Mass Purification Protocol for Drosophila melanogaster Wing Imaginal Discs: An Alternative to Dissection to Obtain Large Numbers of Disc Cells
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Fly Stocks
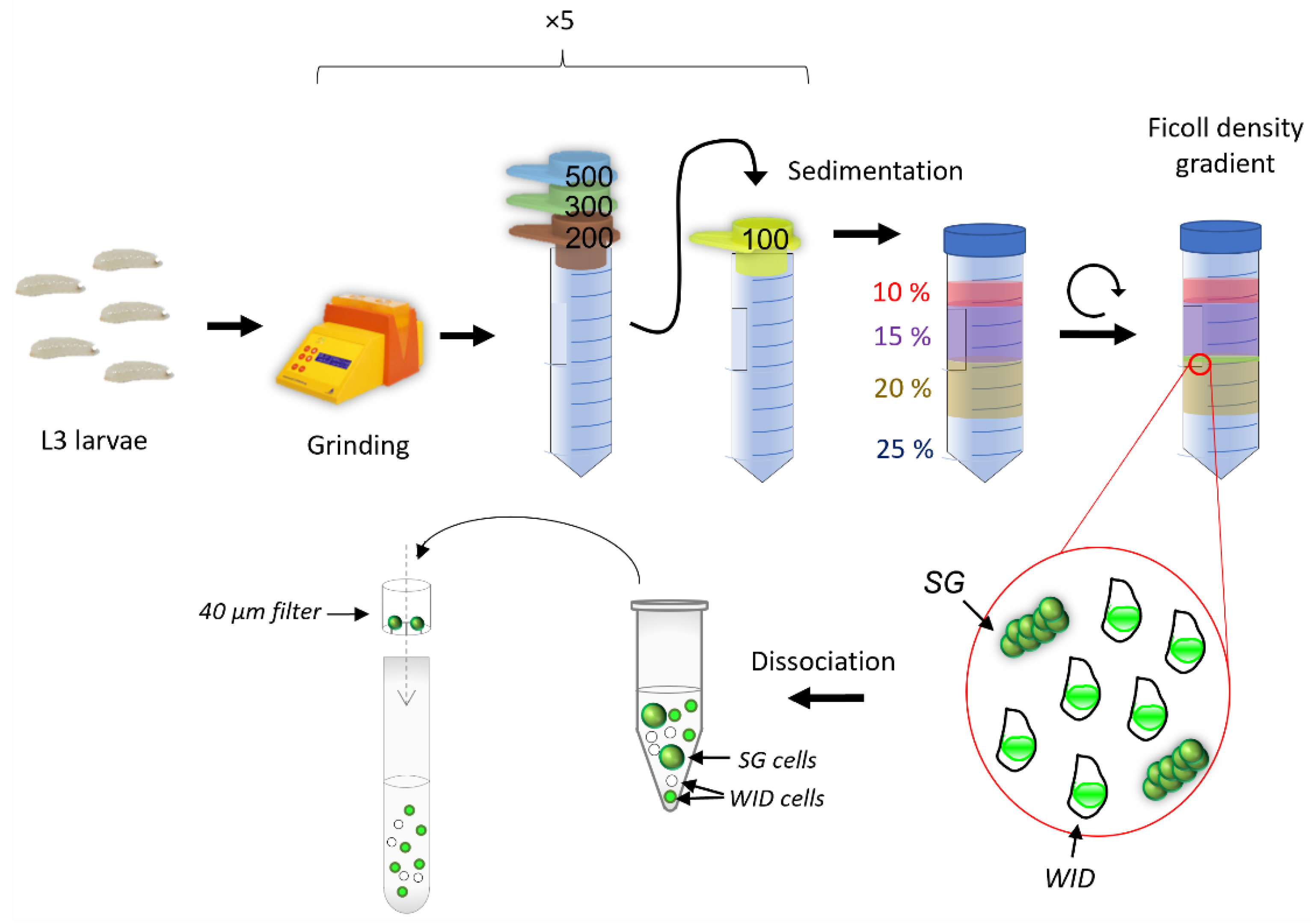
2.2. Protocol for Mass Enrichment of Wing Imaginal Discs
2.3. Immunostaining and Images Acquisition
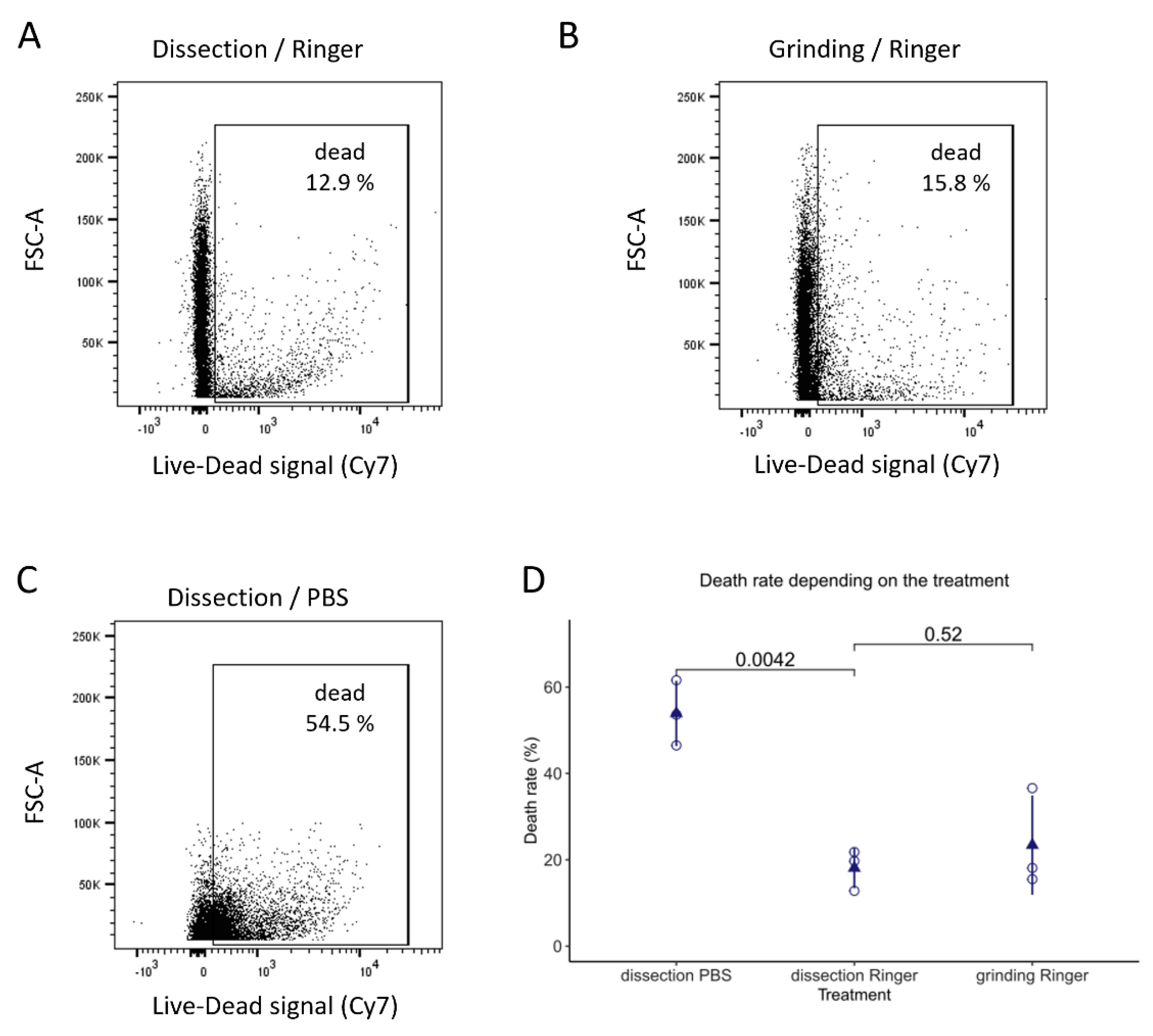
2.4. Flow Cytometry Analysis
3. Results
3.1. Grinding and Buffer Parameters to Release Internal Organs without Damaging Them
3.1.1. Grinding Larvae: A Fine-Tuning between Breaking the Cuticle and Keeping the Organs Undamaged
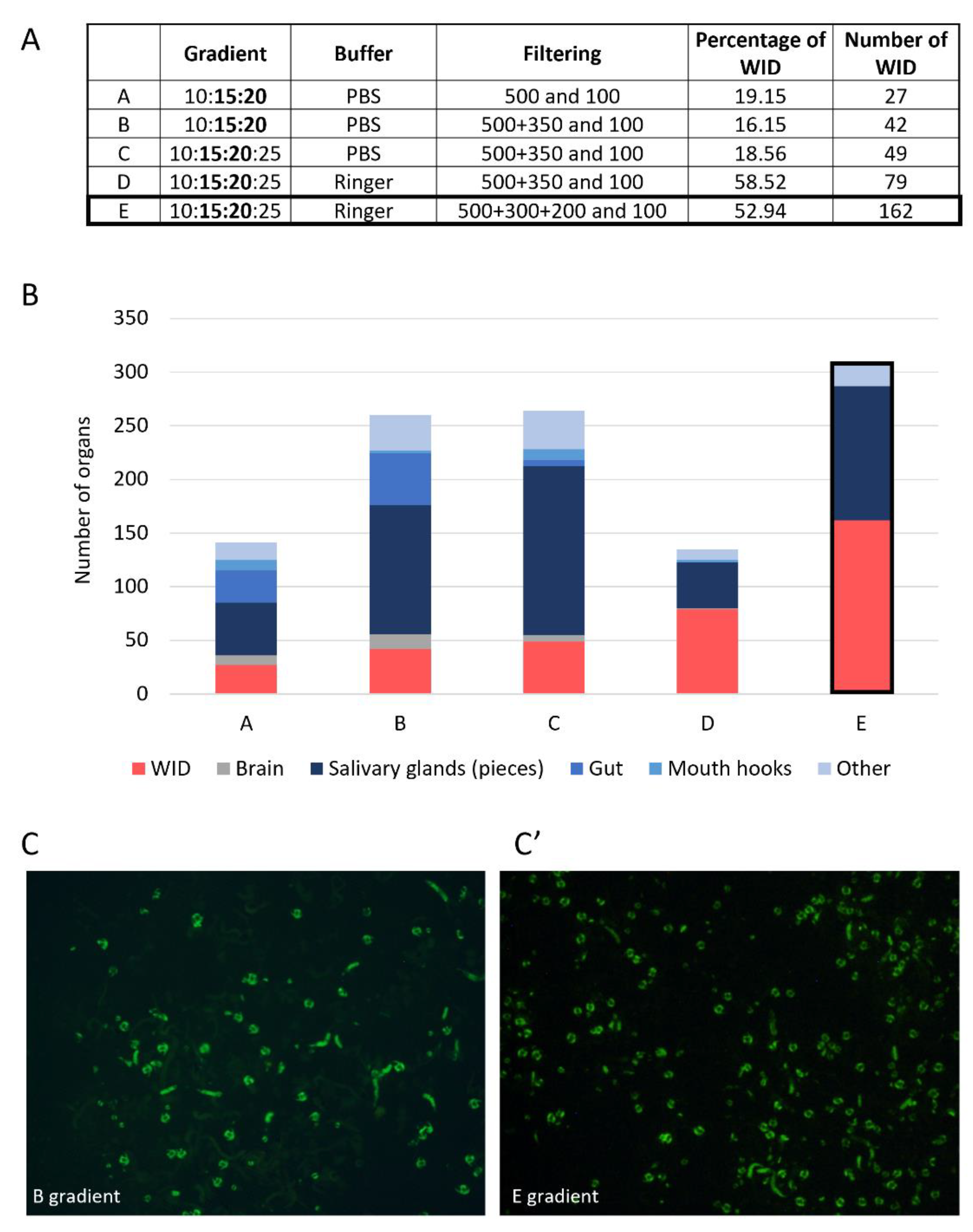
3.1.2. Filtration and Ficoll Gradient: Steps to Separate Organs of Interest from the Others
3.1.3. Finding the Best Buffer for Isolating and Maintaining Intact Imaginal Discs
3.2. Final Protocol (See Supplementary File S1 for Details)
4. Discussion
- (i)
- We used around 600 larvae as an input to obtain, on average, 140 wing imaginal discs. Depending on the researcher, this amount can be dissected in about two to three hours. However, if more wing imaginal discs are needed, the dissection time increases proportionally. One of the advantages of the mass enrichment protocol is that it can process several samples simultaneously by using parallel filtration montages and doing the filtration and centrifugations (sedimentation and Ficoll density gradient) at the same time (Figure 1). Therefore, its duration is only slightly impacted and should not take more than three hours.
- (ii)
- When high amounts of organs are needed, multiple researchers usually dissect them together. As each researcher has their own method and ease of dissection, which may be influenced by the time spent dissecting, this may pose reproducibility problems. The enrichment protocol is standardized and can be performed entirely by one researcher, reducing reproducibility issues, which constitutes another advantage over manual dissection.
- (iii)
- Since dissection allows only one sample to be processed at a time, this induces a delay in the treatment of each condition. Our protocol does not present this disadvantage, as several samples can be treated almost simultaneously. Thus, it allows a more homogenous treatment between conditions.
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brand, A.H.; Perrimon, N. Targeted Gene Expression as a Means of Altering Cell Fates and Generating Dominant Phenotypes. Development 1993, 118, 401–415. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.S.; Luo, L. A Protocol for Mosaic Analysis with a Repressible Cell Marker (MARCM) in Drosophila. Nat. Protoc. 2006, 1, 2583–2589. [Google Scholar] [CrossRef]
- Celniker, S.E.; Rubin, G.M. The Drosophila melanogaster Genome. Annu. Rev. Genom. Hum. Genet. 2003, 4, 89–117. Available online: https://www.annualreviews.org/doi/10.1146/annurev.genom.4.070802.110323 (accessed on 22 July 2022). [CrossRef] [PubMed]
- Beira, J.V.; Paro, R. The Legacy of Drosophila Imaginal Discs. Chromosoma 2016, 125, 573. [Google Scholar] [CrossRef] [PubMed]
- Gou, J.; Stotsky, J.A.; Othmer, H.G. Growth Control in the Drosophila Wing Disk. WIREs Syst. Biol. Med. 2020, 12, e1478. [Google Scholar] [CrossRef]
- Denton, D.; O’Keefe, L.; Kumar, S. Chapter Fifteen—Drosophila as a Model to Understand Autophagy Deregulation in Human Disorders. In Progress in Molecular Biology and Translational Science; Martinez, A.B., Galluzzi, L., Eds.; Autophagy in Health and Disease; Academic Press: Cambridge, MA, USA, 2020; Volume 172, pp. 375–409. [Google Scholar]
- Morata, G.; Calleja, M. Cell Competition and Tumorigenesis in the Imaginal Discs of Drosophila. Semin. Cancer Biol. 2020, 63, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Lewis, E.B. A Gene Complex Controlling Segmentation in Drosophila. Nature 1978, 276, 565–570. [Google Scholar] [CrossRef]
- Justice, R.W.; Zilian, O.; Woods, D.F.; Noll, M.; Bryant, P.J. The Drosophila Tumor Suppressor Gene Warts Encodes a Homolog of Human Myotonic Dystrophy Kinase and Is Required for the Control of Cell Shape and Proliferation. Genes Dev. 1995, 9, 534–546. [Google Scholar] [CrossRef]
- Loubiere, V.; Delest, A.; Schuettengruber, B.; Martinez, A.-M.; Cavalli, G. Chromatin Immunoprecipitation Experiments from Whole Drosophila Embryos or Larval Imaginal Discs. Bio-Protocol 2017, 7, e2327. [Google Scholar] [CrossRef]
- Diaz-Garcia, S.; Ahmed, S.; Baonza, A. Analysis of the Function of Apoptosis during Imaginal Wing Disc Regeneration in Drosophila melanogaster. PLoS ONE 2016, 11, e0165554. [Google Scholar] [CrossRef]
- Clavier, A.; Baillet, A.; Rincheval-Arnold, A.; Coléno-Costes, A.; Lasbleiz, C.; Mignotte, B.; Guénal, I. The Pro-Apoptotic Activity of Drosophila Rbf1 Involves DE2F2-Dependent Downregulation of Diap1 and Buffy MRNA. Cell Death Dis. 2014, 5, e1405. [Google Scholar] [CrossRef] [PubMed]
- Clavier, A.; Rincheval-Arnold, A.; Baillet, A.; Mignotte, B.; Guénal, I. Two Different Specific JNK Activators Are Required to Trigger Apoptosis or Compensatory Proliferation in Response to Rbf1 in Drosophila. Cell Cycle 2016, 15, 283–294. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fristrom, J.W.; Mitchell, H.K. The Preparative Isolation of Imaginal Discs from Larvae of Drosophila melanogaster. J. Cell Biol. 1965, 27, 445–448. [Google Scholar] [CrossRef] [PubMed]
- Zweidler, A.; Cohen, L.H. Large-Scale Isolation and Fractionation of Organs of Drosophila melanogaster Larvae. J. Cell Biol. 1971, 51, 240–248. [Google Scholar] [CrossRef] [PubMed]
- Marty, F.; Rockel-Bauer, C.; Simigdala, N.; Brunner, E.; Basler, K. Large-Scale Imaginal Disc Sorting: A Protocol for “Omics”-Approaches. Methods 2014, 68, 260–264. [Google Scholar] [CrossRef]
- de Noiron, J.; Hoareau, M.; Colin, J.; Guénal, I. Apoptosis Quantification in Tissue: Development of a Semi-Automatic Protocol and Assessment of Critical Steps of Image Processing. Biomolecules 2021, 11, 1523. [Google Scholar] [CrossRef]
- Pfeiffer, B.D.; Ngo, T.-T.B.; Hibbard, K.L.; Murphy, C.; Jenett, A.; Truman, J.W.; Rubin, G.M. Refinement of Tools for Targeted Gene Expression in Drosophila. Genetics 2010, 186, 735–755. [Google Scholar] [CrossRef]
- Hrdlicka, L.; Gibson, M.; Kiger, A.; Micchelli, C.; Schober, M.; Schöck, F.; Perrimon, N. Analysis of Twenty-Four Gal4 Lines in Drosophila melanogaster. Genesis 2002, 34, 51–57. [Google Scholar] [CrossRef]
- Tomizawa, M.; Tsumaki, K.; Sone, M. Characterization of the Activity of β-Galactosidase from Escherichia Coli and Drosophila melanogaster in Fixed and Non-Fixed Drosophila Tissues. Biochim. Open 2016, 3, 1–7. [Google Scholar] [CrossRef]
- Ephrussi, B.; Beadle, G.W. A Technique of Transplantation for Drosophila. Am. Nat. 1936, 70, 218–225. [Google Scholar] [CrossRef]
- Sarkissian, T.; Timmons, A.; Arya, R.; Abdelwahid, E.; White, K. Detecting Apoptosis in Drosophila Tissues and Cells. Methods 2014, 68, 89–96. [Google Scholar] [CrossRef] [PubMed]
- Brun, S.; Rincheval-Arnold, A.; Colin, J.; Risler, Y.; Mignotte, B.; Guénal, I. The Myb-Related Gene Stonewall Induces Both Hyperplasia and Cell Death in Drosophila: Rescue of Fly Lethality by Coexpression of Apoptosis Inducers. Cell Death Differ. 2006, 13, 1752–1762. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Iyer, E.P.R.; Iyer, S.C.; Sulkowski, M.J.; Cox, D.N. Isolation and Purification of Drosophil Peripheral Neurons by Magnetic Bead Sorting. J. Vis. Exp. JoVE 2009, 34, e1599. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hoareau, M.; de Noiron, J.; Colin, J.; Guénal, I. Mass Purification Protocol for Drosophila melanogaster Wing Imaginal Discs: An Alternative to Dissection to Obtain Large Numbers of Disc Cells. Biology 2022, 11, 1384. https://doi.org/10.3390/biology11101384
Hoareau M, de Noiron J, Colin J, Guénal I. Mass Purification Protocol for Drosophila melanogaster Wing Imaginal Discs: An Alternative to Dissection to Obtain Large Numbers of Disc Cells. Biology. 2022; 11(10):1384. https://doi.org/10.3390/biology11101384
Chicago/Turabian StyleHoareau, Marion, Juliette de Noiron, Jessie Colin, and Isabelle Guénal. 2022. "Mass Purification Protocol for Drosophila melanogaster Wing Imaginal Discs: An Alternative to Dissection to Obtain Large Numbers of Disc Cells" Biology 11, no. 10: 1384. https://doi.org/10.3390/biology11101384
APA StyleHoareau, M., de Noiron, J., Colin, J., & Guénal, I. (2022). Mass Purification Protocol for Drosophila melanogaster Wing Imaginal Discs: An Alternative to Dissection to Obtain Large Numbers of Disc Cells. Biology, 11(10), 1384. https://doi.org/10.3390/biology11101384





