Quorum Sensing in ESKAPE Bugs: A Target for Combating Antimicrobial Resistance and Bacterial Virulence
Simple Summary
Abstract
1. Introduction
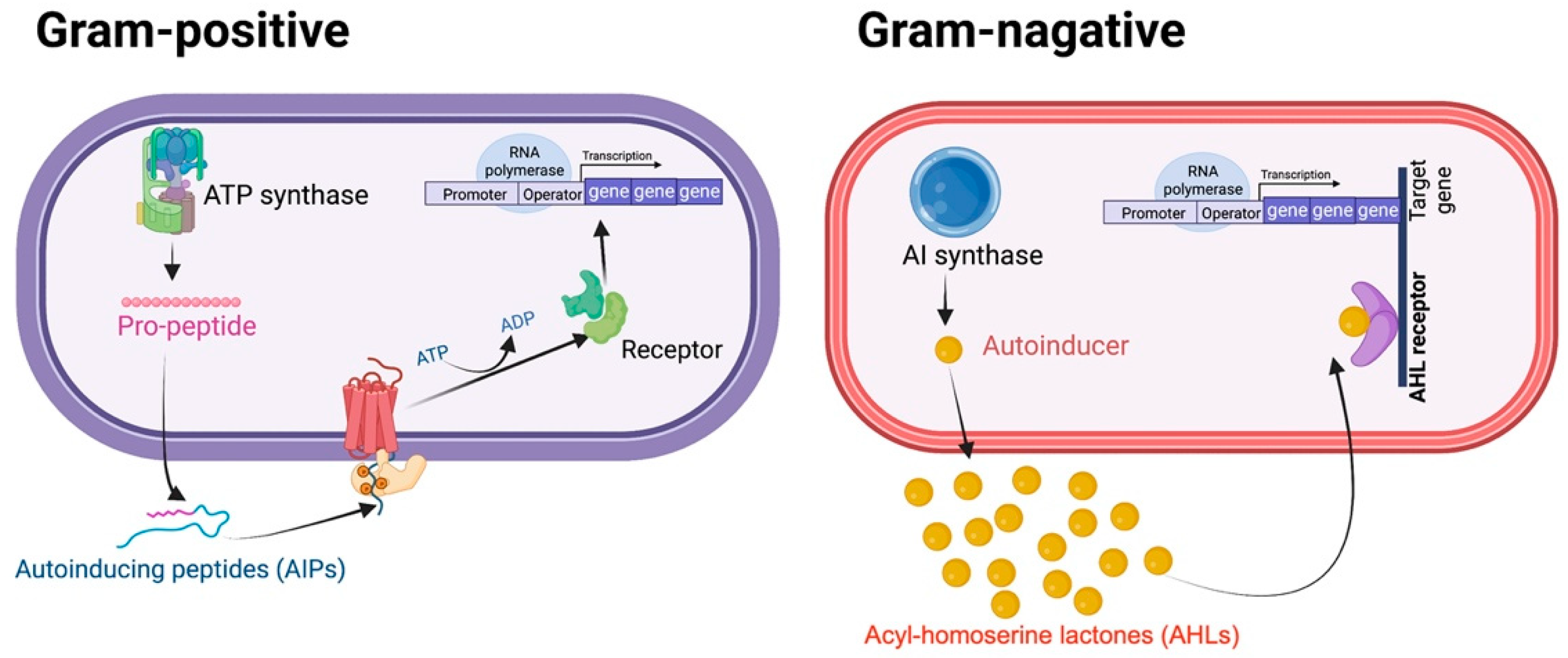
2. QS-Mediated Drug Resistance and Bacterial Virulence in ESKAPE Pathogens
2.1. Enterococcus spp.
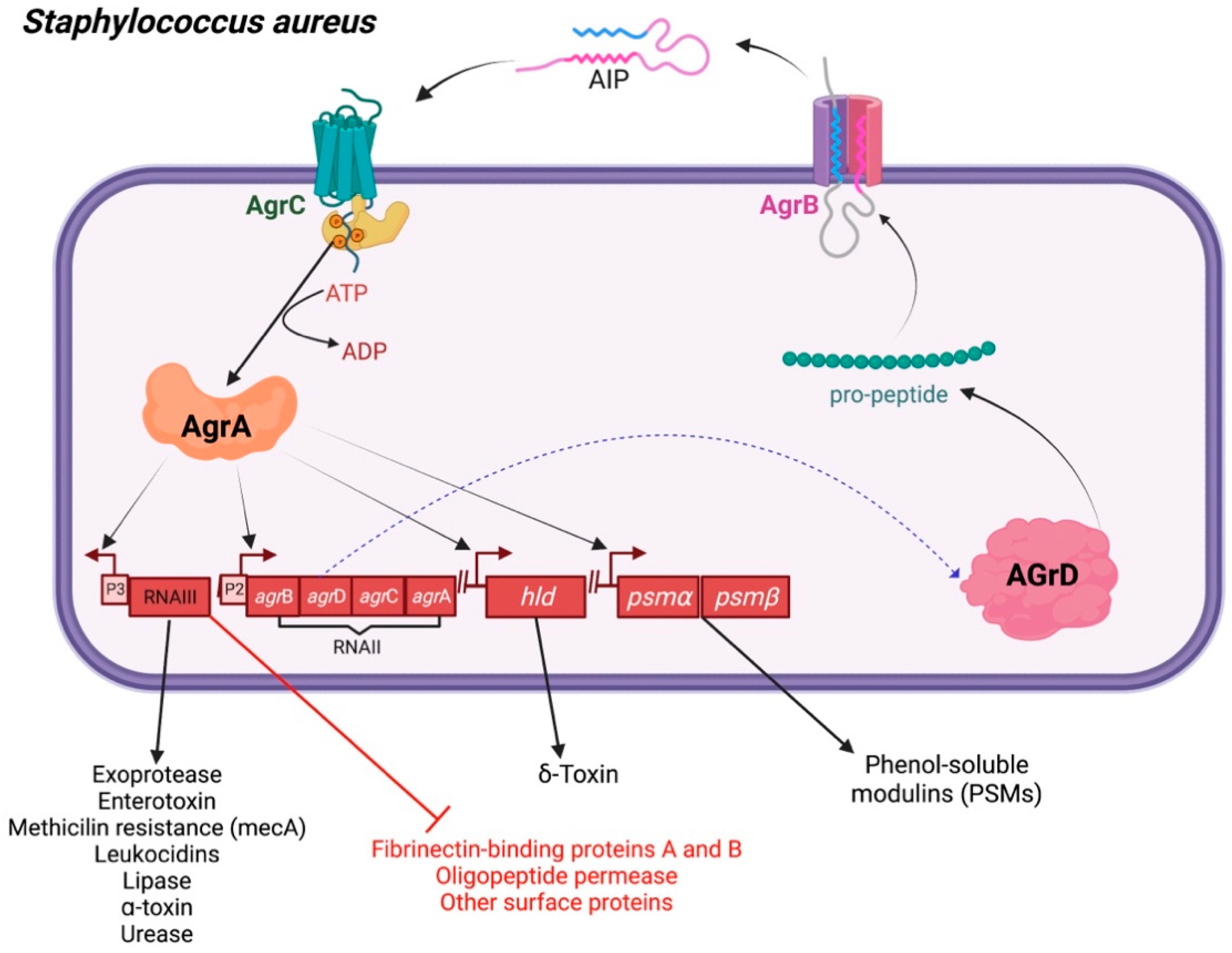
2.2. Staphylococcus aureus
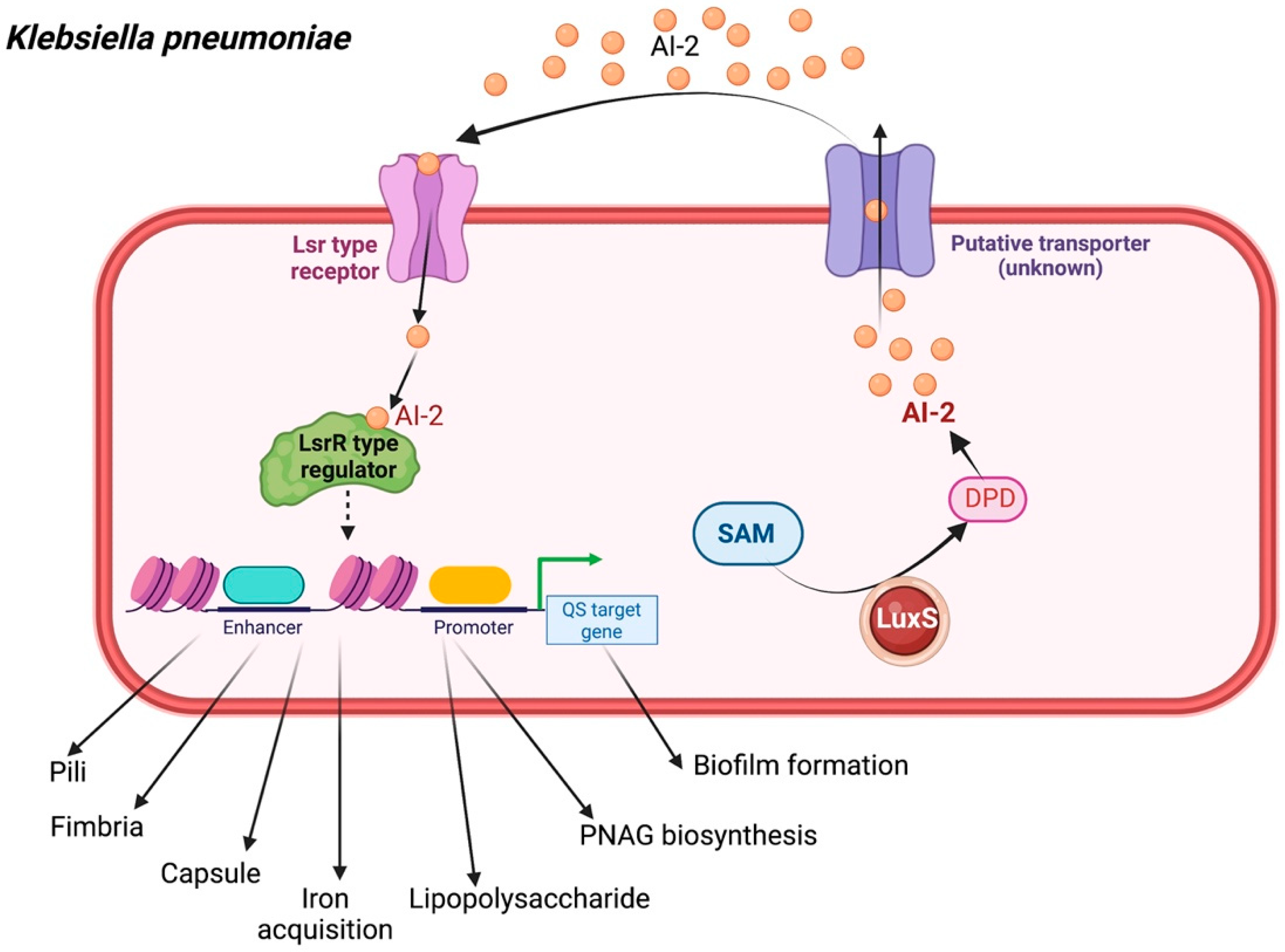
2.3. Klebsiella pneumoniae
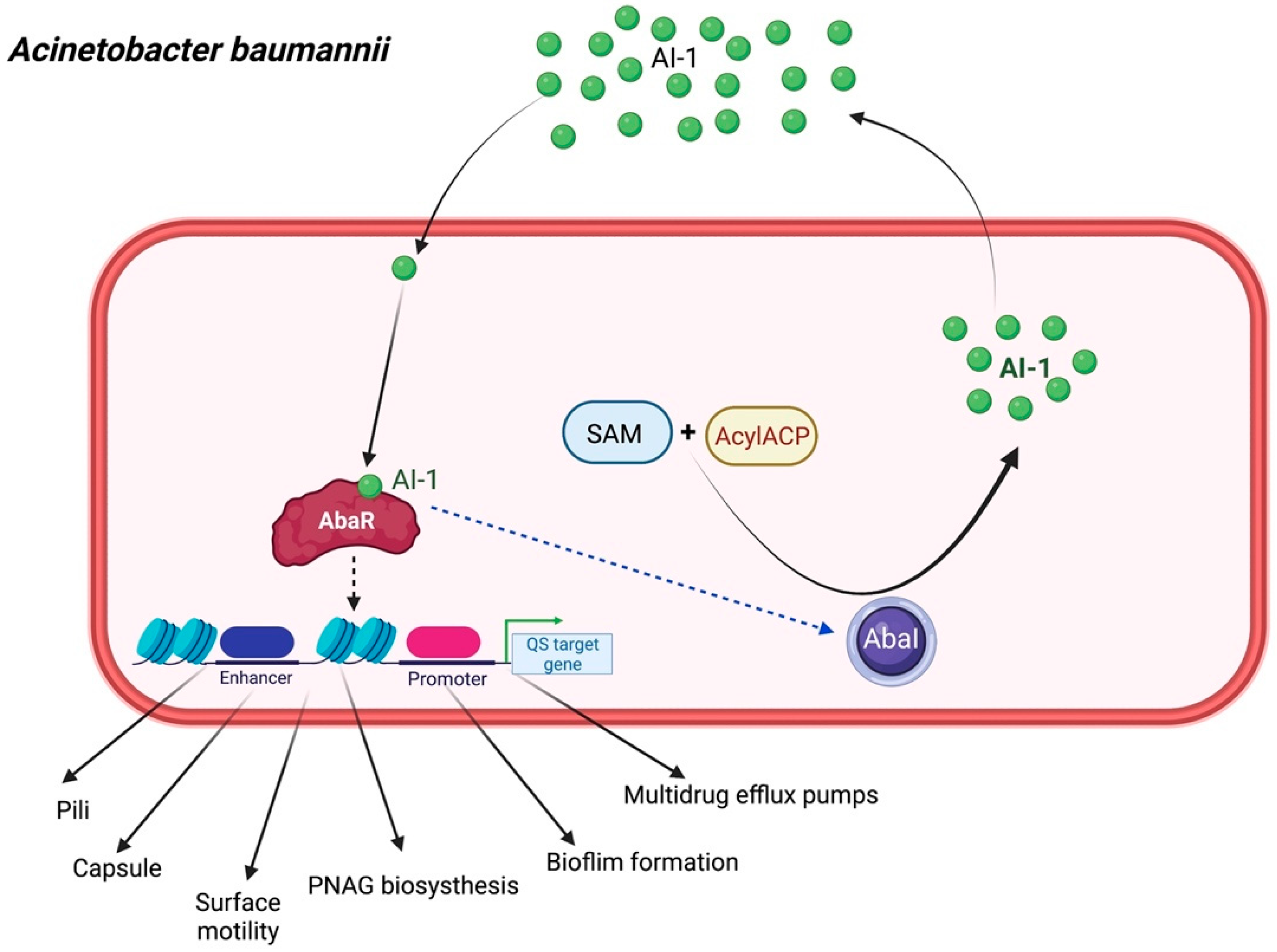
2.4. Acinetobacter baumannii
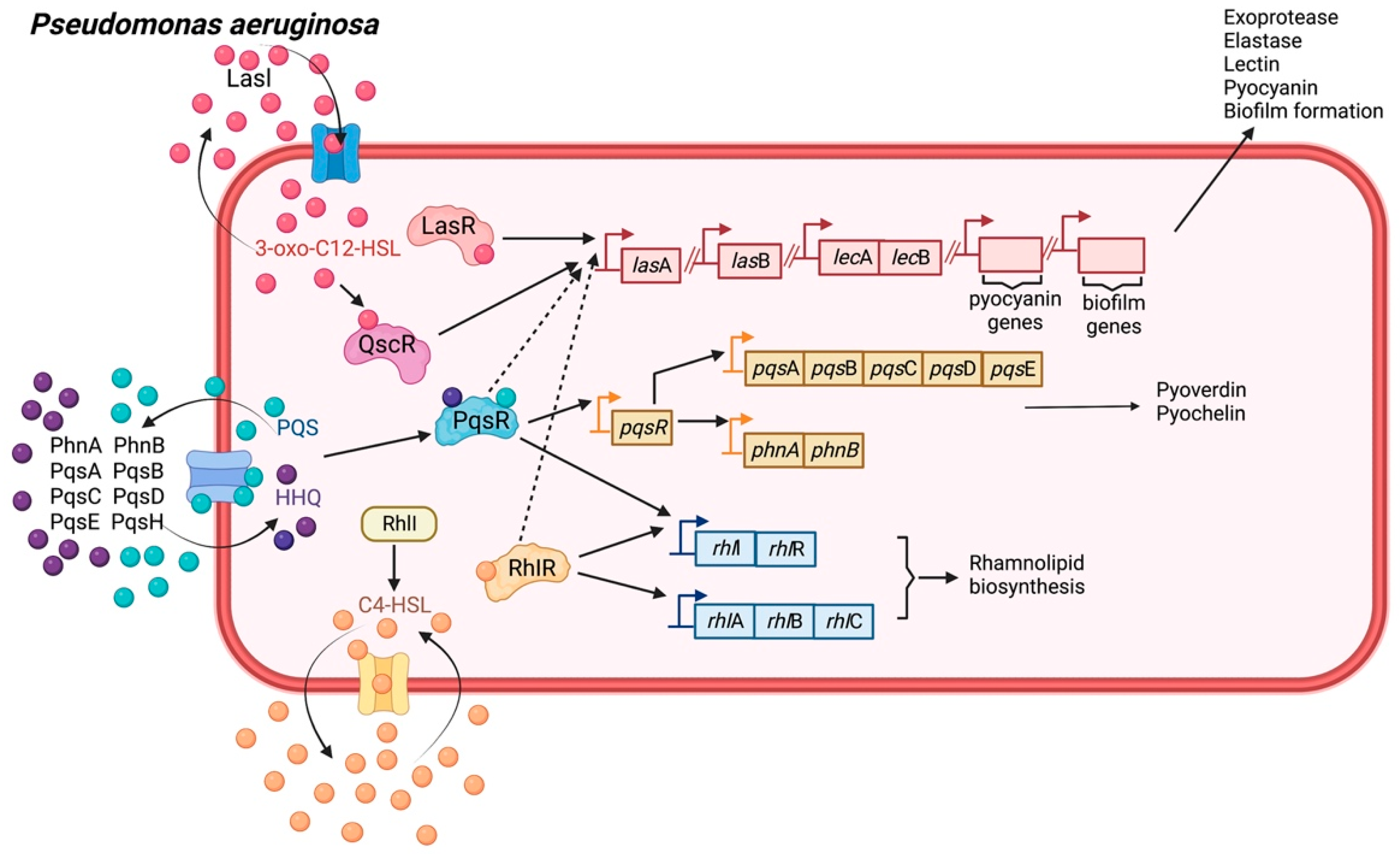
2.5. Pseudomonas aeruginosa
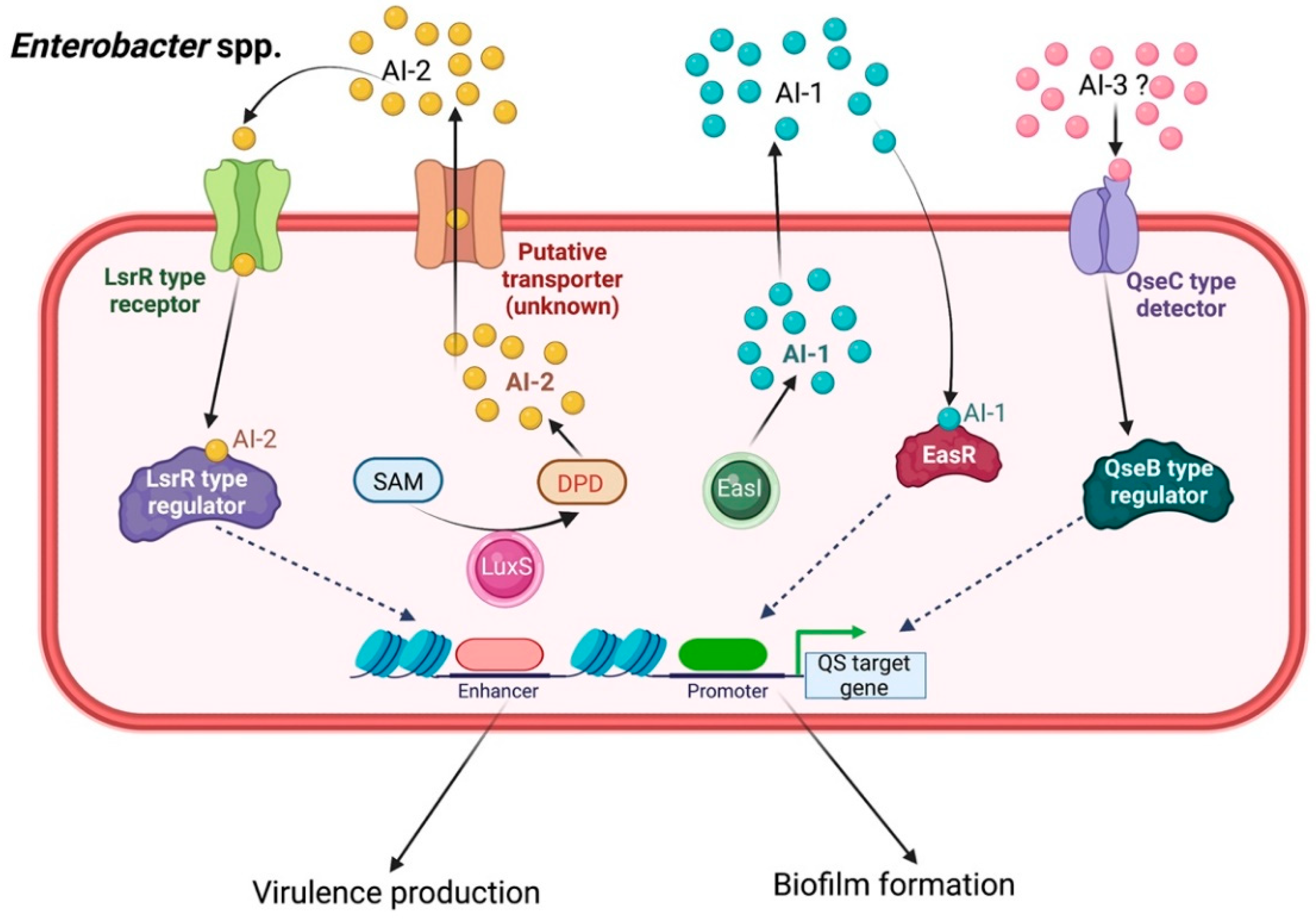
2.6. Enterobacter spp.
3. Therapeutic Approaches Targeting QS Systems Counteract Drug Resistance and Virulence in ESKAPE Bugs
3.1. Targeting AI Synthase
3.2. Sequestration of QS Ligands
3.3. Blocking of QS Transcriptional Regulators
3.4. Alternative Approach for Inhibiting QS Using Probiotics
3.5. Alternative Approach for Inhibiting QS Using Plant Extracts
4. Clinical Applications and Future Perspectives
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Bhargava, N.; Sharma, P.; Capalash, N. Quorum sensing in Acinetobacter: An emerging pathogen. Crit. Rev. Microbiol. 2010, 36, 349–360. [Google Scholar] [CrossRef]
- Stacy, D.M.; Welsh, M.A.; Rather, P.N.; Blackwell, H.E. Attenuation of quorum sensing in the pathogen Acinetobacter baumannii using non-native N-Acyl homoserine lactones. ACS Chem. Biol. 2012, 7, 1719–1728. [Google Scholar] [CrossRef]
- Zarrilli, R. Acinetobacter baumannii virulence determinants involved in biofilm growth and adherence to host epithelial cells. Virulence 2016, 7, 367–368. [Google Scholar] [CrossRef] [PubMed]
- Colquhoun, J.M.; Rather, P.N. Insights into mechanisms of biofilm formation in Acinetobacter baumannii and implications for uropathogenesis. Front. Cell. Infect. Microbiol. 2020, 10, 253. [Google Scholar] [CrossRef]
- Boucher, H.W.; Talbot, G.H.; Bradley, J.S.; Edwards, J.E.; Gilbert, D.; Rice, L.B.; Bartlett, J. Bad bugs, no drugs: No ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 2009, 48, 1–12. [Google Scholar] [CrossRef]
- Borges, A.; Simões, M. Quorum sensing inhibition by marine bacteria. Mar. Drugs 2019, 17, 427. [Google Scholar] [CrossRef]
- Theuretzbacher, U.; Outterson, K.; Engel, A.; Karlén, A. The global preclinical antibacterial pipeline. Nat. Rev. Microbiol. 2020, 18, 275–285. [Google Scholar] [CrossRef] [PubMed]
- Santajit, S.; Indrawattana, N. Mechanisms of antimicrobial resistance in ESKAPE pathogens. Biomed. Res. Int. 2016, 2016, 2475067. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.H.; Wang, L.H.; Xu, J.L.; Zhang, H.B.; Zhang, X.F.; Zhang, L.H. Quenching quorum-sensing-dependent bacterial infection by an N-acyl homoserine lactonase. Nature 2001, 411, 813–817. [Google Scholar] [CrossRef]
- Tommasi, R.; Brown, D.G.; Walkup, G.K.; Manchester, J.I.; Miller, A.A. ESKAPEing the labyrinth of antibacterial discovery. Nat. Rev. Drug Discov. 2015, 14, 529–542. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.B.; Bassler, B.L. Quorum sensing in bacteria. Annu. Rev. Microbiol. 2001, 55, 165–199. [Google Scholar] [CrossRef] [PubMed]
- Shaaban, M.; Elgaml, A.; Habib, E.S.E. Biotechnological applications of quorum sensing inhibition as novel therapeutic strategies for multidrug resistant pathogens. Microb. Pathog. 2019, 127, 138–143. [Google Scholar] [CrossRef] [PubMed]
- Diggle, S.P.; Griffin, A.S.; Campbell, G.S.; West, S.A. Cooperation and conflict in quorum-sensing bacterial populations. Nature 2007, 450, 411–414. [Google Scholar] [CrossRef] [PubMed]
- Castillo-Juárez, I.; Maeda, T.; Mandujano-Tinoco, E.A.; Tomás, M.; Pérez-Eretza, B.; García-Contreras, S.J.; García-Contreras, R. Role of quorum sensing in bacterial infections. World J. Clin. Cases 2015, 3, 575–598. [Google Scholar] [CrossRef] [PubMed]
- Egland, K.A.; Greenberg, E.P. Quorum sensing in Vibrio fischeri: Analysis of the LuxR DNA binding region by alanine-scanning mutagenesis. J. Bacteriol. 2001, 183, 382–386. [Google Scholar] [CrossRef] [PubMed]
- Clewell, D.B.; Weaver, K.E.; Dunny, G.M.; Coque, T.M.; Francia, M.V.; Hayes, F. Extrachromosomal and mobile elements in enterococci: Transmission, maintenance, and epidemiology. In Enterococci: From Commensals to Leading Causes of Drug Resistant Infection; Gilmore, M.S., Clewell, D.B., Ike, Y., Shankar, N., Eds.; Massachusetts Eye and Ear Infirmary: Boston, MA, USA, 2014; pp. 309–320. [Google Scholar]
- Dunny, G.M. Enterococcal sex pheromones: Signaling, social behavior, and evolution. Annu. Rev. Genet. 2013, 47, 457–482. [Google Scholar] [CrossRef] [PubMed]
- Varahan, S.; Harms, N.; Gilmore, M.S.; Tomich, J.M.; Hancock, L.E. An ABC transporter is required for secretion of peptide sex pheromones in Enterococcus faecalis. MBio 2014, 5, e01726-14. [Google Scholar] [CrossRef] [PubMed]
- An, F.Y.; Clewell, D.B. Identification of the cAD1 sex pheromone precursor in Enterococcus faecalis. J. Bacteriol. 2002, 184, 1880–1887. [Google Scholar] [CrossRef] [PubMed]
- Haas, W.; Shepard, B.D.; Gilmore, M.S. Two-component regulator of Enterococcus faecalis cytolysin responds to quorum-sensing autoinduction. Nature 2002, 415, 84–87. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Li, W.; Hou, B.; Zhang, C. Quorum sensing LuxS/autoinducer-2 inhibits Enterococcus faecalis biofilm formation ability. J. Appl. Oral Sci. 2018, 26, 1–8. [Google Scholar] [CrossRef]
- Oli, A.K.; Javaregowda, P.K.; Jain, A.; Kelmani, C.R. Mechanism Involved in Biofilm Formation of Enterococcus Faecalis. Available online: https://www.intechopen.com/online-first/81571 (accessed on 20 January 2022).
- Rutherford, S.T.; Bassler, B.L. Bacterial quorum sensing: Its role in virulence and possibilities for its control. Cold Spring Harb. Perspect. Med. 2012, 2, a012427. [Google Scholar] [CrossRef] [PubMed]
- Lyon, G.J.; Wright, J.S.; Muir, T.W.; Novick, R.P. Key determinants of receptor activation in the agr autoinducing peptides of Staphylococcus aureus. Biochemistry 2002, 41, 10095–10104. [Google Scholar] [CrossRef] [PubMed]
- Murray, E.J.; Williams, P. Detection of agr-type autoinducing peptides produced by Staphylococcus aureus. In Quorum Sensing; Humana Press: New York, NY, USA, 2018; pp. 89–96. [Google Scholar]
- Kirchdoerfer, R.N.; Garner, A.L.; Flack, C.E.; Mee, J.M.; Horswill, A.R.; Janda, K.D.; Wilson, I.A. Structural basis for ligand recognition and discrimination of a quorum-quenching antibody. J. Biol. Chem. 2011, 286, 17351–17358. [Google Scholar] [CrossRef]
- Ma, R.; Qiu, S.; Jiang, Q.; Sun, H.; Xue, T.; Cai, G.; Sun, B. AI-2 quorum sensing negatively regulates rbf expression and biofilm formation in Staphylococcus aureus. Int. J. Med. Microbiol. 2017, 307, 257–267. [Google Scholar] [CrossRef] [PubMed]
- Fuqua, C. The QscR quorum-sensing regulon of Pseudomonas aeruginosa: An orphan claims its identity. J. Bacteriol. 2006, 188, 3169–3171. [Google Scholar] [CrossRef] [PubMed]
- Patankar, A.V.; González, J.E. Orphan LuxR regulators of quorum sensing. FEMS Microbiol. Rev. 2009, 33, 739–756. [Google Scholar] [CrossRef] [PubMed]
- Pacheco, T.; Gomes, A.É.I.; Siqueira, N.M.G.; Assoni, L.; Darrieux, M.; Venter, H.; Ferraz, L.F.C. Sdia, a quorum-sensing regulator, suppresses fimbriae expression, biofilm formation, and quorum-sensing signaling molecules production in Klebsiella pneumoniae. Front. Microbiol. 2021, 12, 597735. [Google Scholar] [CrossRef]
- Balestrino, D.; Haagensen, J.A.; Rich, C.; Forestier, C. Characterization of type 2 quorum sensing in Klebsiella pneumoniae and relationship with biofilm formation. J. Bacteriol. 2005, 187, 2870–2880. [Google Scholar] [CrossRef]
- Yin, W.F.; Purmal, K.; Chin, S.; Chan, X.Y.; Koh, C.L.; Sam, C.K.; Chan, K.G. N-acyl homoserine lactone production by Klebsiella pneumoniae isolated from human tongue surface. Sensors 2012, 12, 3472–3483. [Google Scholar] [CrossRef]
- Herzberg, M.; Kaye, I.K.; Peti, W.; Wood, T.K. YdgG (TqsA) controls biofilm formation in Escherichia coli K-12 through autoinducer 2 transport. J. Bacteriol. 2006, 188, 587–598. [Google Scholar] [CrossRef] [PubMed]
- Schauder, S.; Bassler, B.L. The languages of bacteria. Genes Dev. 2001, 15, 1468–1480. [Google Scholar] [CrossRef] [PubMed]
- De Keersmaecker, S.C.; Sonck, K.; Vanderleyden, J. Let LuxS speak up in AI-2 signaling. Trends Microbiol. 2006, 14, 114–119. [Google Scholar] [CrossRef]
- Pereira, C.S.; Thompson, J.A.; Xavier, K.B. AI-2-mediated signalling in bacteria. FEMS Microbiol. Rev. 2013, 37, 156–181. [Google Scholar] [CrossRef] [PubMed]
- Niu, C.; Clemmer, K.M.; Bonomo, R.A.; Rather, P.N. Isolation and characterization of an autoinducer synthase from Acinetobacter baumannii. J. Bacteriol. 2008, 190, 3386–3392. [Google Scholar] [CrossRef] [PubMed]
- Saipriya, K.; Swathi, C.H.; Ratnakar, K.S.; Sritharan, V. Quorum-sensing system in Acinetobacter baumannii: A potential target for new drug development. J. Appl. Microbiol. 2020, 128, 15–27. [Google Scholar] [CrossRef] [PubMed]
- Subhadra, B.; Oh, M.H.; Choi, C.H. Quorum sensing in Acinetobacter: With special emphasis on antibiotic resistance, biofilm formation and quorum quenching. AIMS Microbiol. 2016, 2, 27–41. [Google Scholar] [CrossRef]
- Choi, A.H.; Slamti, L.; Avci, F.Y.; Pier, G.B.; Maira-Litrán, T. The pgaABCD locus of Acinetobacter baumannii encodes the production of poly-β-1-6-N-acetylglucosamine, which is critical for biofilm formation. J. Bacteriol. 2009, 191, 5953–5963. [Google Scholar] [CrossRef]
- Boşgelmez-Tınaz, G.; Ulusoy, S.; Arıdoğan, B.; Eroğlu, F.; Kaya, S. N-butanoyl-L-homoserine lactone (BHL) deficient Pseudomonas aeruginosa isolates from an intensive care unit. Microbiol. Res. 2005, 160, 399–403. [Google Scholar] [CrossRef] [PubMed]
- Chan, M.C.; Chiu, S.K.; Hsueh, P.R.; Wang, N.C.; Wang, C.C.; Fang, C.T. Risk factors for healthcare-associated extensively drug-resistant Acinetobacter baumannii infections: A case-control study. PLoS ONE 2014, 9, e85973. [Google Scholar] [CrossRef] [PubMed]
- López-Martín, M.; Dubern, J.F.; Alexander, M.R.; Williams, P. Abam regulates quorum sensing, biofilm formation, and virulence in Acinetobacter baumannii. J. Bacteriol. 2021, 203, e00635-20. [Google Scholar] [CrossRef] [PubMed]
- Michalska, A.D.; Sacha, P.T.; Kaczynska, K.; Tryniszewska, E.A. The diversity of aminoglycoside-modifying enzymes among ESBL-positive proteus mirabilis clinical strains. Medtube Sci. 2014, 4, 16–20. [Google Scholar]
- Nemec, A.; Maixnerová, M.; van der Reijden, T.J.; Van den Broek, P.J.; Dijkshoorn, L. Relationship between the AdeABC efflux system gene content, netilmicin susceptibility and multidrug resistance in a genotypically diverse collection of Acinetobacter baumannii strains. J. Antimicrob. Chemother. 2007, 60, 483–489. [Google Scholar] [CrossRef] [PubMed]
- Yoon, E.J.; Courvalin, P.; Grillot-Courvalin, C. RND-type efflux pumps in multidrug-resistant clinical isolates of Acinetobacter baumannii: Major role for AdeABC overexpression and AdeRS mutations. Antimicrob. Agents Chemother. 2013, 57, 2989–2995. [Google Scholar] [CrossRef] [PubMed]
- Modarresi, F.; Azizi, O.; Shakibaie, M.R.; Motamedifar, M.; Mosadegh, E.; Mansouri, S. Iron limitation enhances acyl homoserine lactone (AHL) production and biofilm formation in clinical isolates of Acinetobacter baumannii. Virulence 2015, 6, 152–161. [Google Scholar] [CrossRef] [PubMed]
- Eze, E.C.; Chenia, H.Y.; El Zowalaty, M.E. Acinetobacter baumannii biofilms: Effects of physicochemical factors, virulence, antibiotic resistance determinants, gene regulation, and future antimicrobial treatments. Infect. Drug Resist. 2018, 11, 2277–2299. [Google Scholar] [CrossRef] [PubMed]
- Gallagher, L.A.; McKnight, S.L.; Kuznetsova, M.S.; Pesci, E.C.; Manoil, C. Functions required for extracellular quinolone signaling by Pseudomonas aeruginosa. J. Bacteriol. 2002, 184, 6472–6480. [Google Scholar] [CrossRef] [PubMed]
- Daniels, R.; Vanderleyden, J.; Michiels, J. Quorum sensing and swarming migration in bacteria. FEMS Microbiol. Rev. 2004, 28, 261–289. [Google Scholar] [CrossRef] [PubMed]
- Hodgkinson, J.T.; Welch, M.; Spring, D.R. Learning the language of bacteria. ACS Chem. Biol. 2007, 2, 715–717. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Lequette, Y.; Greenberg, E.P. Activity of purified QscR, a Pseudomonas aeruginosa orphan quorum-sensing transcription factor. Mol. Microbiol. 2006, 59, 602–609. [Google Scholar] [CrossRef]
- Maseda, H.; Sawada, I.; Saito, K.; Uchiyama, H.; Nakae, T.; Nomura, N. Enhancement of the mexAB-oprM efflux pump expression by a quorum-sensing autoinducer and its cancellation by a regulator, MexT, of the mexEF-oprN efflux pump operon in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2004, 48, 1320–1328. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Kim, Y.U.; Koh, B.H.; Hwang, S.S.; Kim, S.H.; Lépine, F.; Cho, Y.H.; Lee, G.R. HHQ and PQS, two Pseudomonas aeruginosa quorum-sensing molecules, down-regulate the innate immune responses through the nuclear factor-κB pathway. Immunology 2010, 129, 578–588. [Google Scholar] [CrossRef] [PubMed]
- Lazar, V.; Holban, A.M.; Curutiu, C.; Chifiriuc, M.C. Modulation of quorum sensing and biofilms in less investigated gram-negative ESKAPE pathogens. Front. Microbiol. 2021, 12, 2072. [Google Scholar] [CrossRef] [PubMed]
- Lau, Y.Y.; Sulaiman, J.; Chen, J.W.; Yin, W.F.; Chan, K.G. Quorum sensing activity of Enterobacter asburiae isolated from lettuce leaves. Sensors 2013, 13, 14189–14199. [Google Scholar] [CrossRef] [PubMed]
- Shankar, M.; Ponraj, P.; Illakkiam, D.; Rajendhran, J.; Gunasekaran, P. Inactivation of the transcriptional regulator-encoding gene sdiA enhances rice root colonization and biofilm formation in Enterobacter cloacae GS1. J. Bacteriol. 2013, 195, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Lau, Y.Y.; How, K.Y.; Yin, W.F.; Chan, K.G. Functional characterization of quorum sensing LuxR-type transcriptional regulator, EasR in Enterobacter asburiae strain L1. PeerJ 2020, 8, e10068. [Google Scholar] [CrossRef]
- Rezzonico, F.; Smits, T.H.; Duffy, B. Detection of AI-2 receptors in genomes of Enterobacteriaceae suggests a role of type-2 quorum sensing in closed ecosystems. Sensors 2012, 12, 6645–6665. [Google Scholar] [CrossRef]
- Tay, S.B.; Yew, W.S. Development of quorum-based anti-virulence therapeutics targeting Gram-negative bacterial pathogens. Int. J. Mol. Sci. 2013, 14, 16570–16599. [Google Scholar] [CrossRef]
- Reading, N.C.; Sperandio, V. Quorum sensing: The many languages of bacteria. FEMS Microbiol. Lett. 2006, 254, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Shih, P.C.; Huang, C.T. Effects of quorum-sensing deficiency on Pseudomonas aeruginosa biofilm formation and antibiotic resistance. J. Antimicrob. Chemother. 2002, 49, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Ganesh, P.S.; Rai, V.R. Attenuation of quorum-sensing-dependent virulence factors and biofilm formation by medicinal plants against antibiotic resistant Pseudomonas aeruginosa. J. Tradit. Complement. Med. 2018, 8, 170–177. [Google Scholar] [CrossRef] [PubMed]
- Parsek, M.R.; Val, D.L.; Hanzelka, B.L.; Cronan, J.E., Jr.; Greenberg, E.P. Acyl homoserine-lactone quorum-sensing signal generation. Proc. Natl. Acad. Sci. USA 1999, 96, 4360–4365. [Google Scholar] [CrossRef]
- Rasmussen, T.B.; Givskov, M. Quorum-sensing inhibitors as anti-pathogenic drugs. Int. J. Med. Microbiol. 2006, 296, 149–161. [Google Scholar] [CrossRef] [PubMed]
- Priyadarshi, A.; Kim, E.E.; Hwang, K.Y. Structural insights into Staphylococcus aureus enoyl-ACP reductase (FabI), in complex with NADP and triclosan. Proteins Struct. Funct. Genet. 2010, 78, 480–486. [Google Scholar] [CrossRef] [PubMed]
- Hraiech, S.; Hiblot, J.; Lafleur, J.; Lepidi, H.; Papazian, L.; Rolain, J.M.; Raoult, D.; Elias, M.; Silby, M.W.; Bzdrenga, J.; et al. Inhaled lactonase reduces Pseudomonas aeruginosa quorum sensing and mortality in rat pneumonia. PLoS ONE 2014, 9, e107125. [Google Scholar] [CrossRef]
- Guendouze, A.; Plener, L.; Bzdrenga, J.; Jacquet, P.; Rémy, B.; Elias, M.; Lavigne, J.P.; Daudé, D.; Chabrière, E. Effect of quorum quenching lactonase in clinical isolates of Pseudomonas aeruginosa and comparison with quorum sensing inhibitors. Front. Microbiol. 2017, 8, 227. [Google Scholar] [CrossRef]
- Fan, X.; Liang, M.; Wang, L.; Chen, R.; Li, H.; Liu, X. Aii810, a novel cold-adapted N-acylhomoserine lactonase discovered in a metagenome, can strongly attenuate Pseudomonas aeruginosa virulence factors and biofilm formation. Front. Microbiol. 2017, 8, 1950. [Google Scholar] [CrossRef]
- Sakr, M.M.; Aboshanab, K.M.; Elkhatib, W.F.; Yassien, M.A.; Hassouna, N.A. Overexpressed recombinant quorum quenching lactonase reduces the virulence, motility and biofilm formation of multidrug-resistant Pseudomonas aeruginosa clinical isolates. Appl. Microbiol. Biotechnol. 2018, 102, 10613–10622. [Google Scholar] [CrossRef]
- Schipper, C.; Hornung, C.; Bijtenhoorn, P.; Quitschau, M.; Grond, S.; Streit, W.R. Metagenome-derived clones encoding two novel lactonase family proteins involved in biofilm inhibition in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 2009, 75, 224–233. [Google Scholar] [CrossRef]
- Dong, W.; Zhu, J.; Guo, X.; Kong, D.; Zhang, Q.; Zhou, Y.; Ruan, Z. Characterization of AiiK, an AHL lactonase, from Kurthia huakui LAM0618T and its application in quorum quenching on Pseudomonas aeruginosa PAO1. Sci. Rep. 2018, 8, 6013. [Google Scholar] [CrossRef]
- Grover, N.; Plaks, J.G.; Summers, S.R.; Chado, G.R.; Schurr, M.J.; Kaar, J.L. Acylase-containing polyurethane coatings with anti-biofilm activity. Biotechnol. Bioeng. 2016, 113, 2535–2543. [Google Scholar] [CrossRef]
- Lee, J.; Lee, I.; Nam, J.; Hwang, D.S.; Yeon, K.M.; Kim, J. Immobilization and stabilization of acylase on carboxylated polyaniline nanofibers for highly effective antifouling application via quorum quenching. ACS Appl. Mater. Interfaces 2017, 9, 15424–15432. [Google Scholar] [CrossRef] [PubMed]
- Sio, C.F.; Otten, L.G.; Cool, R.H.; Diggle, S.P.; Braun, P.G.; Bos, R.; Daykin, M.; Caámara, M.; Williams, P.; Quax, W.J. Quorum quenching by an N-acyl-homoserine lactone acylase from Pseudomonas aeruginosa PAO1. Infect. Immun. 2006, 74, 1673–1682. [Google Scholar] [CrossRef] [PubMed]
- Bijtenhoorn, P.; Mayerhofer, H.; Müller-Dieckmann, J.; Utpatel, C.; Schipper, C.; Hornung, C.; Szesny, M.; Grond, S.; Thürmer, A.; Brzuszkiewicz, E.; et al. A novel metagenomic short-chain dehydrogenase/reductase attenuates Pseudomonas aeruginosa biofilm formation and virulence on Caenorhabditis elegans. PLoS ONE 2011, 6, e26278. [Google Scholar] [CrossRef]
- Zhang, X.; Ou-Yang, S.; Wang, J.; Liao, L.; Wu, R.; Wei, J. Construction of antibacterial surface via layer-by-layer method. Curr. Pharm. Des. 2018, 24, 926–935. [Google Scholar] [CrossRef] [PubMed]
- Pustelny, C.; Albers, A.; Büldt-Karentzopoulos, K.; Parschat, K.; Chhabra, S.R.; Cámara, M.; Fetzner, S. Dioxygenase-mediated quenching of quinolone-dependent quorum sensing in Pseudomonas aeruginosa. Chem. Biol. 2009, 16, 1259–1267. [Google Scholar] [CrossRef]
- Kaufmann, G.F.; Park, J.; Mee, J.M.; Ulevitch, R.J.; Janda, K.D. The quorum quenching antibody RS2-1G9 protects macrophages from the cytotoxic effects of the Pseudomonas aeruginosa quorum sensing signalling molecule N-3-oxo-dodecanoyl-homoserine lactone. Mol. Immunol. 2008, 45, 2710–2714. [Google Scholar] [CrossRef] [PubMed]
- Koul, S.; Prakash, J.; Mishra, A.; Kalia, V.C. Potential emergence of multi-quorum sensing inhibitor resistant (MQSIR) bacteria. Indian J. Microbiol. 2016, 56, 1–18. [Google Scholar] [CrossRef]
- Santajit, S.; Seesuay, W.; Mahasongkram, K.; Sookrung, N.; Pumirat, P.; Ampawong, S.; Reamtong, O.; Chongsa-Nguan, M.; Chaicumpa, W.; Indrawattana, N. Human single-chain variable fragments neutralize Pseudomonas aeruginosa quorum sensing molecule, 3O-C12-HSL, and prevent cells from the HSL-mediated apoptosis. Front. Microbiol. 2020, 11, 1172. [Google Scholar] [CrossRef]
- Park, J.; Jagasia, R.; Kaufmann, G.F.; Mathison, J.C.; Ruiz, D.I.; Moss, J.A.; Meijler, M.M.; Ulevitch, R.J.; Janda, K.D. Infection control by antibody disruption of bacterial quorum sensing signaling. Chem. Biol. 2007, 14, 1119–1127. [Google Scholar] [CrossRef]
- Balaban, N.; Rasooly, A. Staphylococcal enterotoxins. Int. J. Food Microbiol. 2000, 61, 1–10. [Google Scholar] [CrossRef]
- Suresh, M.K.; Biswas, R.; Biswas, L. An update on recent developments in the prevention and treatment of Staphylococcus aureus biofilms. Int. J. Med. Microbiol. 2019, 309, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Weng, L.X.; Yang, Y.X.; Zhang, Y.Q.; Wang, L.H. A new synthetic ligand that activates QscR and blocks antibiotic-tolerant biofilm formation in Pseudomonas aeruginosa. Appl. Microbiol. Biotechnol. 2014, 98, 2565–2572. [Google Scholar] [CrossRef]
- O’Loughlin, C.T.; Miller, L.C.; Siryaporn, A.; Drescher, K.; Semmelhack, M.F.; Bassler, B.L. A quorum-sensing inhibitor blocks Pseudomonas aeruginosa virulence and biofilm formation. Proc. Natl. Acad. Sci. USA 2013, 110, 17981–17986. [Google Scholar] [CrossRef] [PubMed]
- Geske, G.D.; O’Neill, J.C.; Blackwell, H.E. N-phenylacetanoyl-L-homoserine lactones can strongly antagonize or superagonize quorum sensing in Vibrio fischeri. ACS Chem. Biol. 2007, 2, 315–319. [Google Scholar] [CrossRef] [PubMed]
- Nait Chabane, Y.; Mlouka, M.B.; Alexandre, S.; Nicol, M.; Marti, S.; Pestel-Caron, M.; Vila, J.; Jouenne, T.; Dé, E. Virstatin inhibits biofilm formation and motility of Acinetobacter baumannii. BMC Microbiol. 2014, 14, 62. [Google Scholar] [CrossRef]
- Valdez, J.C.; Peral, M.C.; Rachid, M.; Santana, M.; Perdigon, G. Interference of Lactobacillus plantarum with Pseudomonas aeruginosa in vitro and in infected burns: The potential use of probiotics in wound treatment. Clin. Microbiol. Infect. 2005, 11, 472–479. [Google Scholar] [CrossRef]
- Cui, T.; Bai, F.; Sun, M.; Lv, X.; Li, X.; Zhang, D.; Du, H. Lactobacillus crustorum ZHG 2-1 as novel quorum-quenching bacteria reducing virulence factors and biofilms formation of Pseudomonas aeruginosa. LWT 2020, 117, 108696. [Google Scholar] [CrossRef]
- Rana, S.; Bhawal, S.; Kumari, A.; Kapila, S.; Kapila, R. pH-dependent inhibition of AHL-mediated quorum sensing by cell-free supernatant of lactic acid bacteria in Pseudomonas aeruginosa PAO1. Microb. Pathog. 2020, 142, 104105. [Google Scholar] [CrossRef]
- Li, J.; Wang, W.; Xu, S.X.; Magarvey, N.A.; McCormick, J.K. Lactobacillus reuteri-produced cyclic dipeptides quench agr-mediated expression of toxic shock syndrome toxin-1 in staphylococci. Proc. Natl. Acad. Sci. USA 2011, 108, 3360–3365. [Google Scholar] [CrossRef]
- Yan, X.; Gu, S.; Cui, X.; Shi, Y.; Wen, S.; Chen, H.; Ge, J. Antimicrobial, anti-adhesive and anti-biofilm potential of biosurfactants isolated from Pediococcus acidilactici and Lactobacillus plantarum against Staphylococcus aureus CMCC26003. Microb. Pathog. 2019, 127, 12–20. [Google Scholar] [CrossRef]
- Bouyahya, A.; Chamkhi, I.; Balahbib, A.; Rebezov, M.; Shariati, M.A.; Wilairatana, P.; Mubarak, M.S.; Benali, T.; El Omari, N. Mechanisms, anti-quorum-sensing actions, and clinical trials of medicinal plant bioactive compounds against bacteria: A comprehensive review. Molecules 2022, 27, 1484. [Google Scholar] [CrossRef]
- Packiavathy, I.A.S.V.; Agilandeswari, P.; Musthafa, K.S.; Pandian, S.K.; Ravi, A.V. Antibiofilm and quorum sensing inhibitory potential of Cuminum cyminum and its secondary metabolite methyl Eugenol against gram negative bacterial pathogens. Food Res. Int. 2012, 8, 85–92. [Google Scholar] [CrossRef]
- Zhou, L.; Zheng, H.; Tang, Y.; Yu, W.; Gong, Q. Eugenol inhibits quorum sensing at sub-inhibitory concentrations. Biotechnol. Lett. 2013, 35, 631–637. [Google Scholar] [CrossRef] [PubMed]
- Rathinam, P.; Kumar, H.S.V.; Viswanathan, P. Eugenol exhibits anti-virulence properties by competitively binding to quorum sensing receptors. Biofouling 2017, 33, 624–639. [Google Scholar] [CrossRef] [PubMed]
- Lou, Z.; Letsididi, K.S.; Yu, F.; Pei, Z.; Wang, H.; Letsididi, R. Inhibitive effect of Eugenol and its nanoemulsion on quorum sensing–mediated virulence factors and biofilm formation by Pseudomonas aeruginosa. J. Food Prot. 2019, 82, 379–389. [Google Scholar] [CrossRef]
- Al-Shabib, N.A.; Husain, F.M.; Ahmad, I.; Baig, M.H. Eugenol inhibits quorum sensing and biofilm of toxigenic MRSA strains isolated from food handlers employed in Saudi Arabia. Biotechnol. Biotechnol. Equip. 2017, 11, 387–396. [Google Scholar] [CrossRef]
- Tapia-Rodriguez, M.R.; Hernandez-Mendoza, A.; Gonzalez-Aguilar, G.A.; Martinez Tellez, M.A.; Martins, C.M.; Ayala-Zavala, J.F. Carvacrol as potential quorum sensing inhibitor of Pseudomonas aeruginosa and biofilm production on stainless steel surfaces. Food Control 2017, 75, 255–261. [Google Scholar] [CrossRef]
- Tapia-Rodriguez, M.R.; Bernal-Mercado, A.T.; Gutierrez-Pacheco, M.M.; Vazquez-Armenta, F.J.; Hernandez-Mendoza, A.; Gonzalez-Aguilar, G.A.; Martinez-Tellez, M.A.; Nazzaro, F.; Ayala-Zavala, J.F. Virulence of Pseudomonas aeruginosa exposed to Carvacrol: Alterations of the quorum sensing at enzymatic and gene levels. J. Cell Commun. Signal. 2019, 13, 531–537. [Google Scholar] [CrossRef]
- Pejin, B.; Ciric, A.; Glamoclija, J.; Nikolic, M.; Sokovic, M. In vitro anti quorum sensing activity of Phytol. Nat. Prod. Res. 2015, 29, 374–377. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, R.; Devi, K.R.; Kannappan, A.; Pandian, S.K.; Ravi, A.V. Pipervtle and its bioactive metabolite Phytol mitigates quorum sensing mediated virulence factors and biofilm of nosocomial pathogen Serratia marcescens in vitro. J. Ethnopharmacol. 2016, 193, 592–603. [Google Scholar] [CrossRef]
- Amaya, S.; Pereira, J.A.; Borkosky, S.A.; Valdez, J.C.; Bardón, A.; Arena, M.E. Inhibition of quorum sensing in Pseudomonas aeruginosa by Sesquiterpene lactones. Phytomedicine 2012, 19, 1173–1177. [Google Scholar] [CrossRef] [PubMed]
- Rasamiravaka, T.; Vandeputte, O.M.; Pottier, L.; Huet, J.; Rabemanantsoa, C.; Kiendrebeogo, M.; Andriantsimahavandy, A.; Rasamindrakotroka, A.; Stévigny, C.; Duez, P.; et al. Pseudomonas aeruginosa biofilm formation and persistence, along with the production of quorum sensing-dependent virulence factors, are disrupted by a triterpenoid coumarate ester isolated from Dalbergia trichocarpa, a Tropical Legume. PLoS ONE 2015, 10, e0132791. [Google Scholar] [CrossRef] [PubMed]
- Alves, S.; Duarte, A.; Sousa, S.; Domingues, F.C. Study of the major essential oil compounds of Coriandrum sativum against Acinetobacter baumannii and the effect of Linalool on adhesion, biofilms and quorum sensing. Biofouling 2016, 32, 155–165. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Vega, P.; Xu, Y.; Chen, C.-Y.; Irudayaraj, J. Exploring the anti-quorum sensing activity of ad-limonene nanoemulsion for Escherichia coli O157:H7. J. Biomed. Mater. Res. A 2018, 106, 1979–1986. [Google Scholar] [CrossRef]
- Rémy, B.; Plener, L.; Decloquement, P.; Armstrong, N.; Elias, M.; Daudé, D.; Chabrière, É. Lactonase specificity is key to quorum quenching in Pseudomonas aeruginosa. Front. Microbiol. 2020, 11, 762. [Google Scholar] [CrossRef]
- Chow, J.Y.; Yang, Y.; Tay, S.B.; Chua, K.L.; Yew, W.S. Disruption of biofilm formation by the human pathogen Acinetobacter baumannii using engineered quorum-quenching lactonases. Antimicrob. Agents Chemother. 2014, 58, 1802–1805. [Google Scholar] [CrossRef]
- Witzgall, F.; Depke, T.; Hoffmann, M.; Empting, M.; Brönstrup, M.; Müller, R.; Blankenfeldt, W. The alkylquinolone repertoire of Pseudomonas aeruginosa is linked to structural flexibility of the FabH-like 2-heptyl-3-hydroxy-4 (1H)-quinolone (PQS) biosynthesis enzyme PqsBC. Chem. Biochem. 2018, 19, 1531–1544. [Google Scholar]
- Paczkowski, J.E.; Mukherjee, S.; McCready, A.R.; Cong, J.P.; Aquino, C.J.; Kim, H.; Henke, B.R.; Smith, C.D.; Bassler, B.L. Flavonoids suppress Pseudomonas aeruginosa virulence through allosteric inhibition of quorum-sensing receptors. J. Biol. Chem. 2017, 292, 4064–4076. [Google Scholar] [CrossRef]
- Plaza-Diaz, J.; Ruiz-Ojeda, F.J.; Gil-Campos, M.; Gil, A. Mechanisms of action of probiotics. Adv. Nutr. 2019, 10, S49–S66. [Google Scholar] [CrossRef]
- Pejin, B.; Ciric, A.; Glamoclija, J.; Nikolic, M.; Stanimirovic, B.; Sokovic, M. Quercetin potently reduces biofilm formation of the strain Pseudomonas aeruginosa PAO1 in vitro. Curr. Pharm. Biotechnol. 2015, 16, 733–737. [Google Scholar] [CrossRef] [PubMed]
- Gopu, V.; Meena, C.K.; Shetty, P.H. Quercetin influences Quorum Sensing in Food Borne Bacteria: In-Vitro and In-Silico Evidence. PLoS ONE 2015, 10, e0134684. [Google Scholar] [CrossRef] [PubMed]
- Erdönmez, D.; Rad, A.Y.; Aksöz, N.; Erdönmez, D.; Rad, A.Y.; Aksöz, N. Anti-Quorum Sensing Potential of Antioxidant Quercetin and Resveratrol. Braz. Arch. Biol. Technol. 2018, 61, e18160756. [Google Scholar] [CrossRef]
- Ouyang, J.; Feng, W.; Lai, X.; Chen, Y.; Zhang, X.; Rong, L.; Sun, F.; Chen, Y. Quercetin Inhibits Pseudomonas aeruginosa Biofilm Formation via the Vfr-Mediated LasIR System. Microb. Pathog. 2020, 149, 104291. [Google Scholar] [CrossRef]
- Sedlmayer, F.; Woischnig, A.K.; Unterreiner, V.; Fuchs, F.; Baeschlin, D.; Khanna, N.; Fussenegger, M. 5-Fluorouracil blocks quorum-sensing of biofilm-embedded methicillin-resistant Staphylococcus aureus in mice. Nucleic Acids Res. 2021, 49, e73. [Google Scholar] [CrossRef] [PubMed]
- Walz, J.M.; Avelar, R.L.; Longtine, K.J.; Carter, K.L.; Mermel, L.A.; Heard, S.O.; 5-FU Catheter Study Group. Anti-infective external coating of central venous catheters: A randomized, noninferiority trial comparing 5-fluorouracil with chlorhexidine/silver sulfadiazine in preventing catheter colonization. Crit. Care Med. 2010, 38, 2095–2102. [Google Scholar]
- Van Delden, C.; Köhler, T.; Brunner-Ferber, F.; François, B.; Carlet, J.; Pechère, J.C. Azithromycin to prevent Pseudomonas aeruginosa ventilator-associated pneumonia by inhibition of quorum sensing: A randomized controlled trial. Intensive Care Med. 2012, 38, 1118–1125. [Google Scholar] [CrossRef] [PubMed]

| Strategies | Anti-QS Agents | Modes of Action | Effect on ESKAPE Organisms | References |
|---|---|---|---|---|
| Inhibition of AI synthases | Sinefungin | Structural analogues of S-adenosyl methionine (SAM) and acyl-carrier protein (ACP), the substrates of AHL synthases | Prevent bacterial infection and diminish QS-mediated virulence factors by blocking P. aeruginosa AHL synthesis | [64,65] |
| Butyryl-SAM | ||||
| L/D-S-adenosylhomocysteine | ||||
| Triclosan | Reduction of the establishment of enoyl-ACP reductase precursors (FabI) | Decrease S. aureus AHL production | [66] | |
| Targeting of QS Ligands | AHL lactonases (such as SsoPox, lactonase Aii810, QQ lactonase enzyme AHL-1, a novel lactonase cloned by bpiB01 and bpiB04 and lactonase AiiK) | Hydrolysis of the AHL lactone ring to form the consequent N-acyl homoserine | Lessen the extracellular proteases and pyocyanin biosynthesis, rhamnolipids, swarming motility and biofilm production and prevent bacterial infection of P. aeruginosa | [67,68,69,70,71,72] |
| Acylases (i.e., N-acyl homoserine lactone acylase PA2385, acylase (EC.3.5.1.14) | Degradation of the AHL amide bond and generation of the corresponding free fatty acid and a lactone ring | Decrease elastase, pyocyanin synthesis and biofilm biomass and formation in P. aeruginosa | [73,74,75] | |
| Oxidoreductases (e.g., BpiB09) | Oxidation and consequent inhibition of signal QS molecules | Slow down bacterial motility and reduce biofilm formation and pyocyanin production in P. aeruginosa; hinder bacterial biofilm development and decrease the growth rate of K. pneumoniae | [76,77] | |
| 3-Hydroxy-2-methyl-4(1H)-quinolone 2, 4-dioxygenase | Catalysis of the conversion of PQS to N-octanoylanthranilic acid and carbon monoxide | Hamper lectin A, pyocyanin and rhamnolipid biosynthesis of P. aeruginosa | [78] | |
| Quorum quenching antibody, RS2-1G9 | Hydrolysis of 3-oxo-C12-HSL | Inhibit the activation of the mitogen- activated protein kinase p38 and protect against the cytotoxic effects of P. aeruginosa on macrophages generated from murine bone marrow | [79] | |
| XYD-11G2 antibody | Hydrolysis of 3-oxo-C12-HSL | Conquest the bacterial QS signals of P. aeruginosa | [80] | |
| Human single-chain variable fragments | Hydrolysis of 3-oxo-C12-HSL | Prevent mammalian cell apoptosis triggered by P. aeruginosa | [81] | |
| AP4-24H11 antibody | Targeting of autoinducing peptide-4 | Induce the protective properties of S. aureus-produced abscess in vivo | [82] | |
| Synthetic RIP | Targeting of autoinducing peptide-4 | Diminish S. aureus infections in vivo | [83] | |
| Blockade of QS Transcriptional Regulators | Flavonoids | Allosteric blockade of the AI-binding receptors LasR and RhlR | Modulate the transcription of QS-controlled target promoters and limit the synthesis of virulence factors in P. aeruginosa | [84] |
| N-decanoyl-L-homoserine benzyl ester | Repression of the quorum sensing control repressor | Impair the production of biofilms, swarming activity, and protease and elastase enzymes in P. aeruginosa | [85] | |
| Meta-bromo-thiolactone | Allosteric blockade of the AI-binding receptors LasR and RhlR | Decrease pyocyanin synthesis and biofilm development in P. aeruginosa | [86] | |
| A4, 4-bromophenyl-PHL B7, 4-iodo PHL C10 and 3-nitro PHL C14 | Blockade of the AI-binding receptors, including TraR, LasR, and LuxR | Suppress the development of virulence factors in P. aeruginosa | [87] | |
| Virstatin | Inhibition of the expression of the anoR gene | Prevent bacterial movement and biofilm formation in A. baumannii | [88] | |
| Probiotics | L. plantarum PA 100 | Blockade of the function and inhibition of the synthesis of acyl homoserine lactones | Diminish biofilm production and elastase activity in P. aeruginosa | [89] |
| C. crustorum ZHG 2-1 | Degradation of C4-HSL and 3-oxo-C12-HSL | Suppress virulence factors (chitinase and protease), reduce swarming and swimming motilities, and inhibit biofilm formation in P. aeruginosa | [90] | |
| Cell-free acidic supernatants L. lactis NCDC 309, L. rhamnosus MTCC 5897, L. rhamnosus MTCC 5857, L. fermentum MTCC 5898, L. acidophilus NCDC 15, L. delbrueckii subsp. lactis, L. plantarum NCDC 372 | Destruction of C4-HSL and 3-oxo-C12-HSL | Inhibit biofilm formation, elastase, and expression of lasI and rhlI in P. aeruginosa | [91] | |
| L. reuteri RC-14 | Inhibition of arg gene expression by bioactive cyclic dipeptides (known as 2,5-diketo-piperazines, or DKPs) | Neutralise S. aureus MN8 toxin TSST-1 synthesis (toxic shock syndrome) | [92] | |
| L. plantarum, P. acidilactici | Downregulation of genes including cidA, icaA, dltB, agrA, sortaseA, and sarA | Suppress the formation of S. aureus biofilm | [93] | |
| Plant extracts | Eugenol | Suppression the expression of las and pqs systems | Prevent biofilm formation of P. aeruginosa PAO1. | [94,95,96,97] |
| Reduction in the level of QS synthase genes, including lasI, rhlI, and rhlA, | Inhibit biofilm growth and regressed virulence production (including pyocyanin, pyocyanin, and elastase) of P. aeruginosa PAO1 | [98] | ||
| Unknown | Limit the production of protease and pigments in MRSA | [99] | ||
| Carvacrol (2-methyl-5-(1-methylethyl)-phenol) | Blocking lasI and lasR expression | Lower the biofilm development and bacterial motility of P. aeruginosa | [100,101] | |
| Phytol | Unknown | Suppress flagella mobilization, restricts the formation of pyocyanin, and inhibits the establishment of the biofilm in P. aeruginosa PAO1 | [102,103] | |
| Sesquiterpene lactones | Unknown | Diminish the QS mediators in P. aeruginosa ATCC 27853 | [104] | |
| Oleanolic aldehyde coumarate | Downregulation of lasI/lasR, rhlI/rhlR, and gacA expression | Decrease the P. aeruginosa ‘s biofilm biogenesis | [105] | |
| Linalool | Unknown | Prevent the establishment of A. baumannii‘s biofilms and alter this strain’s surface adhesion. | [106,107] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santajit, S.; Sookrung, N.; Indrawattana, N. Quorum Sensing in ESKAPE Bugs: A Target for Combating Antimicrobial Resistance and Bacterial Virulence. Biology 2022, 11, 1466. https://doi.org/10.3390/biology11101466
Santajit S, Sookrung N, Indrawattana N. Quorum Sensing in ESKAPE Bugs: A Target for Combating Antimicrobial Resistance and Bacterial Virulence. Biology. 2022; 11(10):1466. https://doi.org/10.3390/biology11101466
Chicago/Turabian StyleSantajit, Sirijan, Nitat Sookrung, and Nitaya Indrawattana. 2022. "Quorum Sensing in ESKAPE Bugs: A Target for Combating Antimicrobial Resistance and Bacterial Virulence" Biology 11, no. 10: 1466. https://doi.org/10.3390/biology11101466
APA StyleSantajit, S., Sookrung, N., & Indrawattana, N. (2022). Quorum Sensing in ESKAPE Bugs: A Target for Combating Antimicrobial Resistance and Bacterial Virulence. Biology, 11(10), 1466. https://doi.org/10.3390/biology11101466






