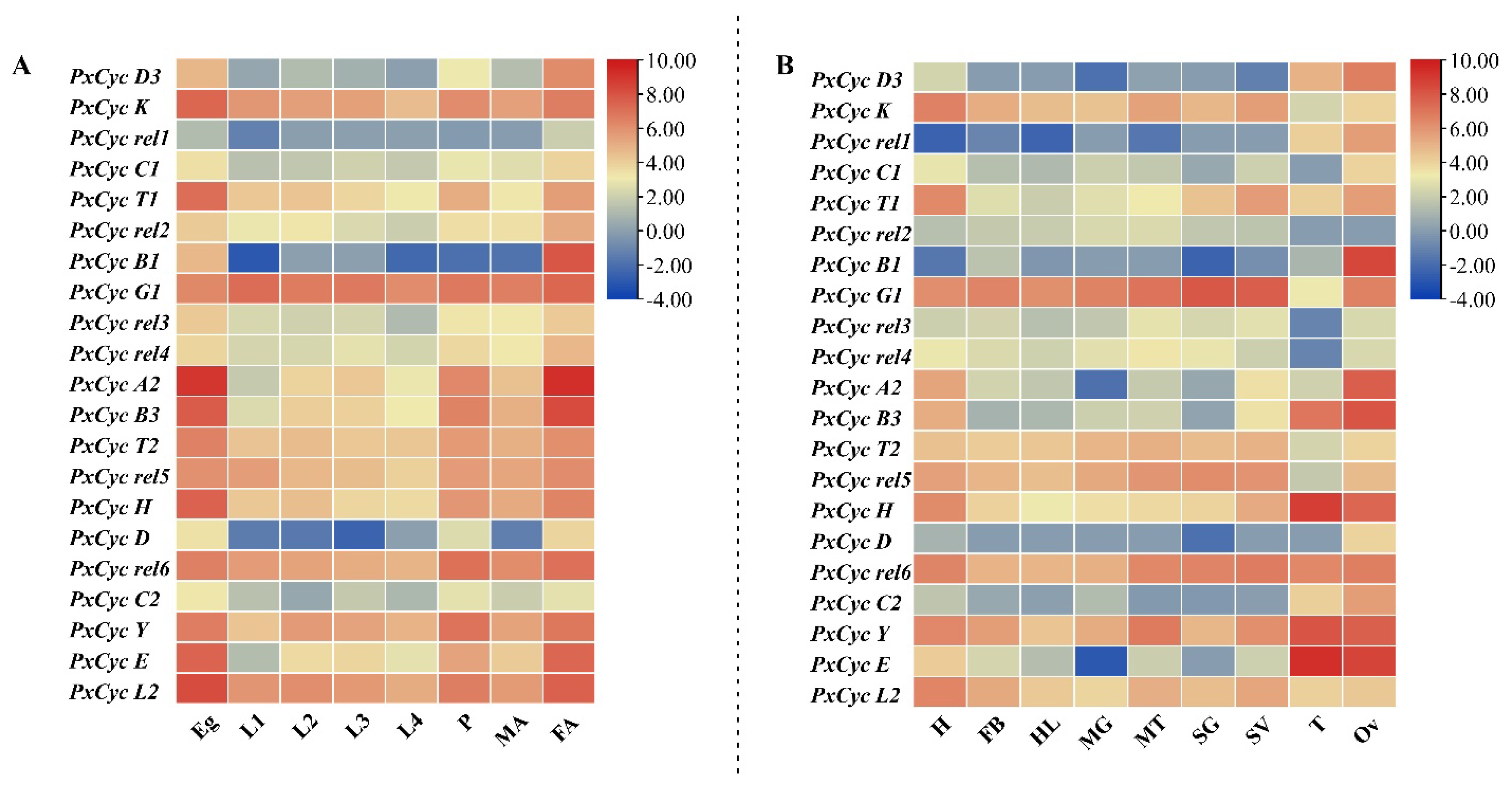
3.6. Transcriptome-Based Expression Profiling of Cyclin-like Genes in P. xylostella
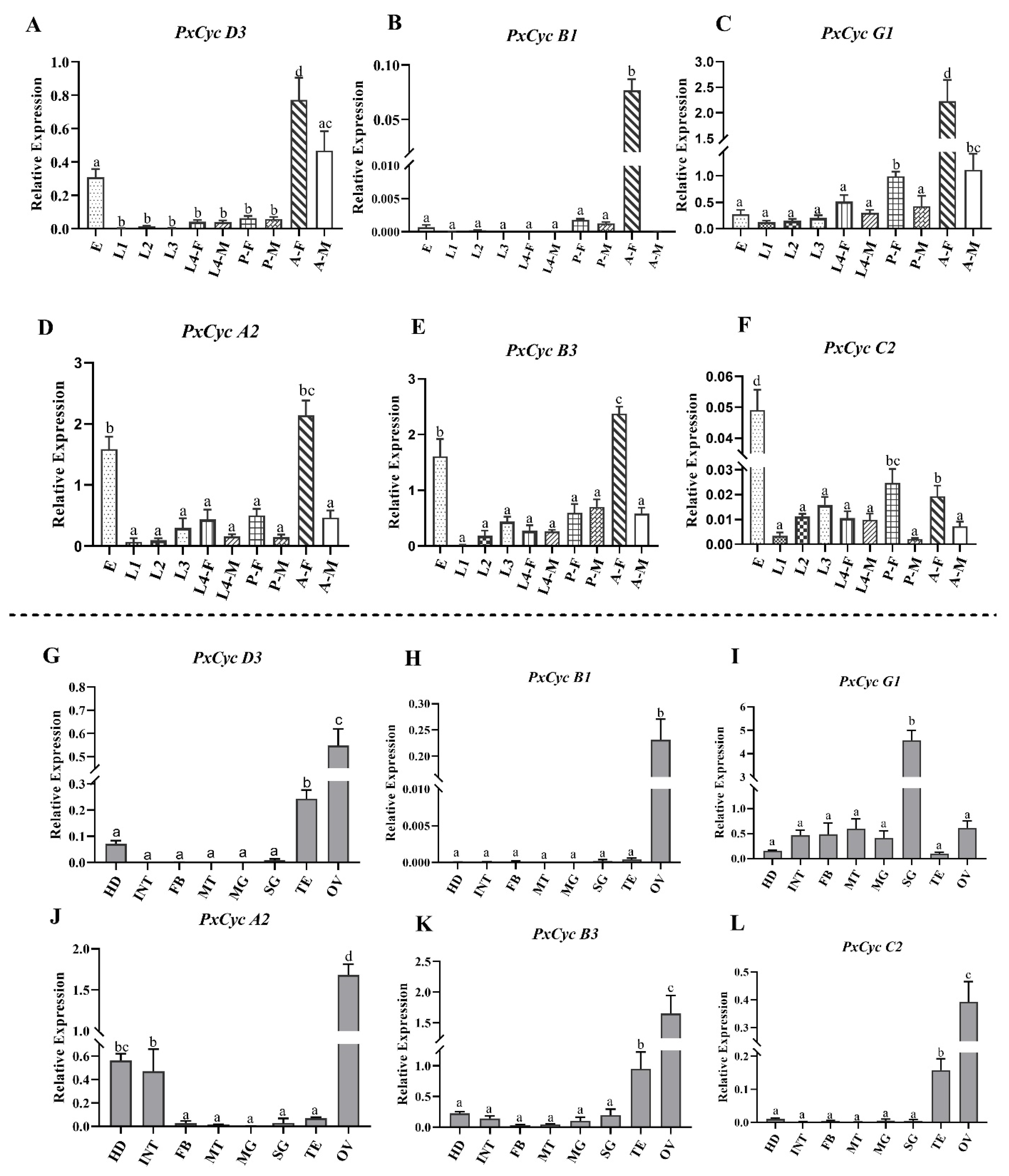
3.7. Validation of Cyclin Gene Expressions through RT-qPCR
3.8. RNAi of PxCyc B1 Gene
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Meng, J.; Peng, M.; Yang, J.; Zhao, Y.; Hu, J.; Zhu, Y.; He, H. Genome-wide analysis of the cyclin gene family and their expression profile in Medicago truncatula. Int. J. Mol. Sci. 2020, 21, 9430. [Google Scholar] [CrossRef] [PubMed]
- Nigg, E.A. Cyclin-dependent protein kinases: Key regulators of the eukaryotic cell cycle. BioEssays 1995, 17, 471–480. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.G.; Walker, C.L. Cyclins and cell cycle checkpoints. Annu. Rev. Pharmacol. Toxicol. 1999, 39, 295–312. [Google Scholar] [CrossRef] [PubMed]
- Gunbin, K.V.; Suslov, V.V.; Turnaev, I.I.; Afonnikov, D.A.; Kolchanov, N.A. Molecular evolution of cyclin proteins in animals and fungi. BMC Evol. Biol. 2011, 11, 224. [Google Scholar] [CrossRef]
- Murray, A.W. Recycling the cell cycle: Cyclins revisited. Cell 2004, 116, 221–234. [Google Scholar] [CrossRef]
- Sherr, C.J.; Roberts, J.M. Living with or without cyclins and cyclin-dependent kinases. Genes Dev. 2004, 18, 2699–2711. [Google Scholar] [CrossRef]
- Evans, T.; Rosenthal, E.T.; Youngblom, J.; Distel, D.; Hunt, T. Cyclin: A protein specified by maternal mRNA in sea urchin eggs that is destroyed at each cleavage division. Cell 1983, 33, 389–396. [Google Scholar] [CrossRef]
- Renaudin, J.P.; Colasanti, J.; Rime, H.; Yuan, Z.; Sundaresan, V. Cloning of four cyclins from maize indicates that higher plants have three structurally distinct groups of mitotic cyclins. Proc. Natl. Acad. Sci. USA 1994, 91, 7375–7379. [Google Scholar] [CrossRef]
- Errico, A.; Deshmukh, K.; Tanaka, Y.; Pozniakovsky, A.; Hunt, T. Identification of substrates for cyclin dependent kinases. Adv. Enzym. Regul. 2010, 50, 375–399. [Google Scholar] [CrossRef] [PubMed]
- Satyanarayana, A.; Kaldis, P. Mammalian cell-cycle regulation: Several Cdks, numerous cyclins and diverse compensatory mechanisms. Oncogene 2009, 28, 2925–2939. [Google Scholar] [CrossRef]
- Coqueret, O. Linking cyclins to transcriptional control. Gene 2002, 299, 35–55. [Google Scholar] [CrossRef]
- Debat, V.; Bloyer, S.; Faradji, F.; Gidaszewski, N.; Navarro, N.; Orozco-Terwengel, P.; Ribeiro, V.; Schlötterer, C.; Deutsch, J.S.; Peronnet, F. Developmental stability: A major role for cyclin G in Drosophila melanogaster. PLoS Genet. 2011, 7, e1002314. [Google Scholar] [CrossRef]
- Nugent, J.; Alfa, C.E.; Young, T.; Hyams, J.S. Conserved structural motifs in cyclins identified by sequence analysis. J. Cell Sci. 1991, 99, 669–674. [Google Scholar] [CrossRef] [PubMed]
- Horne, M.C.; Goolsby, G.L.; Donaldson, K.L.; Tran, D.; Neubauer, M.; Wahl, A.F. Cyclin G1 and cyclin G2 comprise a new family of cyclins with contrasting tissue-specific and cell cycle-regulated expression (∗). J. Biol. Chem. 1996, 271, 6050–6061. [Google Scholar] [CrossRef] [PubMed]
- Lehner, C.F.; O’Farrell, P.H. The roles of Drosophila cyclins A and B in mitotic control. Cell 1990, 61, 535–547. [Google Scholar] [CrossRef]
- Jacobs, H.W.; Knoblich, J.A.; Lehner, C.F. Drosophila cyclin B3 is required for female fertility and is dispensable for mitosis like Cyclin B. Genes Dev. 1998, 12, 3741–3751. [Google Scholar] [CrossRef]
- Xie, X.J.; Hsu, F.N.; Gao, X.; Xu, W.; Ni, J.Q.; Xing, Y.; Huang, L.; Hsiao, H.C.; Zheng, H.; Wang, C.; et al. CDK8-cyclin C mediates nutritional regulation of developmental transitions through the ecdysone receptor in Drosophila. PLoS Biol. 2015, 13, e1002250. [Google Scholar] [CrossRef]
- Faradji, F.; Bloyer, S.; Dardalhon-Cuménal, D.; Randsholt, N.B.; Peronnet, F. Drosophila melanogaster cyclin G coordinates cell growth and cell proliferation. Cell Cycle 2011, 10, 805–818. [Google Scholar] [CrossRef]
- Pan, M.; Hong, K.; Chen, X.; Pan, C.; Chen, X.; Kuang, X.; Lu, C. BmCyclin B and BmCyclin B3 are required for cell cycle progression in the silkworm, Bombyx mori. Sci. China Life Sci. 2013, 56, 360–365. [Google Scholar] [CrossRef]
- Li, Z.; Feng, X.; Liu, S.S.; You, M.; Furlong, M.J. Biology, ecology, and management of the diamondback moth in China. Annu. Rev. Entomol. 2016, 61, 277–296. [Google Scholar] [CrossRef]
- Fu, S.; Liu, Z.; Chen, J.; Sun, G.; Jiang, Y.; Li, M.; Xiong, L.; Chen, S.; Zhou, Y.; Asad, M.; et al. Silencing arginine kinase/integrin β(1) subunit by transgenic plant expressing dsRNA inhibits the development and survival of Plutella xylostella. Pest Manag. Sci. 2020, 76, 1761–1771. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wu, Y. High levels of resistance to chlorantraniliprole evolved in field populations of Plutella xylostella. J. Econ. Entomol. 2012, 105, 1019–1023. [Google Scholar] [CrossRef] [PubMed]
- Jiang, T.; Wu, S.; Yang, T.; Zhu, C.; Gao, C. Monitoring field populations of Plutella xylostella (Lepidoptera: Plutellidae) for resistance to eight insecticides in China. Fla. Entomol. 2015, 98, 65–73. [Google Scholar] [CrossRef]
- Asad, M.; Liu, D.; Li, J.; Chen, J.; Yang, G. Development of CRISPR/Cas9-mediated gene-drive construct targeting the phenotypic gene in Plutella xylostella. Front. Physiol. 2022, 13, 1244. [Google Scholar] [CrossRef] [PubMed]
- Peng, L.; Wang, L.; Yang, Y.F.; Zou, M.M.; He, W.Y.; Wang, Y.; Wang, Q.; Vasseur, L.; You, M.S. Transcriptome profiling of the Plutella xylostella (Lepidoptera: Plutellidae) ovary reveals genes involved in oogenesis. Gene 2017, 637, 90–99. [Google Scholar] [CrossRef]
- Asad, M.; Munir, F.; Xu, X.; Li, M.; Jiang, Y.; Chu, L.; Yang, G. Functional characterization of the cis-regulatory region for the vitellogenin gene in Plutella xylostella. Insect Mol. Biol. 2020, 29, 137–147. [Google Scholar] [CrossRef]
- Xu, X.; Yang, J.; Harvey-Samuel, T.; Huang, Y.; Asad, M.; Chen, W.; He, W.; Yang, G.; Alphey, L.; You, M. Identification and characterization of the vasa gene in the diamondback moth, Plutella xylostella. Insect Biochem. Mol. Biol. 2020, 122, 103371. [Google Scholar] [CrossRef]
- Guo, A.Y.; Zhu, Q.H.; Chen, X.; Luo, J.C. GSDS: A gene structure display server. Yi Chuan Hereditas 2007, 29, 1023–1026. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- You, M.; Yue, Z.; He, W.; Yang, X.; Yang, G.; Xie, M.; Zhan, D.; Baxter, S.W.; Vasseur, L.; Gurr, G.M.; et al. A heterozygous moth genome provides insights into herbivory and detoxification. Nat. Genet. 2013, 45, 220–225. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Chou, K.C.; Shen, H.B. Cell-PLoc: A package of Web servers for predicting subcellular localization of proteins in various organisms. Nat. Protoc. 2008, 3, 153–162. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the ExPASy server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar]
- Farré, D.; Roset, R.; Huerta, M.; Adsuara, J.E.; Roselló, L.; Albà, M.M.; Messeguer, X. Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic Acids Res. 2003, 31, 3651–3653. [Google Scholar] [CrossRef]
- Mei, Y.; Jing, D.; Tang, S.; Chen, X.; Chen, H.; Duanmu, H.; Cong, Y.; Chen, M.; Ye, X.; Zhou, H.; et al. InsectBase 2.0: A comprehensive gene resource for insects. Nucleic Acids Res. 2022, 50, D1040–D1045. [Google Scholar] [CrossRef] [PubMed]
- Rehmsmeier, M.; Steffen, P.; Hochsmann, M.; Giegerich, R. Fast and effective prediction of microRNA/target duplexes. RNA 2004, 10, 1507–1517. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Ma, X.L.; He, W.Y.; Wang, P.; You, M.S. Cell lines from diamondback moth exhibiting differential susceptibility to baculovirus infection and expressing midgut genes. Insect Sci. 2019, 26, 251–262. [Google Scholar] [CrossRef]
- Kwon, D.H.; Park, J.H.; Ashok, P.A.; Lee, U.; Lee, S.H. Screening of target genes for RNAi in Tetranychus urticae and RNAi toxicity enhancement by chimeric genes. Pestic. Biochem. Physiol. 2016, 130, 1–7. [Google Scholar] [CrossRef]
- Patil, A.A.; Tatsuke, T.; Mon, H.; Lee, J.M.; Morokuma, D.; Hino, M.; Kusakabe, T. Characterization of Armitage and Yb containing granules and their relationship to nuage in ovary-derived cultured silkworm cell. Biochem. Biophys. Res. Commun. 2017, 490, 134–140. [Google Scholar] [CrossRef]
- Lucas, K.J.; Zhao, B.; Liu, S.; Raikhel, A.S. Regulation of physiological processes by microRNAs in insects. Curr. Opin. Insect Sci. 2015, 11, 1–7. [Google Scholar] [CrossRef]
- Zhang, Q.; Dou, W.; Taning, C.N.T.; Smagghe, G.; Wang, J.-J. Regulatory roles of microRNAs in insect pests: Prospective targets for insect pest control. Curr. Opin. Biotechnol. 2021, 70, 158–166. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Dou, W.; Pan, D.; Chen, E.-H.; Niu, J.-Z.; Smagghe, G.; Wang, J.-J. Genome-wide analysis of microRNAs in relation to pupariation in oriental fruit fly. Front. Physiol. 2019, 10, 301. [Google Scholar] [CrossRef] [PubMed]
- Obaya, A.J.; Sedivy, J.M. Regulation of cyclin-Cdk activity in mammalian cells. Cell. Mol. Life Sci. CMLS 2002, 59, 126–142. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Chen, F.; Yang, X.; Xu, W.; Xie, J.; Yu, L. Phylogenetic analysis of CDK and cyclin proteins in premetazoan lineages. BMC Evol Biol 2014, 14, 10. [Google Scholar] [CrossRef] [PubMed]
- Inzé, D.; De Veylder, L. Cell cycle regulation in plant development. Annu. Rev. Genet. 2006, 40, 77–105. [Google Scholar] [CrossRef]
- Ren, D.; Song, J.; Ni, M.; Kang, L.; Guo, W. Regulatory mechanisms of cell polyploidy in insects. Front. Cell Dev. Biol. 2020, 8, 361. [Google Scholar] [CrossRef]
- Wang, G.; Kong, H.; Sun, Y.; Zhang, X.; Zhang, W.; Altman, N.; DePamphilis, C.W.; Ma, H. Genome-wide analysis of the cyclin family in Arabidopsis and comparative phylogenetic analysis of plant cyclin-like proteins. Plant Physiol. 2004, 135, 1084–1099. [Google Scholar] [CrossRef]
- La, H.; Li, J.; Ji, Z.; Cheng, Y.; Li, X.; Jiang, S.; Venkatesh, P.N.; Ramachandran, S. Genome-wide analysis of cyclin family in rice (Oryza Sativa L.). Mol. Genet. Genom. 2006, 275, 374–386. [Google Scholar] [CrossRef]
- Hu, X.; Cheng, X.; Jiang, H.; Zhu, S.; Cheng, B.; Xiang, Y. Genome-wide analysis of cyclins in maize (Zea mays). Genet. Mol. Res. 2010, 9, 1490–1503. [Google Scholar] [CrossRef]
- Zhang, T.; Wang, X.; Lu, Y.; Cai, X.; Ye, Z.; Zhang, J. Genome-wide analysis of the cyclin gene family in tomato. Int. J. Mol. Sci. 2013, 15, 120–140. [Google Scholar] [CrossRef]
- Plowman, G.D.; Sudarsanam, S.; Bingham, J.; Whyte, D.; Hunter, T. The protein kinases of Caenorhabditis elegans: A model for signal transduction in multicellular organisms. Proc. Natl. Acad. Sci. USA 1999, 96, 13603–13610. [Google Scholar] [CrossRef] [PubMed]
- Pines, J. Cyclins and cyclin-dependent kinases: A biochemical view. Biochem. J. 1995, 308 Pt 3, 697–711. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Meyer, A.N.; Donoghue, D.J. Nuclear localization of cyclin B1 mediates its biological activity and is regulated by phosphorylation. Proc. Natl. Acad. Sci. USA 1997, 94, 502–507. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Kornbluth, S. All aboard the cyclin train: Subcellular trafficking of cyclins and their CDK partners. Trends Cell Biol. 1999, 9, 207–210. [Google Scholar] [CrossRef]
- Wittkopp, P.J. Evolution of cis-regulatory sequence and function in Diptera. Heredity 2006, 97, 139–147. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Yang, X.; Sun, X.; Li, X.; Liu, Z.; Yin, Q.; Ma, L.; Zhou, D.; Sun, Y.; Shen, B.; et al. Transcription factor FTZ-F1 regulates mosquito cuticular protein CPLCG5 conferring resistance to pyrethroids in Culex pipiens pallens. Parasites Vectors 2020, 13, 514. [Google Scholar] [CrossRef] [PubMed]
- Piulachs, M.D.; Pagone, V.; Bellés, X. Key roles of the Broad-Complex gene in insect embryogenesis. Insect Biochem. Mol. Biol. 2010, 40, 468–475. [Google Scholar] [CrossRef]
- Xu, Q.Y.; Meng, Q.W.; Deng, P.; Fu, K.Y.; Guo, W.C.; Li, G.Q. Isoform specific roles of Broad-Complex in larval development in Leptinotarsa decemlineata. Insect Mol. Biol. 2019, 28, 420–430. [Google Scholar] [CrossRef]
- Spokony, R.F.; Restifo, L.L. Broad Complex isoforms have unique distributions during central nervous system metamorphosis in Drosophila melanogaster. J. Comp. Neurol. 2009, 517, 15–36. [Google Scholar] [CrossRef]
- Zhen, C.; Yang, H.; Luo, S.; Huang, J.; Wu, J. Broad-complex Z3 contributes to the ecdysone-mediated transcriptional regulation of the vitellogenin gene in Bombus lantschouensis. PLoS ONE 2018, 13, e0207275. [Google Scholar] [CrossRef]
- Zeng, B.; Huang, Y.; Xu, J.; Shiotsuki, T.; Bai, H.; Palli, S.R.; Huang, Y.; Tan, A. The FOXO transcription factor controls insect growth and development by regulating juvenile hormone degradation in the silkworm, Bombyx mori. J. Biol. Chem. 2017, 292, 11659–11669. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Zhou, S.; Wang, J.; Hu, W. microRNA profiles and functions in mosquitoes. PLoS Negl. Trop. Dis. 2018, 12, e0006463. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Liu, Y.; Yan, H.; Li, Y.; Zhang, H.; Xu, J.; Puthiyakunnon, S.; Chen, X. miR-281, an abundant midgut-specific miRNA of the vector mosquito Aedes albopictus enhances dengue virus replication. Parasites Vectors 2014, 7, 488. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Hu, S.; Zhang, H.; Yin, H.; Wang, H.; Zhou, D.; Sun, Y.; Ma, L.; Shen, B.; Zhu, C. MiR-279-3p regulates deltamethrin resistance through CYP325BB1 in Culex pipiens pallens. Parasites Vectors 2021, 14, 528. [Google Scholar] [CrossRef]
- Yang, M.; Wang, Y.; Jiang, F.; Song, T.; Wang, H.; Liu, Q.; Zhang, J.; Zhang, J.; Kang, L. miR-71 and miR-263 jointly regulate target genes chitin synthase and chitinase to control locust molting. PLoS Genet. 2016, 12, e1006257. [Google Scholar] [CrossRef]
- Fox, P.M.; Vought, V.E.; Hanazawa, M.; Lee, M.-H.; Maine, E.M.; Schedl, T. Cyclin E and CDK-2 regulate proliferative cell fate and cell cycle progression in the C. elegans germline. Development 2011, 138, 2223–2234. [Google Scholar] [CrossRef]
- Atikukke, G.; Albosta, P.; Zhang, H.; Finley, R.L. A role for Drosophila Cyclin J in oogenesis revealed by genetic interactions with the piRNA pathway. Mech. Dev. 2014, 133, 64–76. [Google Scholar] [CrossRef]
- Tay, J.; Hodgman, R.; Richter, J.D. The control of cyclin B1 mRNA translation during mouse oocyte maturation. Dev. Biol. 2000, 221, 1–9. [Google Scholar] [CrossRef]
- Asad, M.; Liu, D.; Chen, J.; Yang, G. Applications of gene drive systems for population suppression of insect pests. Bull. Entomol. Res. 2022, 1–10. [Google Scholar] [CrossRef]
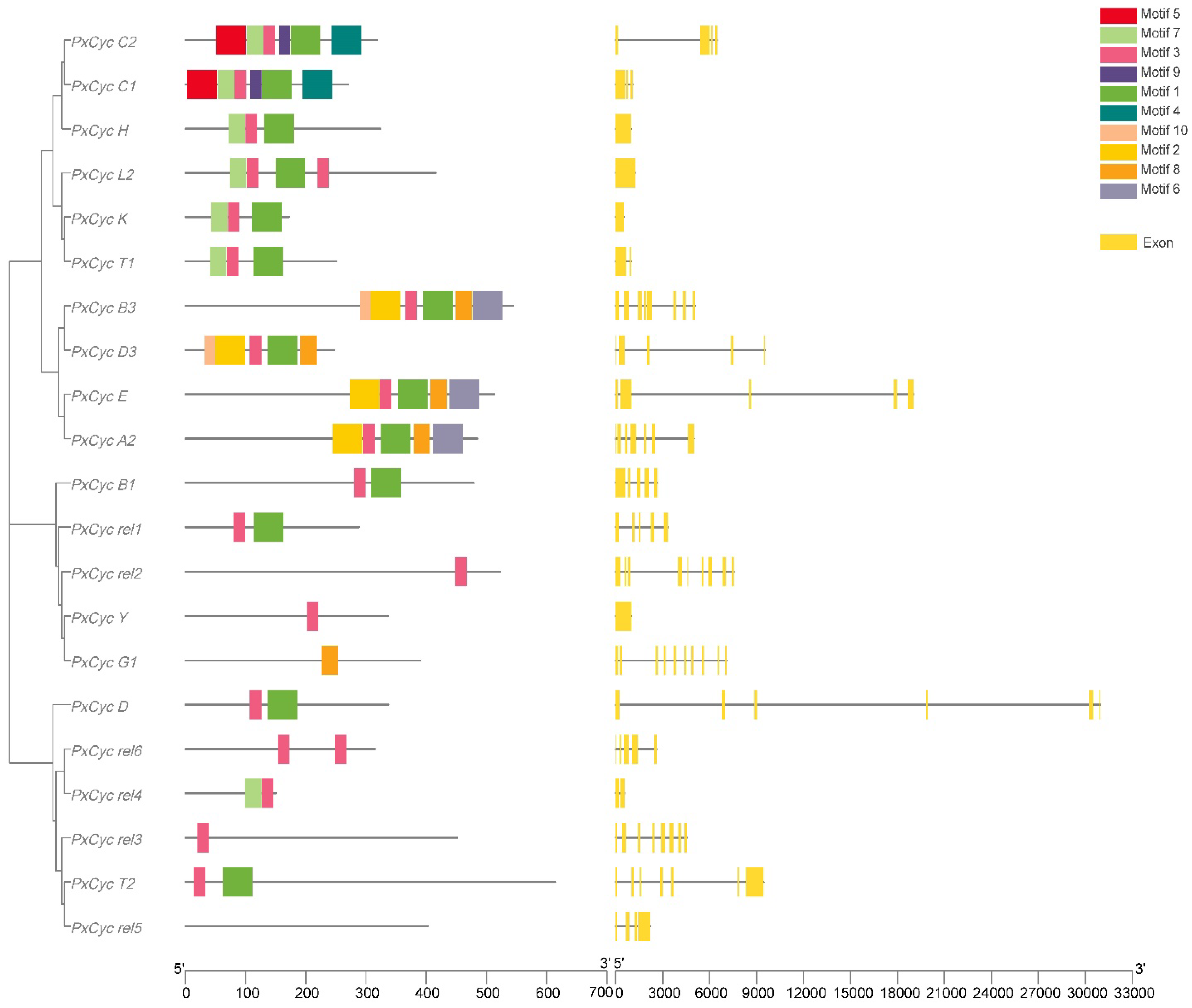
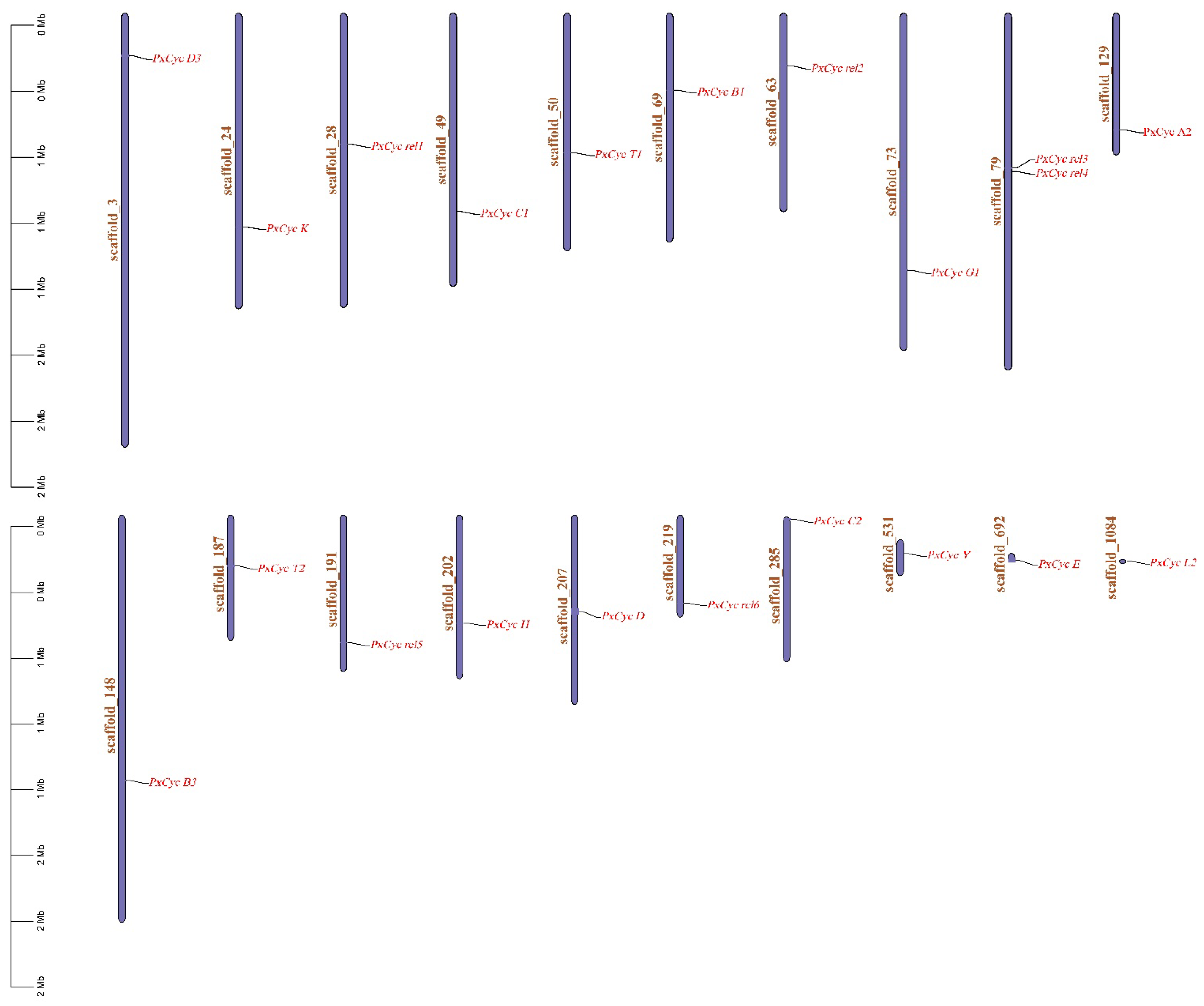
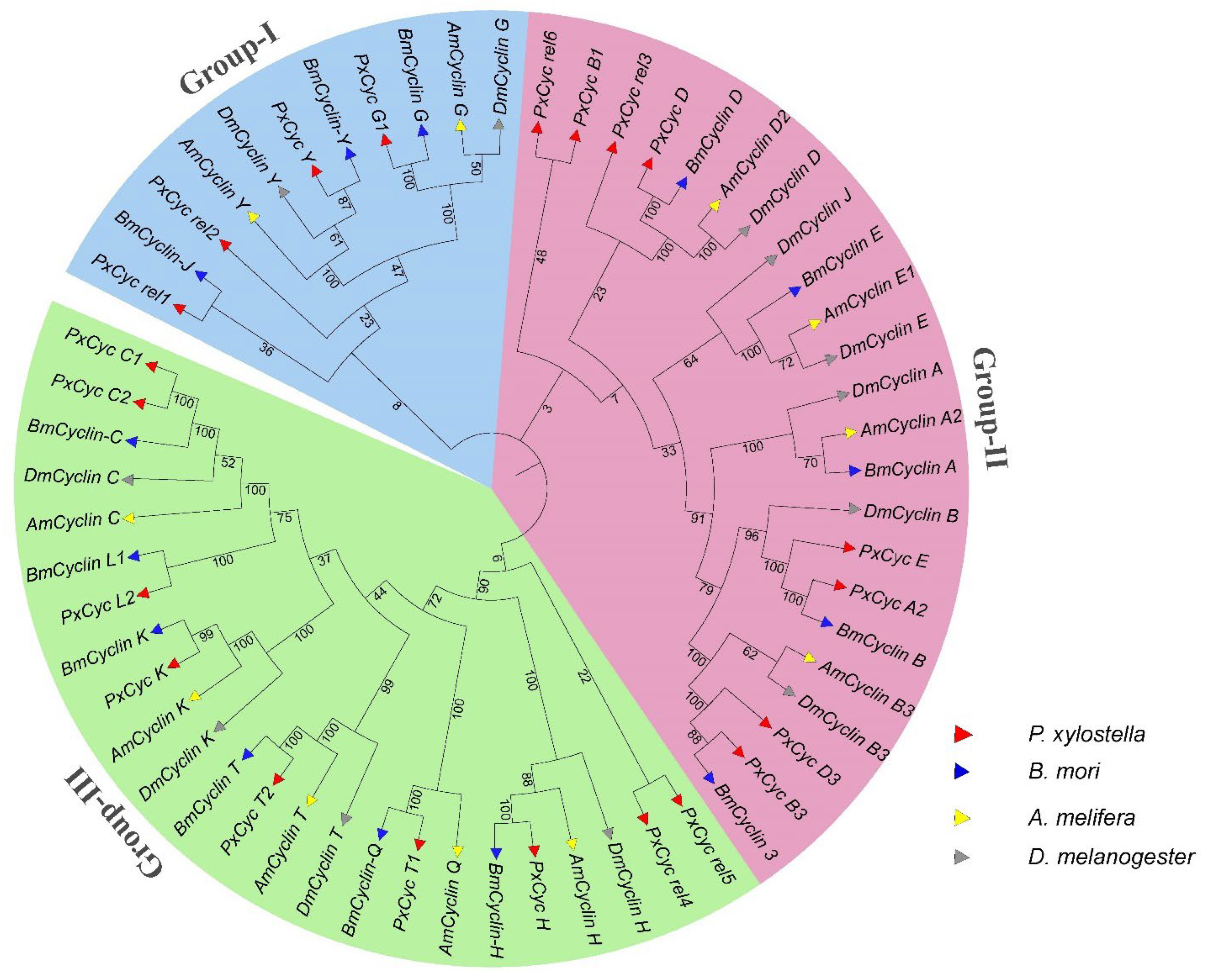
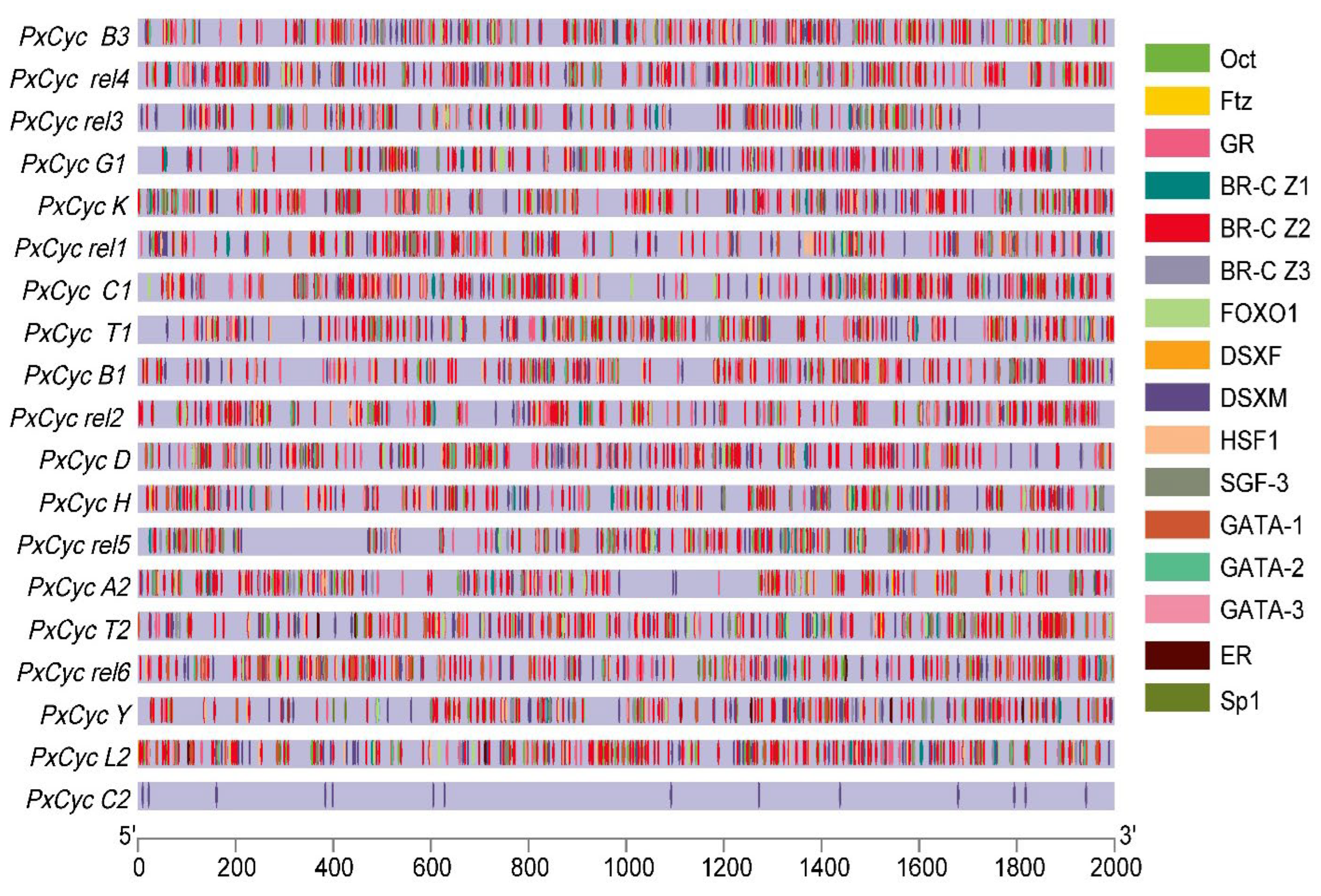


| Gene ID | Name | Position | Gene (bp) | CDS (bp) | Exon | Protein aa | MW (KDa) | PI | Subcellular Localization |
|---|---|---|---|---|---|---|---|---|---|
| Px008345 | PxCyc D3 | scaffold_3:189765..199308 − | 9544 | 744 | 5 | 247 | 28.86152 | 6.9 | Nucleus |
| Px006817 | PxCyc K | scaffold_24:973210..973728 + | 519 | 519 | 1 | 172 | 69.27571 | 9.0 | Nucleus |
| Px007867 | PxCyc rel1 | scaffold_28:594896..598229 − | 3334 | 867 | 5 | 288 | 61.94629 | 9.0 | Nucleus |
| Px012463 | PxCyc C1 | scaffold_49:902075..903146 + | 1072 | 813 | 3 | 270 | 59.41365 | 6.5 | Nucleus. |
| Px012660 | PxCyc T1 | scaffold_50:635009..635991 + | 983 | 756 | 2 | 251 | 57.4899 | 6.6 | Nucleus. |
| Px014457 | PxCyc rel2 | scaffold_63:235869..243413 − | 7545 | 1569 | 9 | 522 | 54.52173 | 9.6 | Nucleus |
| Px015088 | PxCyc B1 | scaffold_69:348318..350973 − | 2656 | 1440 | 5 | 479 | 54.46445 | 9.7 | Nucleus |
| Px015665 | PxCyc G1 | scaffold_73:1165490..1172567 + | 7078 | 1173 | 10 | 390 | 48.74312 | 5.8 | Nucleus |
| Px016300 | PxCyc rel3 | scaffold_79:702120..706664 − | 4545 | 1356 | 8 | 451 | 45.15651 | 6.3 | Cytoplasm/Nucleus |
| Px016302 | PxCyc rel4 | scaffold_79:719945..720507 − | 563 | 453 | 2 | 150 | 43.66896 | 8.1 | Nucleus |
| Px001947 | PxCyc A2 | scaffold_129:528131..533128 + | 4998 | 1458 | 7 | 485 | 38.2414 | 9.0 | Cytoplasm/Nucleus |
| Px003066 | PxCyc B3 | scaffold_148:1207690..1212770 − | 5081 | 1638 | 8 | 545 | 37.60928 | 9.2 | Nucleus |
| Px004935 | PxCyc T2 | scaffold_187:227985..237433 − | 9449 | 1845 | 7 | 614 | 37.60928 | 9.7 | Nucleus |
| Px005179 | PxCyc rel5 | scaffold_191:581647..583871 + | 2225 | 1209 | 4 | 402 | 37.5119 | 5.0 | Cytoplasm/Extracell |
| Px005660 | PxCyc H | scaffold_202:491765..492739 − | 975 | 975 | 1 | 324 | 37.30218 | 6.8 | Cytoplasm |
| Px005787 | PxCyc D | scaffold_207:424017..454987 + | 30,971 | 1014 | 6 | 337 | 34.33513 | 6.5 | Nucleus |
| Px006186 | PxCyc rel6 | scaffold_219:400848..403467 + | 30,971 | 948 | 5 | 315 | 31.59559 | 8.6 | Nucleus |
| Px008001 | PxCyc C2 | scaffold_285:6758..13234 − | 6477 | 957 | 4 | 318 | 31.13283 | 7.6 | Nucleus |
| Px013211 | PxCyc Y | scaffold_531:63257..64267 − | 1011 | 1011 | 1 | 336 | 28.2765 | 5.6 | Nucleus |
| Px015145 | PxCyc E | scaffold_692:22981..42013 + | 19,033 | 1542 | 5 | 513 | 20.30056 | 5.0 | Nucleus |
| Px000785 | PxCyc L2 | scaffold_1084:7808..9058 + | 1251 | 1251 | 1 | 416 | 15.84307 | 9.8 | Nucleus |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Asad, M.; Chen, J.; Liao, J.; Liu, D.; Yu, J.; Yang, G. Genome-Wide Identification, Expression Profiling, and Characterization of Cyclin-like Genes Reveal Their Role in the Fertility of the Diamondback Moth. Biology 2022, 11, 1493. https://doi.org/10.3390/biology11101493
Asad M, Chen J, Liao J, Liu D, Yu J, Yang G. Genome-Wide Identification, Expression Profiling, and Characterization of Cyclin-like Genes Reveal Their Role in the Fertility of the Diamondback Moth. Biology. 2022; 11(10):1493. https://doi.org/10.3390/biology11101493
Chicago/Turabian StyleAsad, Muhammad, Jing Chen, Jianying Liao, Dan Liu, Jiajing Yu, and Guang Yang. 2022. "Genome-Wide Identification, Expression Profiling, and Characterization of Cyclin-like Genes Reveal Their Role in the Fertility of the Diamondback Moth" Biology 11, no. 10: 1493. https://doi.org/10.3390/biology11101493
APA StyleAsad, M., Chen, J., Liao, J., Liu, D., Yu, J., & Yang, G. (2022). Genome-Wide Identification, Expression Profiling, and Characterization of Cyclin-like Genes Reveal Their Role in the Fertility of the Diamondback Moth. Biology, 11(10), 1493. https://doi.org/10.3390/biology11101493






