Genomic Effect of DNA Methylation on Gene Expression in Colorectal Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. TCGA Data Collection and Preprocessing
2.2. Collection of DNA Methylation Probes for Each Gene
2.3. Statistical Machine Learning Model
2.4. Performance Evaluation and Model Selection
2.5. Investigation of the Significance of CpG Sites
2.6. Genomic Annotation for Probes
2.7. Validation Data Analysis
2.8. Survival Analysis
2.9. Identification of Signature Genes and Functional Annotation
3. Results
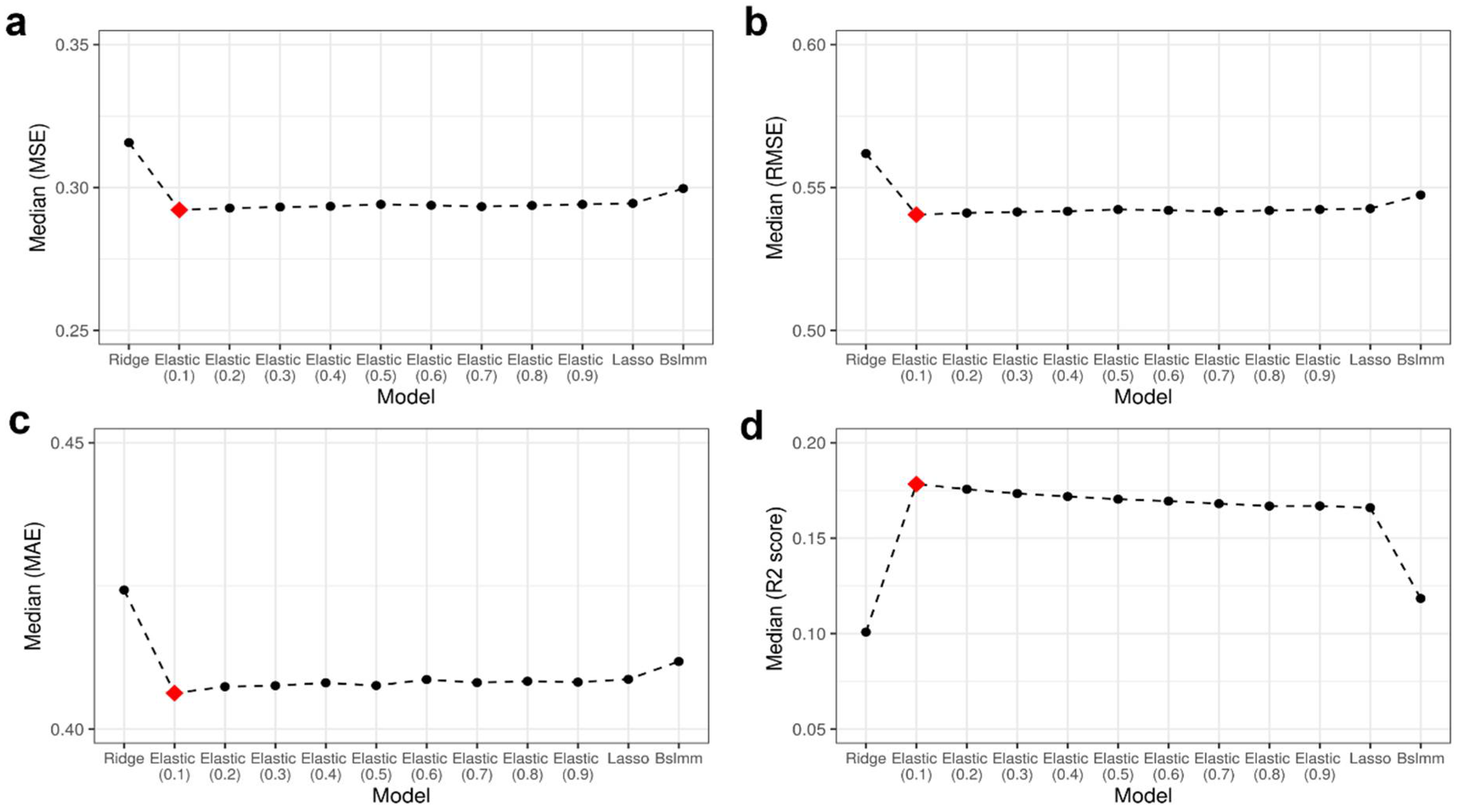
3.1. Best-Performing Model: ElasticNet
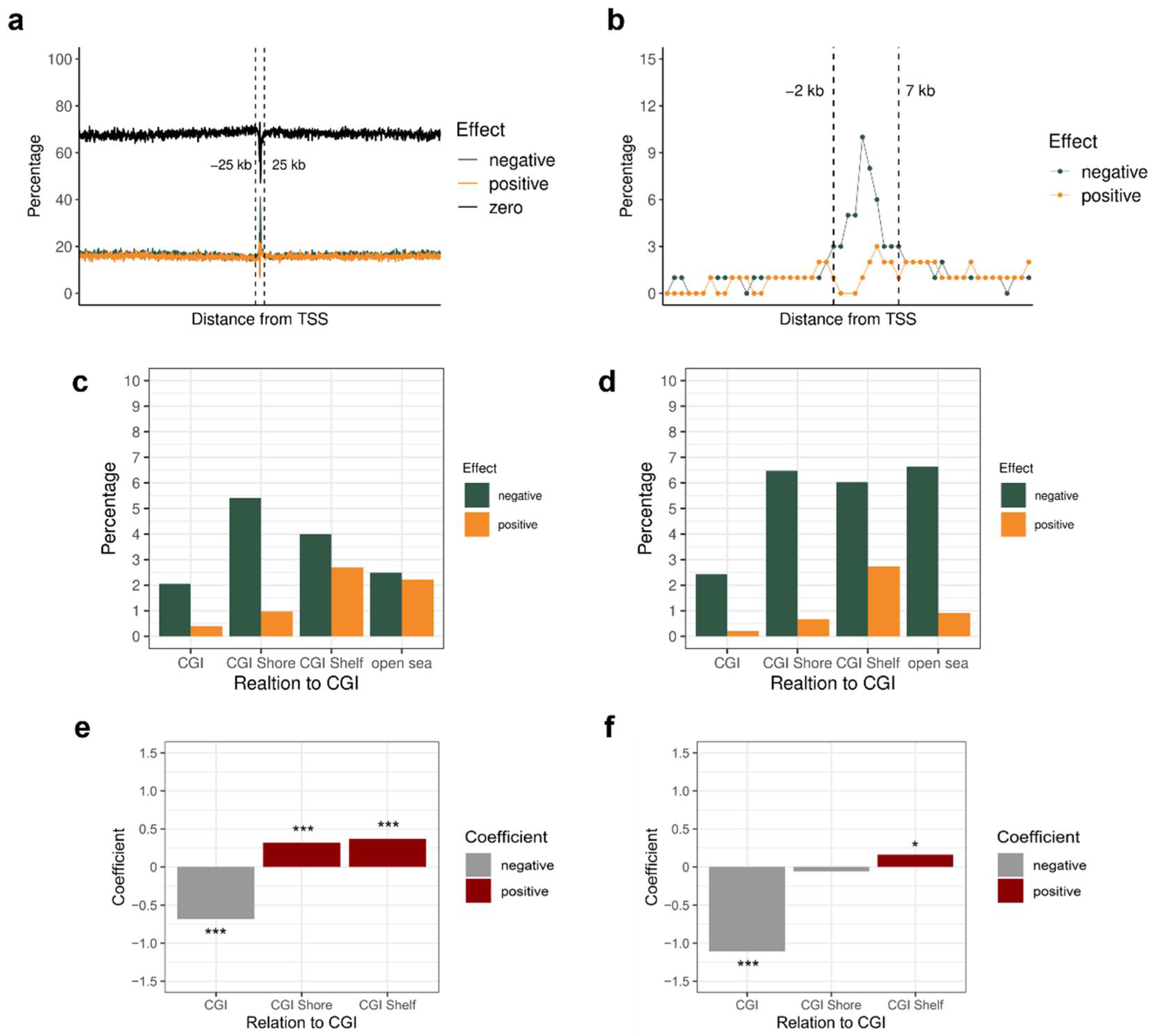
3.2. Importance of DNA Methylation in a Specific Genomic Region
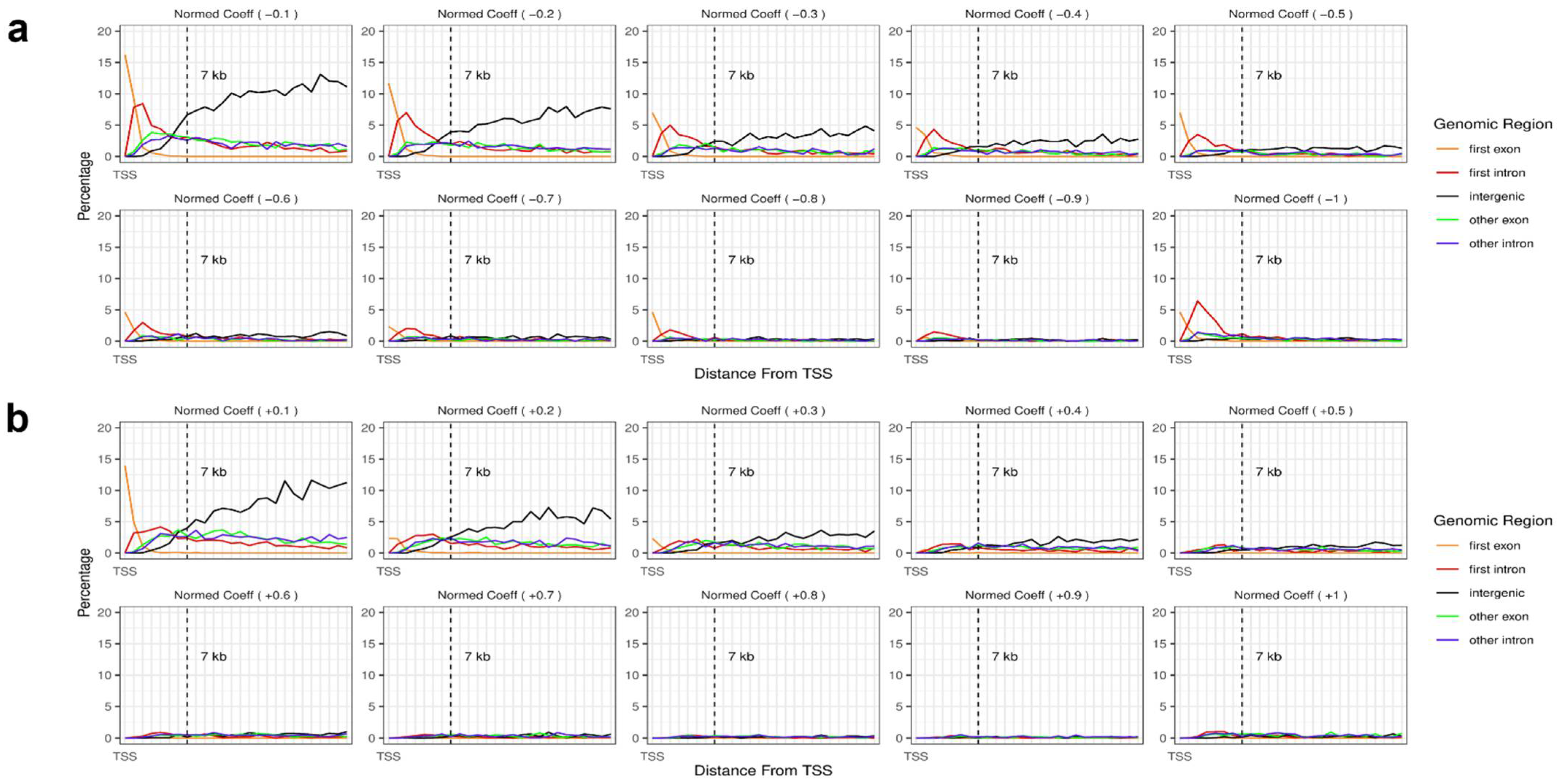
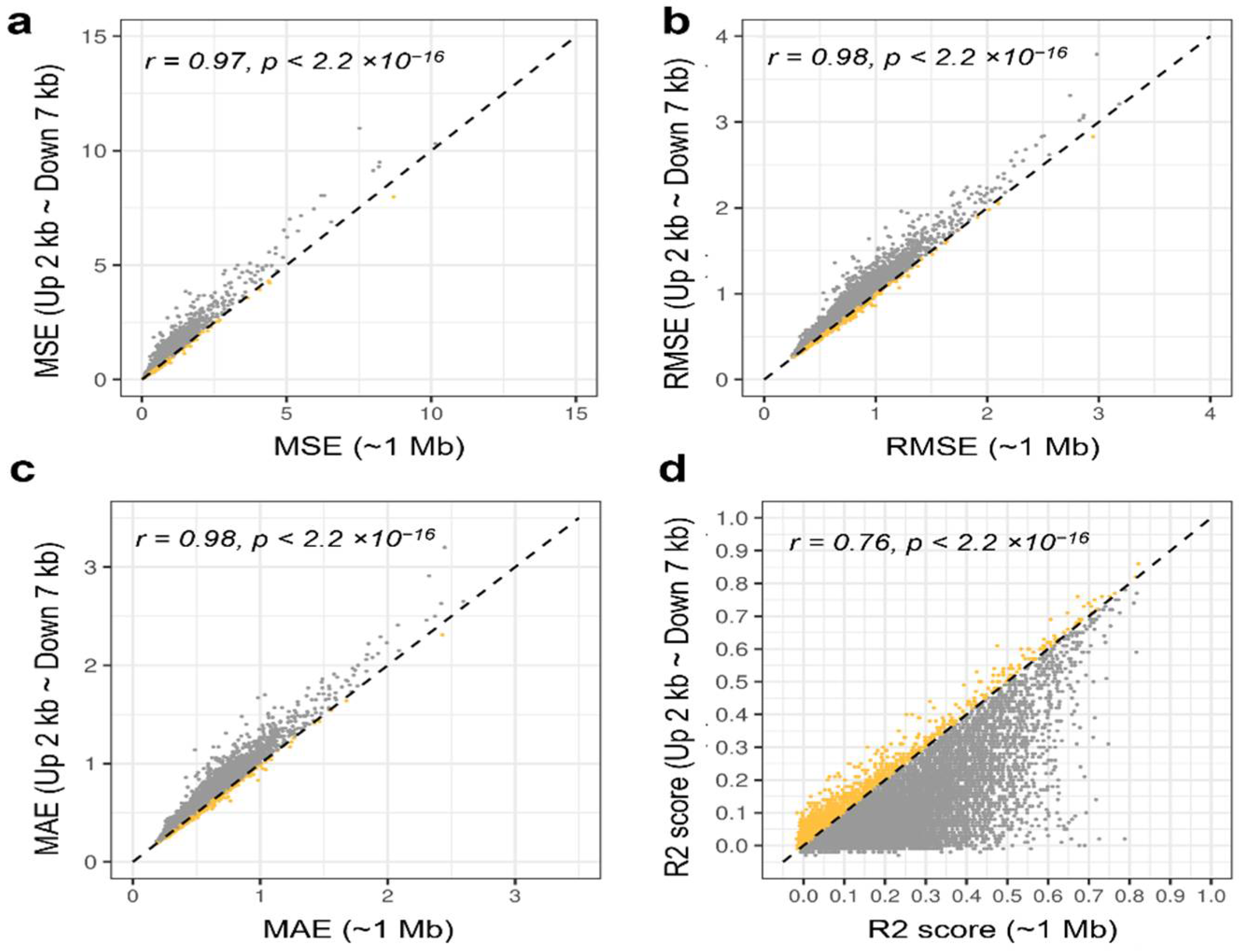
3.3. Prediction with Probes between 2 kb Upstream and 7 kb Downstream of TSS
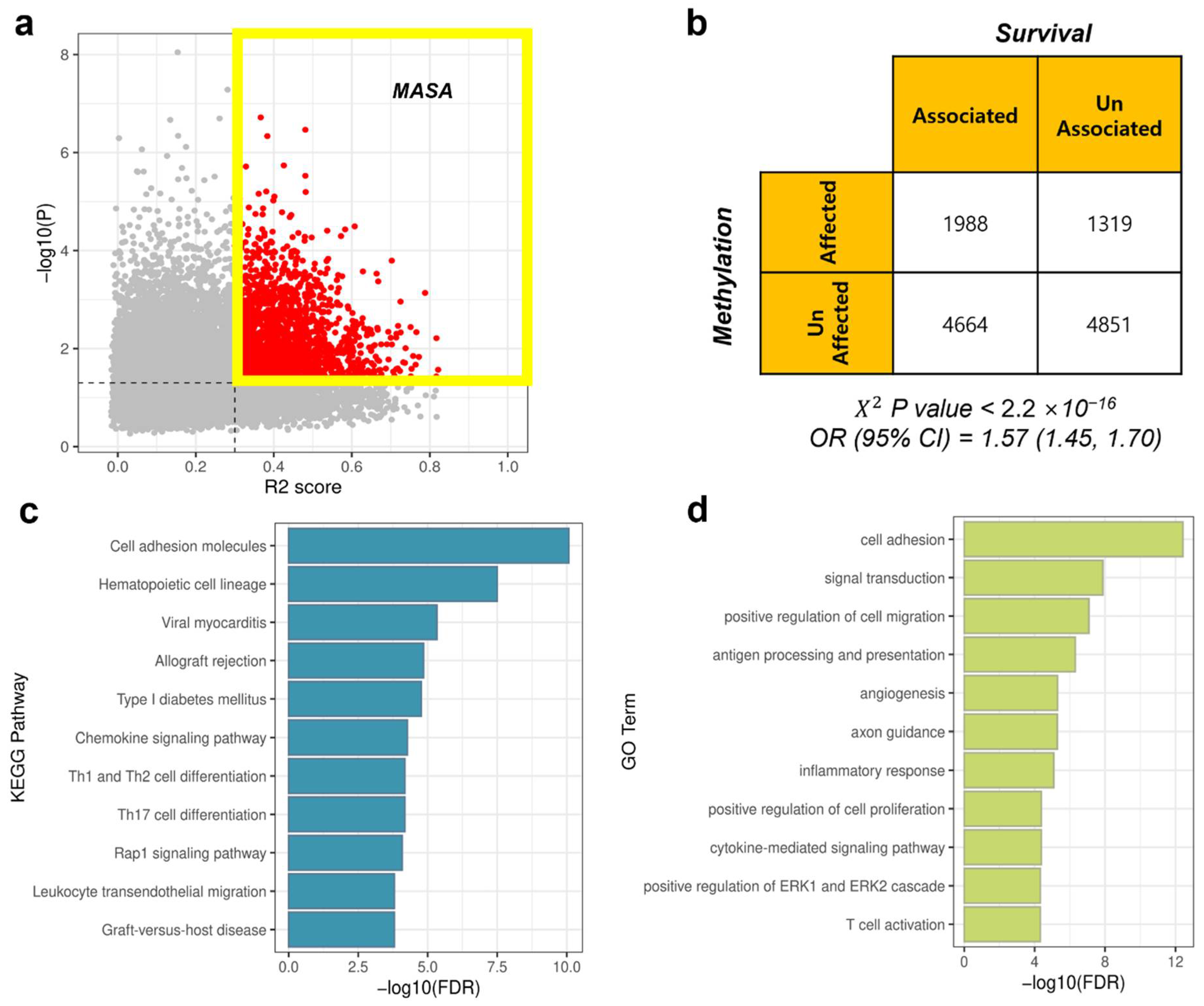
3.4. MASA Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Tapial, S.; Olmedillas-López, S.; Rueda, D.; Arriba, M.; García, J.L.; Vivas, A.; Pérez, J.; Pena-Couso, L.; Olivera, R.; Rodríguez, Y.; et al. Cimp-Positive Status is More Representative in Multiple Colorectal Cancers than in Unique Primary Colorectal Cancers. Sci. Rep. 2019, 9, 10516. [Google Scholar] [CrossRef]
- Ogino, S.; Goel, A. Molecular classification and correlates in colorectal cancer. J. Mol. Diagn. 2008, 10, 13–27. [Google Scholar] [CrossRef]
- Bae, J.M.; Kim, J.H.; Kang, G.H. Molecular Subtypes of Colorectal Cancer and Their Clinicopathologic Features, With an Emphasis on the Serrated Neoplasia Pathway. Arch. Pathol. Lab. Med. 2016, 140, 406–412. [Google Scholar] [CrossRef]
- Nguyen, H.T.; Duong, H.Q. The molecular characteristics of colorectal cancer: Implications for diagnosis and therapy (Review). Oncol. Lett. 2018, 16, 9–18. [Google Scholar] [CrossRef]
- Advani, S.M.; Advani, P.S.; Brown, D.W.; DeSantis, S.M.; Korphaisarn, K.; VonVille, H.M.; Bressler, J.; Lopez, D.S.; Davis, J.S.; Daniel, C.R.; et al. Global differences in the prevalence of the CpG island methylator phenotype of colorectal cancer. BMC Cancer 2019, 19, 964. [Google Scholar] [CrossRef]
- van Rijnsoever, M.; Grieu, F.; Elsaleh, H.; Joseph, D.; Iacopetta, B. Characterisation of colorectal cancers showing hypermethylation at multiple CpG islands. Gut 2002, 51, 797–802. [Google Scholar] [CrossRef]
- Van Rijnsoever, M.; Elsaleh, H.; Joseph, D.; McCaul, K.; Iacopetta, B. CpG island methylator phenotype is an independent predictor of survival benefit from 5-fluorouracil in stage III colorectal cancer. Clin. Cancer Res. 2003, 9, 2898–2903. [Google Scholar]
- Toyota, M.; Ohe-Toyota, M.; Ahuja, N.; Issa, J.P. Distinct genetic profiles in colorectal tumors with or without the CpG island methylator phenotype. Proc. Natl. Acad. Sci. USA 2000, 97, 710–715. [Google Scholar] [CrossRef]
- Moore, L.D.; Le, T.; Fan, G. DNA Methylation and Its Basic Function. Neuropsychopharmacology 2013, 38, 23–38. [Google Scholar] [CrossRef]
- Ehrlich, M. DNA methylation in cancer: Too much, but also too little. Oncogene 2002, 21, 5400–5413. [Google Scholar] [CrossRef]
- Yakoob, J.; Fan, X.G.; Hu, G.L.; Zhang, Z. DNA methylation and carcinogenesis in digestive neoplasms. World J. Gastroenterol. 1998, 4, 174–177. [Google Scholar] [CrossRef]
- Luczak, M.W.; Jagodziński, P.P. The role of DNA methylation in cancer development. Folia Histochem. Cytobiol. 2006, 44, 143–154. [Google Scholar]
- Levenson, V.V. DNA methylation as a universal biomarker. Expert Rev. Mol. Diagn. 2010, 10, 481–488. [Google Scholar] [CrossRef]
- Locke, W.J.; Guanzon, D.; Ma, C.; Liew, Y.J.; Duesing, K.R.; Fung, K.Y.C.; Ross, J.P. DNA Methylation Cancer Biomarkers: Translation to the Clinic. Front. Genet. 2019, 10, 1150. [Google Scholar] [CrossRef]
- Tan, S.; Gui, W.; Wang, S.; Sun, C.; Xu, X.; Liu, L. A methylation-based prognostic model predicts survival in patients with colorectal cancer. J. Gastrointest. Oncol. 2021, 12, 1590–1600. [Google Scholar] [CrossRef]
- Wang, G.; Wang, F.; Meng, Z.; Wang, N.; Zhou, C.; Zhang, J.; Zhao, L.; Wang, G.; Shan, B. Uncovering potential genes in colorectal cancer based on integrated and DNA methylation analysis in the gene expression omnibus database. BMC Cancer 2022, 22, 138. [Google Scholar] [CrossRef]
- Klett, H.; Balavarca, Y.; Toth, R.; Gigic, B.; Habermann, N.; Scherer, D.; Schrotz-King, P.; Ulrich, A.; Schirmacher, P.; Herpel, E.; et al. Robust prediction of gene regulation in colorectal cancer tissues from DNA methylation profiles. Epigenetics 2018, 13, 386–397. [Google Scholar] [CrossRef]
- Liu, Y.-X.; Li, Q.-Z.; Cao, Y.-N. The effect of key DNA methylation in different regions on gene expression in hepatocellular carcinoma. Mol. Omics 2022, 18, 57–70. [Google Scholar] [CrossRef]
- Choi, J.K. Contrasting chromatin organization of CpG islands and exons in the human genome. Genome Biol. 2010, 11, R70. [Google Scholar] [CrossRef]
- Rechache, N.S.; Wang, Y.; Stevenson, H.S.; Killian, J.K.; Edelman, D.C.; Merino, M.; Zhang, L.; Nilubol, N.; Stratakis, C.A.; Meltzer, P.S.; et al. DNA methylation profiling identifies global methylation differences and markers of adrenocortical tumors. J. Clin. Endocrinol. Metab. 2012, 97, E1004–E1013. [Google Scholar] [CrossRef]
- Song, M.-A.; Tiirikainen, M.; Kwee, S.; Okimoto, G.; Yu, H.; Wong, L.L. Elucidating the Landscape of Aberrant DNA Methylation in Hepatocellular Carcinoma. PLoS ONE 2013, 8, e55761. [Google Scholar] [CrossRef]
- Li, S.; Zhang, J.; Huang, S.; He, X. Genome-wide analysis reveals that exon methylation facilitates its selective usage in the human transcriptome. Brief. Bioinform. 2018, 19, 754–764. [Google Scholar] [CrossRef]
- Anastasiadi, D.; Esteve-Codina, A.; Piferrer, F. Consistent inverse correlation between DNA methylation of the first intron and gene expression across tissues and species. Epigenet. Chromatin 2018, 11, 37. [Google Scholar] [CrossRef]
- Brenet, F.; Moh, M.; Funk, P.; Feierstein, E.; Viale, A.J.; Socci, N.D.; Scandura, J.M. DNA Methylation of the First Exon Is Tightly Linked to Transcriptional Silencing. PLoS ONE 2011, 6, e14524. [Google Scholar] [CrossRef]
- Kerachian, M.A.; Javadmanesh, A.; Azghandi, M.; Mojtabanezhad Shariatpanahi, A.; Yassi, M.; Shams Davodly, E.; Talebi, A.; Khadangi, F.; Soltani, G.; Hayatbakhsh, A.; et al. Crosstalk between DNA methylation and gene expression in colorectal cancer, a potential plasma biomarker for tracing this tumor. Sci. Rep. 2020, 10, 2813. [Google Scholar] [CrossRef]
- Kim, S.; Park, H.J.; Cui, X.; Zhi, D. Collective effects of long-range DNA methylations predict gene expressions and estimate phenotypes in cancer. Sci. Rep. 2020, 10, 3920. [Google Scholar] [CrossRef]
- Weinstein, J.N.; Collisson, E.A.; Mills, G.B.; Shaw, K.R.; Ozenberger, B.A.; Ellrott, K.; Shmulevich, I.; Sander, C.; Stuart, J.M. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 2013, 45, 1113–1120. [Google Scholar] [CrossRef]
- Zhou, X.; Carbonetto, P.; Stephens, M. Polygenic modeling with bayesian sparse linear mixed models. PLoS Genet. 2013, 9, e1003264. [Google Scholar] [CrossRef]
- Waldmann, P.; Mészáros, G.; Gredler, B.; Fürst, C.; Sölkner, J. Evaluation of the lasso and the elastic net in genome-wide association studies. Front. Genet. 2013, 4, 270. [Google Scholar] [CrossRef]
- Epstein, B.; Abou-Shanab, R.A.I.; Shamseldin, A.; Taylor, M.R.; Guhlin, J.; Burghardt, L.T.; Nelson, M.; Sadowsky, M.J.; Tiffin, P. Genome-Wide Association Analyses in the Model Rhizobium Ensifer meliloti. mSphere 2018, 3, e00386-18. [Google Scholar] [CrossRef]
- Ishida, S.; Kato, K.; Tanaka, M.; Odamaki, T.; Kubo, R.; Mitsuyama, E.; Xiao, J.-z.; Yamaguchi, R.; Uematsu, S.; Imoto, S.; et al. Genome-wide association studies and heritability analysis reveal the involvement of host genetics in the Japanese gut microbiota. Commun. Biol. 2020, 3, 686. [Google Scholar] [CrossRef]
- Bao, M.; Wang, K. Genome-wide association studies using a penalized moving-window regression. Bioinformatics 2017, 33, 3887–3894. [Google Scholar] [CrossRef]
- Li, W.; Feng, J.; Jiang, T. IsoLasso: A LASSO regression approach to RNA-Seq based transcriptome assembly. J. Comput. Biol. 2011, 18, 1693–1707. [Google Scholar] [CrossRef]
- Torang, A.; Gupta, P.; Klinke, D.J., 2nd. An elastic-net logistic regression approach to generate classifiers and gene signatures for types of immune cells and T helper cell subsets. BMC Bioinform. 2019, 20, 433. [Google Scholar] [CrossRef]
- van Hasselt, J.G.C.; Rahman, R.; Hansen, J.; Stern, A.; Shim, J.V.; Xiong, Y.; Pickard, A.; Jayaraman, G.; Hu, B.; Mahajan, M.; et al. Transcriptomic profiling of human cardiac cells predicts protein kinase inhibitor-associated cardiotoxicity. Nat. Commun. 2020, 11, 4809. [Google Scholar] [CrossRef]
- Acharjee, A.; Finkers, H.J.; Visser, R.; Maliepaard, C.A. Comparison of Regularized Regression Methods for ~Omics Data. Metabolomics 2013, 3, 1. [Google Scholar] [CrossRef]
- Joyce, J.B.; Grant, C.W.; Liu, D.; MahmoudianDehkordi, S.; Kaddurah-Daouk, R.; Skime, M.; Biernacka, J.; Frye, M.A.; Mayes, T.; Carmody, T.; et al. Multi-omics driven predictions of response to acute phase combination antidepressant therapy: A machine learning approach with cross-trial replication. Transl. Psychiatry 2021, 11, 513. [Google Scholar] [CrossRef]
- Liu, J.; Liang, G.; Siegmund, K.D.; Lewinger, J.P. Data integration by multi-tuning parameter elastic net regression. BMC Bioinform. 2018, 19, 369. [Google Scholar] [CrossRef]
- Colaprico, A.; Silva, T.C.; Olsen, C.; Garofano, L.; Cava, C.; Garolini, D.; Sabedot, T.S.; Malta, T.M.; Pagnotta, S.M.; Castiglioni, I.; et al. TCGAbiolinks: An R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016, 44, e71. [Google Scholar] [CrossRef]
- Gao, G.F.; Parker, J.S.; Reynolds, S.M.; Silva, T.C.; Wang, L.-B.; Zhou, W.; Akbani, R.; Bailey, M.; Balu, S.; Berman, B.P.; et al. Before and After: Comparison of Legacy and Harmonized TCGA Genomic Data Commons’ Data. Cell Syst. 2019, 9, 24–34.e10. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2012, 29, 15–21. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 2014, 31, 166–169. [Google Scholar] [CrossRef]
- Bibikova, M.; Barnes, B.; Tsan, C.; Ho, V.; Klotzle, B.; Le, J.M.; Delano, D.; Zhang, L.; Schroth, G.P.; Gunderson, K.L.; et al. High density DNA methylation array with single CpG site resolution. Genomics 2011, 98, 288–295. [Google Scholar] [CrossRef]
- Friedman, J.; Hastie, T.; Tibshirani, R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J. Stat. Softw. 2010, 33, 1–22. [Google Scholar] [CrossRef]
- Zeng, P.; Zhou, X.; Huang, S. Prediction of gene expression with cis-SNPs using mixed models and regularization methods. BMC Genom. 2017, 18, 368. [Google Scholar] [CrossRef]
- Jierula, A.; Wang, S.; OH, T.-M.; Wang, P. Study on Accuracy Metrics for Evaluating the Predictions of Damage Locations in Deep Piles Using Artificial Neural Networks with Acoustic Emission Data. Appl. Sci. 2021, 11, 2314. [Google Scholar] [CrossRef]
- Hyndman, R.J.; Koehler, A.B. Another look at measures of forecast accuracy. Int. J. Forecast. 2006, 22, 679–688. [Google Scholar] [CrossRef]
- Barretina, J.; Caponigro, G.; Stransky, N.; Venkatesan, K.; Margolin, A.A.; Kim, S.; Wilson, C.J.; Lehár, J.; Kryukov, G.V.; Sonkin, D.; et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012, 483, 603–607. [Google Scholar] [CrossRef]
- Iorio, F.; Knijnenburg, T.A.; Vis, D.J.; Bignell, G.R.; Menden, M.P.; Schubert, M.; Aben, N.; Gonçalves, E.; Barthorpe, S.; Lightfoot, H.; et al. A Landscape of Pharmacogenomic Interactions in Cancer. Cell 2016, 166, 740–754. [Google Scholar] [CrossRef]
- Kim, E.; Jung, S.; Park, W.S.; Lee, J.-H.; Shin, R.; Heo, S.C.; Choe, E.K.; Lee, J.H.; Kim, K.; Chai, Y.J. Upregulation of SLC2A3 gene and prognosis in colorectal carcinoma: Analysis of TCGA data. BMC Cancer 2019, 19, 302. [Google Scholar] [CrossRef]
- Liu, L.; Xu, S.; Huang, L.; He, J.; Liu, G.; Ma, S.; Weng, Y.; Huang, S. Systemic immune microenvironment and regulatory network analysis in patients with lung adenocarcinoma. Transl. Cancer Res. 2021, 10, 2859–2872. [Google Scholar] [CrossRef]
- Hur, J.Y.; Lee, H.Y.; Chang, H.J.; Choi, C.W.; Kim, D.H.; Eo, W.K. Preoperative plateletcrit is a Prognostic Biomarker for Survival in Patients with Non-Small Cell Lung Cancer. J. Cancer 2020, 11, 2800–2807. [Google Scholar] [CrossRef]
- Moreaux, J.; Reme, T.; Leonard, W.; Veyrune, J.L.; Requirand, G.; Goldschmidt, H.; Hose, D.; Klein, B. Gene expression-based prediction of myeloma cell sensitivity to histone deacetylase inhibitors. Br. J. Cancer 2013, 109, 676–685. [Google Scholar] [CrossRef]
- Dennis, G., Jr.; Sherman, B.T.; Hosack, D.A.; Yang, J.; Gao, W.; Lane, H.C.; Lempicki, R.A. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003, 4, P3. [Google Scholar] [CrossRef]
- Lee, B.K.; Bhinge, A.A.; Battenhouse, A.; McDaniell, R.M.; Liu, Z.; Song, L.; Ni, Y.; Birney, E.; Lieb, J.D.; Furey, T.S.; et al. Cell-type specific and combinatorial usage of diverse transcription factors revealed by genome-wide binding studies in multiple human cells. Genome Res. 2012, 22, 9–24. [Google Scholar] [CrossRef]
- Harjunpää, H.; Llort Asens, M.; Guenther, C.; Fagerholm, S.C. Cell Adhesion Molecules and Their Roles and Regulation in the Immune and Tumor Microenvironment. Front. Immunol. 2019, 10, 1078. [Google Scholar] [CrossRef]
- Iwasaki, H.; Akashi, K. Hematopoietic developmental pathways: On cellular basis. Oncogene 2007, 26, 6687–6696. [Google Scholar] [CrossRef]
- Nishida, N.; Yano, H.; Nishida, T.; Kamura, T.; Kojiro, M. Angiogenesis in cancer. Vasc. Health Risk Manag. 2006, 2, 213–219. [Google Scholar] [CrossRef]
- Tonini, T.; Rossi, F.; Claudio, P.P. Molecular basis of angiogenesis and cancer. Oncogene 2003, 22, 6549–6556. [Google Scholar] [CrossRef]
- Vekic, B.; Dragojevic-Simic, V.; Jakovljevic, M.; Kalezic, M.; Zagorac, Z.; Dragovic, S.; Zivic, R.; Pilipovic, F.; Simic, R.; Jovanovic, D.; et al. A Correlation Study of the Colorectal Cancer Statistics and Economic Indicators in Selected Balkan Countries. Front. Public Health 2020, 8, 29. [Google Scholar] [CrossRef]
- Bardhan, K.; Liu, K. Epigenetics and colorectal cancer pathogenesis. Cancers 2013, 5, 676–713. [Google Scholar] [CrossRef]
- Patnaik, S.; Anupriya. Drugs Targeting Epigenetic Modifications and Plausible Therapeutic Strategies Against Colorectal Cancer. Front. Pharmacol. 2019, 10, 588. [Google Scholar] [CrossRef]
- Miranda, E.; Destro, A.; Malesci, A.; Balladore, E.; Bianchi, P.; Baryshnikova, E.; Franchi, G.; Morenghi, E.; Laghi, L.; Gennari, L.; et al. Genetic and epigenetic changes in primary metastatic and nonmetastatic colorectal cancer. Br. J. Cancer 2006, 95, 1101–1107. [Google Scholar] [CrossRef]
- Huang, H.; Fu, J.; Zhang, L.; Xu, J.; Li, D.; Onwuka, J.U.; Zhang, D.; Zhao, L.; Sun, S.; Zhu, L.; et al. Integrative Analysis of Identifying Methylation-Driven Genes Signature Predicts Prognosis in Colorectal Carcinoma. Front. Oncol. 2021, 11, 629860. [Google Scholar] [CrossRef]
- Cooper, S.J.; Trinklein, N.D.; Anton, E.D.; Nguyen, L.; Myers, R.M. Comprehensive analysis of transcriptional promoter structure and function in 1% of the human genome. Genome Res. 2006, 16, 1–10. [Google Scholar] [CrossRef]
- Aman Beshir, J.; Kebede, M. In silico analysis of promoter regions and regulatory elements (motifs and CpG islands) of the genes encoding for alcohol production in Saccharomyces cerevisiaea S288C and Schizosaccharomyces pombe 972h. J. Genet. Eng. Biotechnol. 2021, 19, 8. [Google Scholar] [CrossRef]
- Rao, X.; Evans, J.; Chae, H.; Pilrose, J.; Kim, S.; Yan, P.; Huang, R.L.; Lai, H.C.; Lin, H.; Liu, Y.; et al. CpG island shore methylation regulates caveolin-1 expression in breast cancer. Oncogene 2013, 32, 4519–4528. [Google Scholar] [CrossRef]
- Chae, H.; Lee, S.; Nephew, K.P.; Kim, S. Subtype-specific CpG island shore methylation and mutation patterns in 30 breast cancer cell lines. BMC Syst. Biol. 2016, 10, 116. [Google Scholar] [CrossRef]
- Irizarry, R.A.; Ladd-Acosta, C.; Wen, B.; Wu, Z.; Montano, C.; Onyango, P.; Cui, H.; Gabo, K.; Rongione, M.; Webster, M.; et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat. Genet. 2009, 41, 178–186. [Google Scholar] [CrossRef]
- Xu, W.; Xu, M.; Wang, L.; Zhou, W.; Xiang, R.; Shi, Y.; Zhang, Y.; Piao, Y. Integrative analysis of DNA methylation and gene expression identified cervical cancer-specific diagnostic biomarkers. Signal. Transduct. Target. Ther. 2019, 4, 55. [Google Scholar] [CrossRef] [PubMed]
- Jin, W.; Li, Q.Z.; Zuo, Y.C.; Cao, Y.N.; Zhang, L.Q.; Hou, R.; Su, W.X. Relationship Between DNA Methylation in Key Region and the Differential Expressions of Genes in Human Breast Tumor Tissue. DNA Cell Biol. 2019, 38, 49–62. [Google Scholar] [CrossRef] [PubMed]
- Mortezaee, K. Immune escape: A critical hallmark in solid tumors. Life Sci. 2020, 258, 118110. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Strausberg, R.L. Tumor microenvironments, the immune system and cancer survival. Genome Biol. 2005, 6, 211. [Google Scholar] [CrossRef][Green Version]
- Galli, F.; Aguilera, J.V.; Palermo, B.; Markovic, S.N.; Nisticò, P.; Signore, A. Relevance of immune cell and tumor microenvironment imaging in the new era of immunotherapy. J. Exp. Clin. Cancer Res. 2020, 39, 89. [Google Scholar] [CrossRef]
- Neagu, M.; Constantin, C. Signal Transduction in Immune Cells and Protein Kinases. In Protein Kinase-Mediated Decisions between Life and Death; Engin, A.B., Engin, A., Eds.; Springer International Publishing: Cham, Switzerland, 2021; pp. 133–149. [Google Scholar] [CrossRef]
- Kotsias, F.; Cebrian, I.; Alloatti, A. Antigen processing and presentation. Int. Rev. Cell Mol. Biol. 2019, 348, 69–121. [Google Scholar] [CrossRef]
- Lugano, R.; Ramachandran, M.; Dimberg, A. Tumor angiogenesis: Causes, consequences, challenges and opportunities. Cell. Mol. Life Sci. 2020, 77, 1745–1770. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hong, J.; Rhee, J.-K. Genomic Effect of DNA Methylation on Gene Expression in Colorectal Cancer. Biology 2022, 11, 1388. https://doi.org/10.3390/biology11101388
Hong J, Rhee J-K. Genomic Effect of DNA Methylation on Gene Expression in Colorectal Cancer. Biology. 2022; 11(10):1388. https://doi.org/10.3390/biology11101388
Chicago/Turabian StyleHong, Juyeon, and Je-Keun Rhee. 2022. "Genomic Effect of DNA Methylation on Gene Expression in Colorectal Cancer" Biology 11, no. 10: 1388. https://doi.org/10.3390/biology11101388
APA StyleHong, J., & Rhee, J.-K. (2022). Genomic Effect of DNA Methylation on Gene Expression in Colorectal Cancer. Biology, 11(10), 1388. https://doi.org/10.3390/biology11101388




