1. Introduction
Tomato,
Solanum lycopersicum L. (Solanaceae), is a major crop that is widely used as a model for fruit development [
1]. It is attacked by several pathogens including
Alternaria alternata,
Botrytis cinerea,
Clavibacter michiganensis subsp.
michiganensis,
Phytophthora infestans,
Rhizoctonia solani,
Xanthomonas euvesicatoria pv.
perforans, etc. [
2]. These pathogens are significant limiting factors in the production of tomato crops in greenhouses and cause yield losses, especially at the fruiting stage [
3].
Current methods for managing harmful plant pathogens depend mainly on the extensive use of synthetic chemicals. However, the frequent application of chemicals presents long-term risks to human health and the environment [
4]. There is also evidence that pathogens have developed resistance to chemicals [
5]. To avoid these undesirable consequences, current research efforts are heavily focused on the development of less hazardous practices and/or methods for crop protection.
Biological control using beneficial microorganisms is one of the most environmentally sound and economically viable ways to manage plant diseases. These microorganisms aid plants by promoting growth and suppress phytopathogens due to their antagonistic activity [
6]. In the rhizosphere, plant growth-promoting rhizobacteria (PGPR) are important bacterial groups that respond to soil-borne diseases and function as biocontrol agents [
7]. This activity is largely due to their production of antimicrobial compounds and hydrolytic enzymes, which makes them an excellent, environmentally friendly, and sustainable alternative to indiscriminate chemical treatment for controlling plant pathogens. The development of reliable microbial treatments to promote plant growth requires, however, the identification of new beneficial bacterial strains with high antagonistic activity towards phytopathogens. Several mechanisms have been suggested to explain the antagonistic effects of beneficial microorganisms on phytopathogens, including competition for nutrients, minerals, and colonization sites [
8], inhibition of the pathogens via secreted toxins, antibiotics and biosurfactants [
9], and parasitism based on the production of hydrolytic enzymes that degrade pathogens’ cell walls [
10].
Pseudomonas spp. (Pseudomonadaceae) is one of the most abundant genera of beneficial rhizobacteria among the large and heterogeneous bacterial populations in the rhizosphere and has, therefore, attracted growing attention as a source of potential biological control agents [
11,
12]. Except for some human pathogenic strains [
13], the species of
P. aeruginosa is a versatile and ubiquitous bacterium that has been recognized as an active antagonist of several bacterial and fungal plant pathogens, and has potential practical applications in agricultural systems [
14]. This species can produce secondary metabolites such as indole acetic acid (IAA) and siderophores, and can also solubilize phosphate [
15]. Research on its interactions with plant-infecting fungi such as
Alternaria,
Rhizoctonia, and
Sclerotium and oomycetes such as
Pythium and
Phytophthora have demonstrated that its production of phenazines plays an important role in controlling these pathogens [
16]. In addition,
P. aeruginosa isolates have shown antagonistic activity against
Pythium myriotylum and
Phytophthora capsici in black pepper [
17],
Sclerotium rolfsii in cucumber [
18],
Xanthomonas sp. infecting various crops [
19],
P. myriotylum in ginger [
20,
21],
Colletotrichum gloeosporioides in chili [
22],
Ralstonia solanacearum in tomato [
23], and
Fusarium oxysporum in cotton [
24] and wheat [
25].
The present study was conducted to shed light on the plant growth-promoting activity of the recently isolated P. aeruginosa strain FG106 and its antagonistic activity against A. alternata, R. solani, X. euvesicatoria pv. perforans, C. michiganensis subsp. michianensis, P. infestans, P. colocasiae, and B. cinerea. The specific objectives of this work were to (1) determine the effects of the FG106 strain on growth and development of the above-mentioned plant pathogens, (2) evaluate the biochemical and enzymatic activities of P. aeruginosa FG106, (3) investigate the strain’s effects on tomato germination and growth, (4) characterize the interactions between FG106 and selected bacterial and fungal and oomycete pathogens and diseases under greenhouse conditions, and (5) sequence the strain’s genome to identify the genetic origin of its plant growth-promoting activity and phytopathogen antagonism.
2. Materials and Methods
2.1. Isolation of Biocontrol Bacteria
For isolation of bacteria, root samples were randomly collected from healthy tomato roots grown in different parts of Khorasan Razavi, a province of Iran. Samples were immediately put into a paper bag, transferred to the laboratory, and stored at 4 °C. Roots were washed with tap water, then surface sterilized for 2 min in 70% ethanol and 5 min in 3% sodium hypochlorite solution followed by three washes with sterile water. The water from the third wash was used as a control sample. The roots were crushed and shaken for 20 min on a shaker at 220 rpm, then used to generate a serial dilution up to 10
−5. Pseudomonad strains were isolated using the spread plate technique on King’s B medium agar. After assessment for antagonistic interactions with
R. solani, the isolate showing the strongest antifungal effect, i.e., FG106, was selected for further screening and stored in Luria-Bertani (LB) broth (Duchefa Biochemie, Haarlem, The Netherlands) containing 30% glycerol at −80 °C [
26,
27].
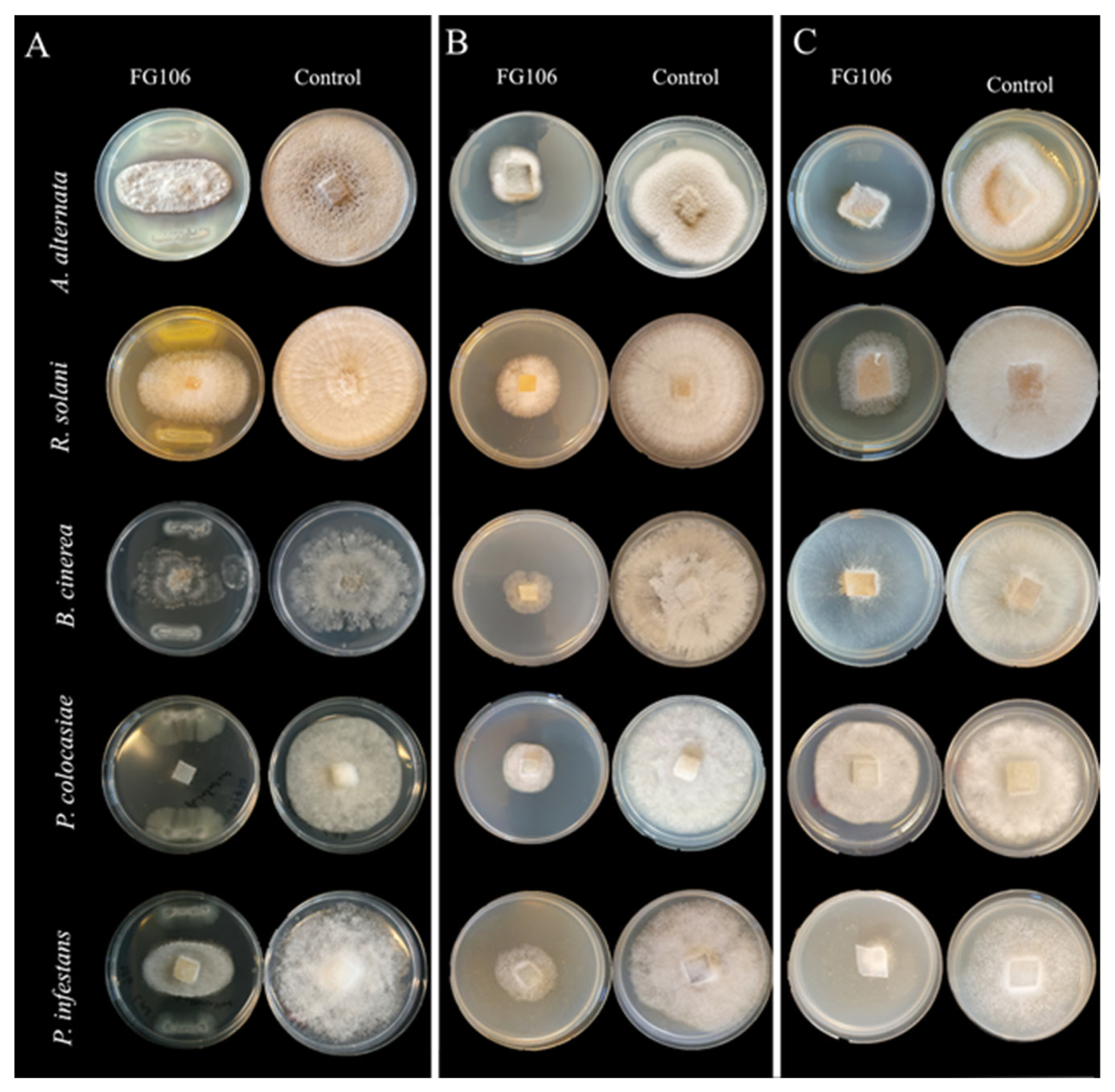
2.2. In Vitro Antagonistic Activity
Strain FG106 was tested against the following plant pathogens to evaluate its antagonistic activity:
A. alternata and
R. solani AG4-HG II,
P. infestans 88069,
P. colocasiae 7290 [
28] and
Botrytis cinerea B05, the Gram-positive bacterium
Clavibacter michiganensis subsp.
michiganensis strain PVCT156.1.1, and the Gram-negative bacterium
Xanthomonas euvesicatoria pv.
perforans strain NCPPB4321. Before dual culture assays, the test pathogen strains were maintained on appropriate media:
R. solani (with a growth period of 7 days) and
A. alternata (14 days) on potato dextrose agar (PDA, Sigma-Aldrich, Germany),
P. infestans on rye agar (14 days), and
P. colocasiae (10 days) and
B. cinerea (7 days) on corn meal agar (CMA, Sigma-Aldrich, Germany).
Dual culture assays were performed in Petri dishes on selective media. The FG106 strain was cultured on either side of the Petri dish, with the initial inoculation being performed 1 cm from the edge of the plate. An agar plug 5 mm in diameter containing the fungal or oomycete pathogen to be tested was then placed in the center of the plate. The resulting plates were incubated at 20 ± 2 °C until the leading edge of the fungus in a control plate containing only the pathogen without FG106 reached the edge of the plate. The antagonistic effect of the bacterial strain on the fungus/oomycete was quantified by computing its inhibition rate as a percentage using below Equation (1) [
27,
29]:
where RI and RC are the minimum distance between the center and the margin of the fungus in the treatment plates and the distance between the center and the margin of the fungus in the control, respectively. This experiment was performed using a completely randomized design with at least 6 replicates per pathogen.
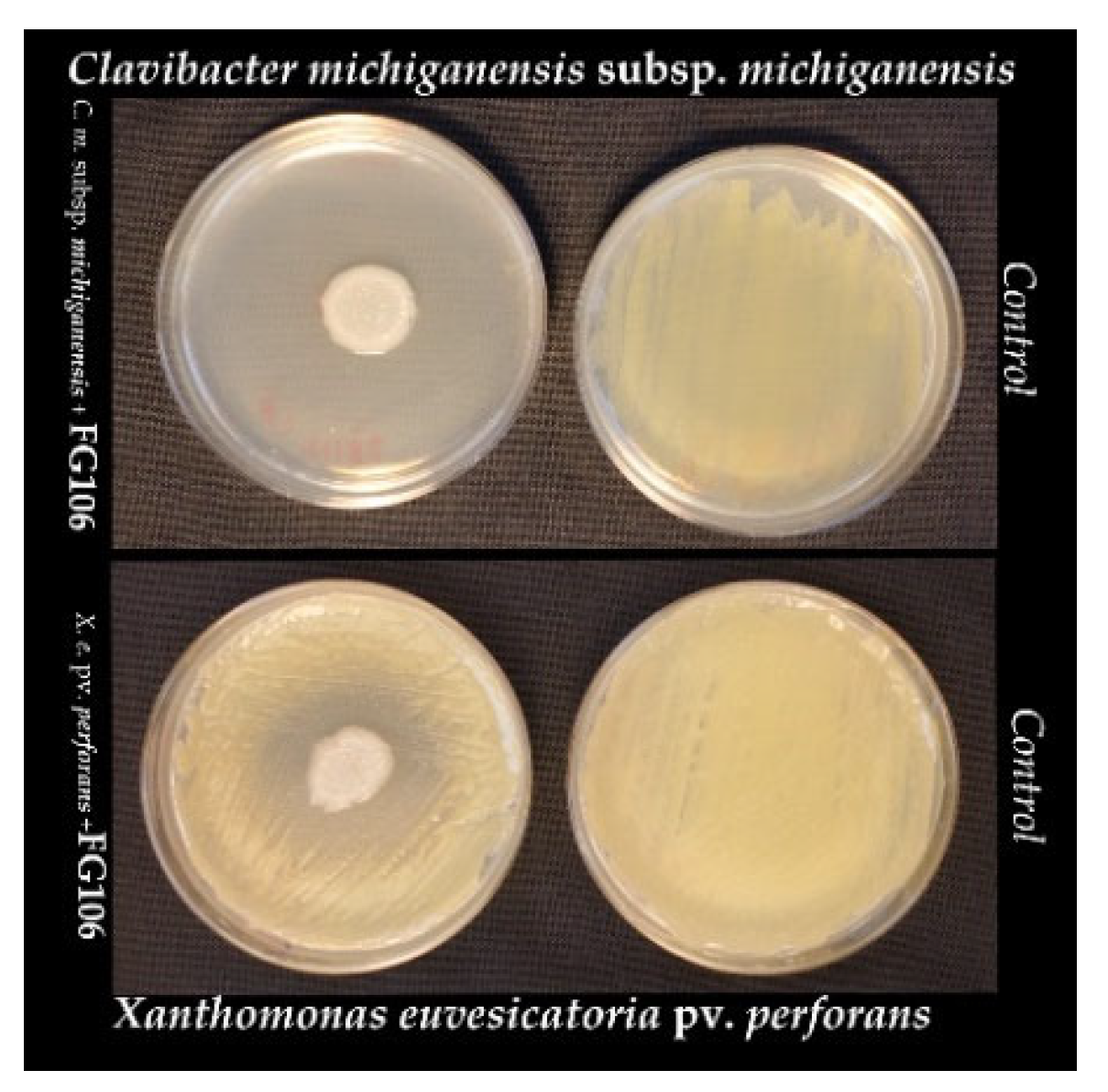
The antagonistic activity of FG106 against the plant pathogenic bacteria
C. m. subsp.
michiganensis and
X. e. pv.
perforans was tested on LB agar plates (6 cm). Suspensions of one pathogenic bacterium in sterile distilled water (OD600 = 0.1) obtained from overnight cultures in nutrient broth (NB) were swabbed uniformly across an LB agar plate, which was then dried and spot-inoculated with an FG106 isolate. The plates were then incubated at 28 °C for 1–5 days. The efficiency of
P. aeruginosa in suppressing the pathogen’s radial growth was determined as described previously [
30]. The antibacterial activity was scored on an arbitrary scale ranging from 0 to 3 based on the scale of growth inhibition around the FG106 colony, where 0 means no halo, 1 refers to halo less than 5 mm in diameter, 2 indicates a 5–10 mm halo, and 3 is a halo larger than 10 mm. Plates inoculated with the pathogens alone were used as positive controls. Three independent replicates were performed for each tested pathogen and control [
31].
2.3. Preliminary Identification of Strain FG106
The DNA of the FG106 strain was isolated using a Quick-DNA Fungal/Bacterial Microprep Kit according to the manufacturer’s recommendations (Zymo Research, Irvine, CA, USA). DNA yield and integrity were measured using a NanoDrop micro photometer (NanoDrop Technologies, South San Francisco, CA, USA) and by agarose gel electrophoresis, respectively. The 16S rRNA gene of FG106 was PCR-amplified using the primer pairs 27F (5′-AGAGTTTGATCMTGGCTCAG-3′) and 907R (5′-CCGTCAATTCMTTTRAGTTT-3′) [
32]. The PCR was performed using 10 ng of FG106 DNA with the following temperature parameters: initial denaturation at 94 °C for 3 min, followed by 35 cycles of 94 °C for 45 s, 50 °C for 30 s, and 72 °C for 30 s, with a final extension step at 72 °C for 5 min. The PCR products were purified using the Qiagen PCR purification kit (Qiagen, UK). The purified PCR products were sequenced for species identification at the Eurofins sequencing facility (Germany). SnapGene software was used (SnapGene, San Diego, CA, USA) to manually analyze and edit the nucleotide sequence obtained from the sequencing platform. The resulting sequence containing the 16S region was searched for matching hits against the National Center for Biotechnology Information (NCBI) GenBank non-redundant nucleotide database (BLASTn) [
33]. Search hits to sequence from records in the database were analyzed for sequence coverage and identity, and the best matched NCBI accession was recorded.
2.4. Production of Volatile and Non-Volatile Metabolites
The antibacterial activity of volatile compounds (VOCs) from the FG106 strain was investigated as described by Nishino [
34]. Briefly, strain FG106 was cultured on a plate with tryptic soy agar (TSA, Sigma-Aldrich, Taufkirchen, Germany) medium, and a 5 mm mycelial plug of one pathogen was separately placed at the center of another plate on specific culture medium. The Petri dishes containing the bacterium and pathogen were then placed face to face, sealed with parafilm, and incubated at 28 °C. A plate containing bacterium-free TSA medium was used as a control.
To investigate the biocontrol effects of non-volatile compounds (NVOCs), the FG106 strain was cultured in tubes containing tryptic soy broth (TSB, Sigma-Aldrich, Taufkirchen, Germany) medium on a shaking incubator (Thermo Scientific, Waltham, MA, USA) for 16 h. The bacterial suspension was then centrifuged at 4200 rpm for 15 min. Two mL of the supernatant was filtered through a 0.22-micron MilliPore (MP) filter, (Sarstedt, Nümbrecht, Germany) supplemented with 18 mL of an appropriate medium in a 9:1 ratio, and placed in a Petri dish. A 5 mm mycelium disk was then placed in the center of the prepared medium. The resulting Petri dishes were maintained at a temperature of 20 °C. The radial growth of the fungal/oomycete colony was measured, and its inhibition percentage was calculated using the previously reported equation [
35,
36]. At least six replicates were performed per tested pathogen.
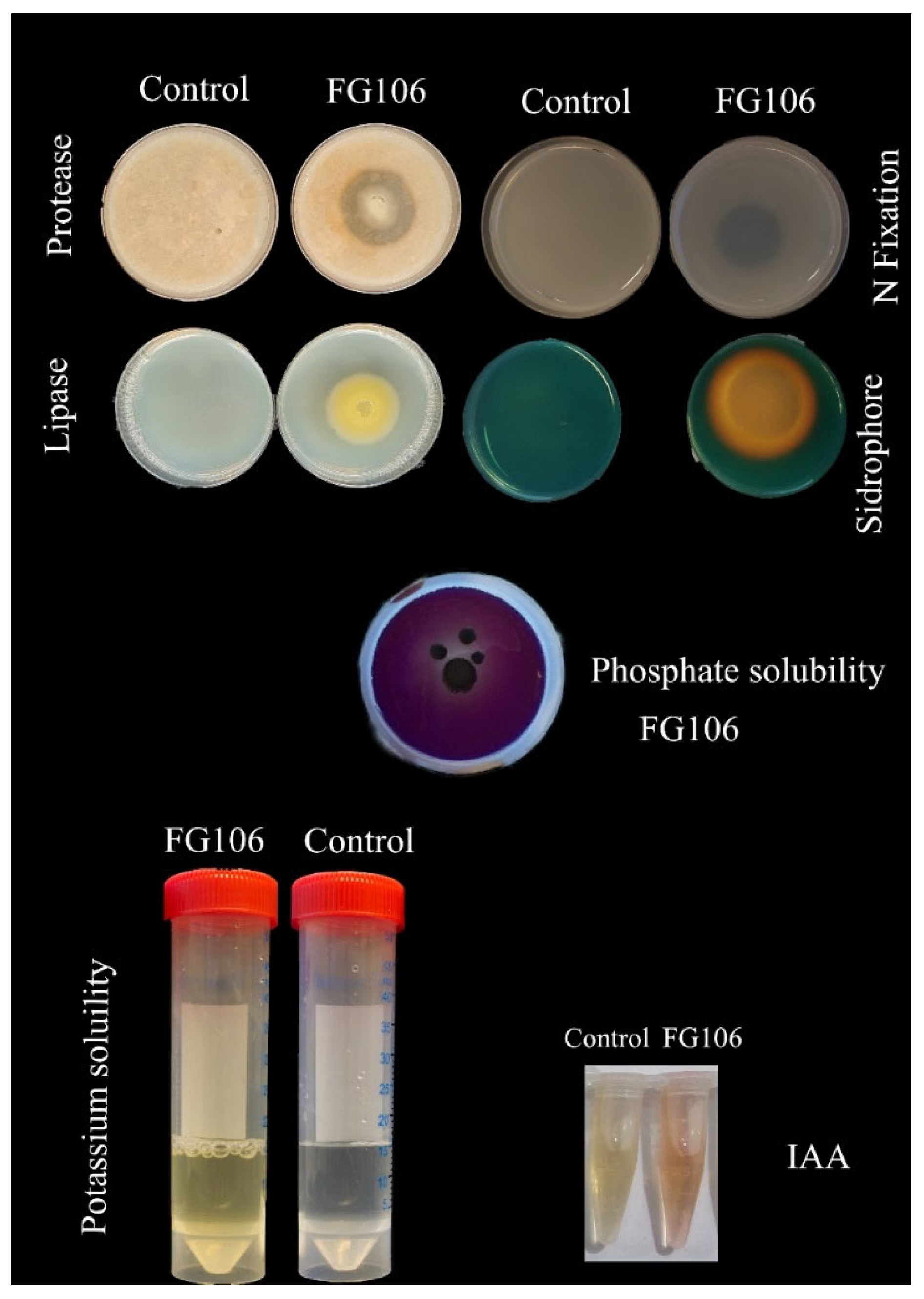
2.5. Evaluation of Hydrolytic Enzyme Activity of FG106
The production of cellulase enzyme activity by the FG106 strain was assessed as previously described [
37,
38], along with protease [
38,
39], chitinase [
40,
41], pectinase [
42], amylase [
27,
43], and lipase production [
44].
2.6. Investigation of Plant Growth Factors
Inorganic phosphate solubilization by strain FG106 was assessed on Pikovskaya agar (Sigma-Aldrich, Germany) with incubation for 7 days at 28 °C [
45], potassium solubilization on Alexandrov medium (Sigma-Aldrich, Taufkirchen, Germany) [
46,
47], and ammonia production on peptone water agar with added Nessler’s reagent (Sigma-Aldrich, Taufkirchen, Germany) [
48]. Medium amended with tryptophan was used to screen for indole acetic acid (IAA) production [
49]. Quantitative determination of IAA was performed using the Acuña method [
50].
2.7. Production of Hydrogen Cyanide (HCN) and Siderophores, and Biofilm Formation
To screen for HCN production, strain FG106 was cultured on glycine-containing King’s B medium that was then pressed against filter paper impregnated with picric acid (Sigma-Aldrich, Taufkirchen, Germany) [
51,
52]. The siderophore assay was performed using Chrome Azurol S medium (Sigma-Aldrich, Taufkirchen, Germany) containing iron chloride [
52,
53]. Biofilm formation was determined in 96 -ell polystyrene microtiter plates (Nunc
™ MicroWell
™ 96-well and flat- bottom microplate, Thermo Fisher Scientific, Waltham, MA, USA) [
54].
2.8. Biosurfactant Production
Sterile distilled water was placed in a Petri dish, and the surface of the dish was completely covered with sunflower oil. Ten microliters of a bacterial supernatant obtained by centrifuging a 10
7 cfu/mL bacterial suspension at 3000 rpm for 10 min was then placed on the oil. It was concluded that the bacterium had the capacity to produce biosurfactants if a clear oil-free zone formed around the bacteria [
55,
56].
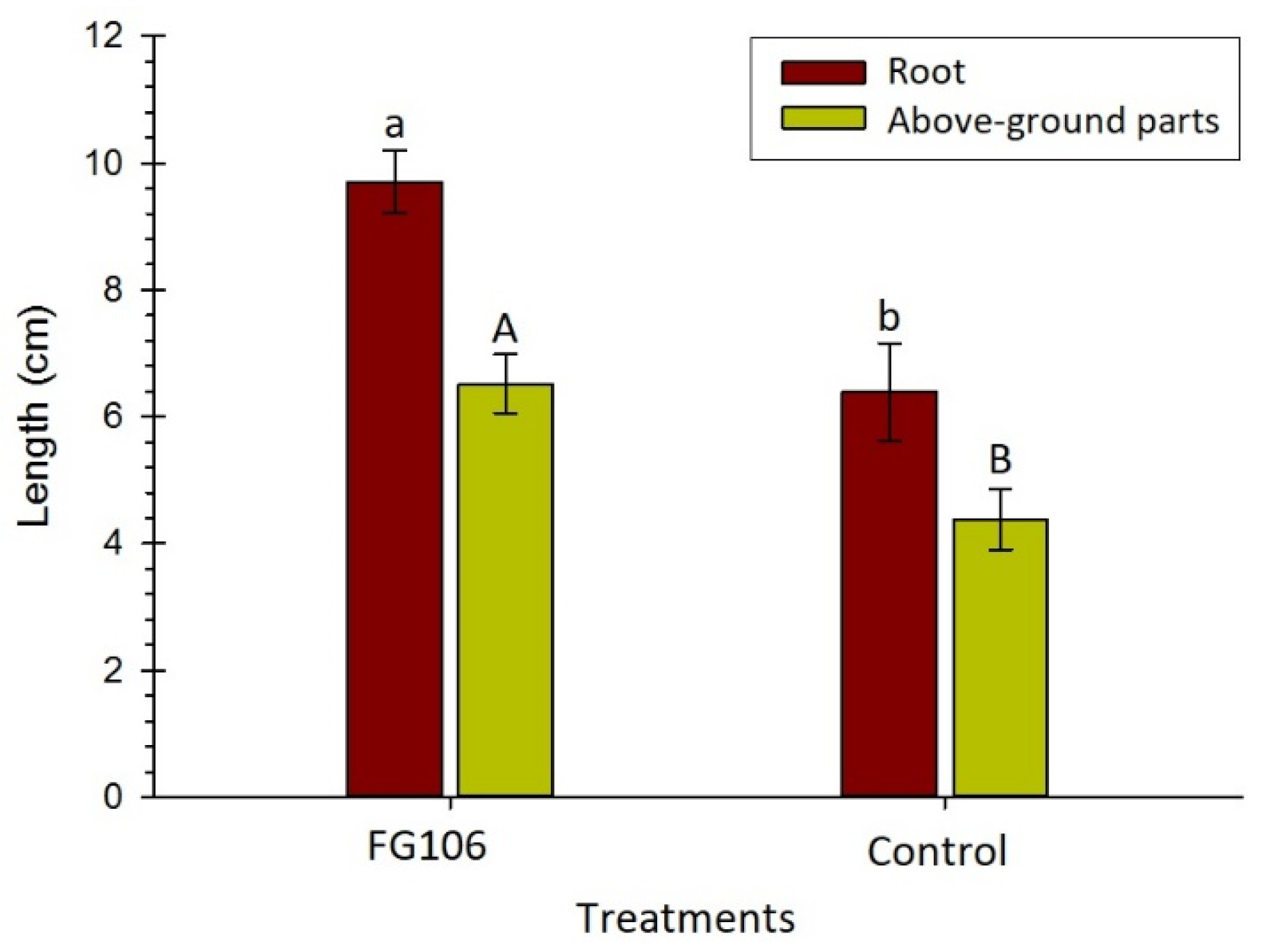
2.9. In Vitro Investigation of the Effect of Strain FG106 on Growth of Tomato Seedlings
Sterile seeds of the tomato cultivar ‘Money Maker’ were soaked in a 2 × 10
7 cfu/mL bacterial suspension for 30 min. After drying, the seeds were transferred to solid 50% Murashige and Skoog (MS) medium (Duchefa Biochemie, Haarlem, The Netherlands). After 14 days, the length of the resulting plants was measured. Control seeds were soaked in 1× phosphate buffered saline (PBS) (PanReac AppliChem, Darmstadt, Germany) [
57].
2.10. Biocontrol Activity Using Detached Leaf Assay (DLA)
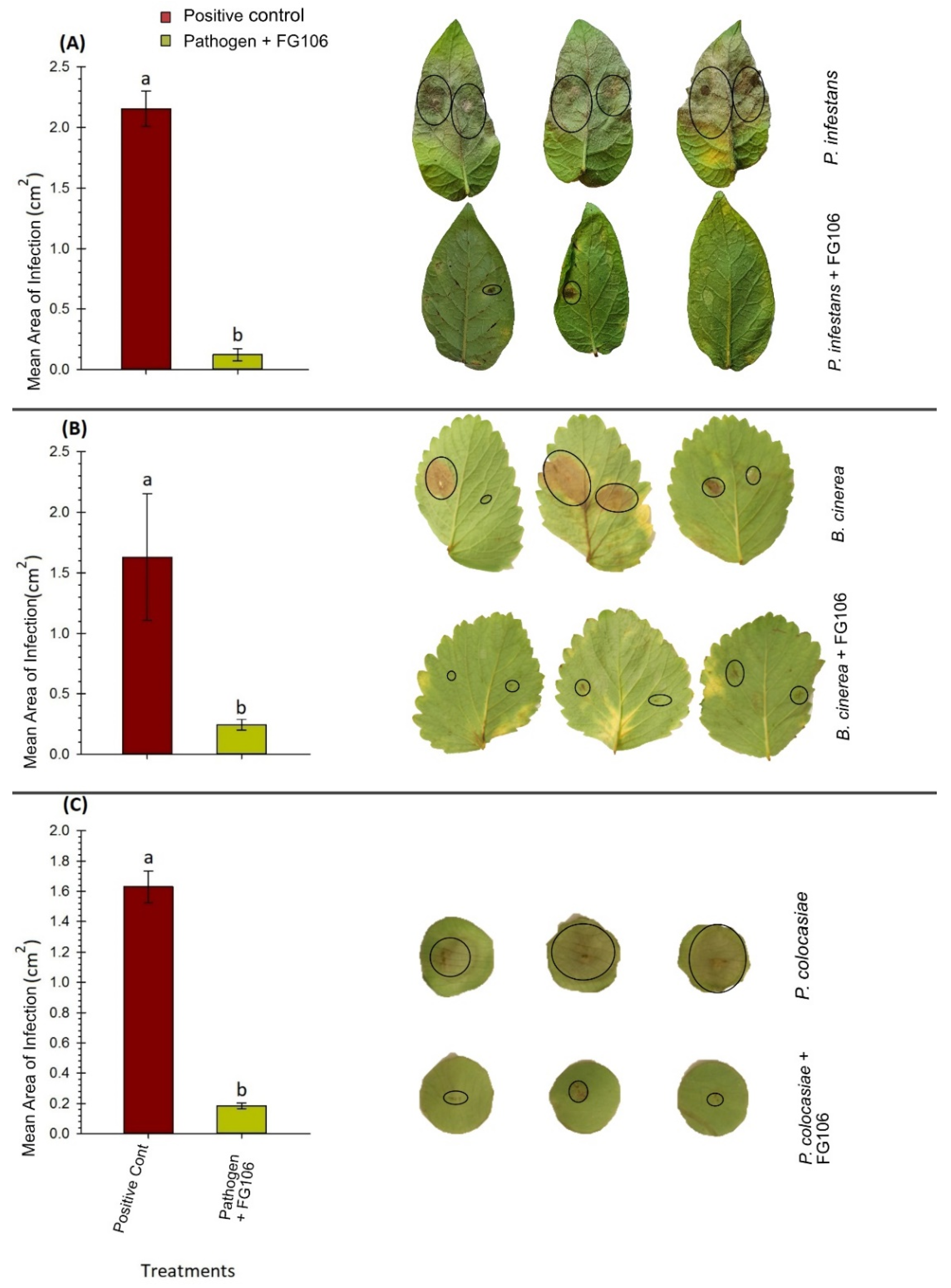
Leaves from 3 to 4 week-old potato and strawberry plants were washed and inoculated with 20 µL of a 2 × 10
7 cfu/mL suspension of strain FG106. After 24 h, 20 μL of a challenge pathogen suspension was added individually; the concentrations of these suspensions were 25,000 sporangia/mL for
P. infestans or 50,000 sporangia/conidia mL for
P. colocasiea or
B. cinerea [
58]. Disease assessment was performed 6 days post-inoculation; the leaf area exhibiting symptoms of infection was measured using the ImageJ software package [
59] and compared to that in control leaves. Control leaves were treated with 20 μL 1× PBS buffer instead of a pathogen suspension. This experiment was repeated three times with 5 leaves per treatment and two spots per leaf [
60,
61].
2.11. Greenhouse Trials
Three important tomato pathogens R. solani, C. m. subsp. michiganensis, and X. e. pv. perforans were selected for greenhouse trials to test the antagonistic potential of FG106.
2.11.1. Rhizoctonia solani
Tomato seeds (
L. esculentum, Mobil) were surface sterilized for 5 min in 2% sodium hypochlorite and rinsed twice with 70% ethanol, followed by three washes with sterile distilled water. Seeds were placed in sterilized plastic pot trays containing perlite soil and then grown for 4 weeks in a growth chamber with daily watering. Roots of 4-week-old seedlings were then dipped in a 2 × 10
8 cfu/mL suspension of strain FG106 for 20 min, while control seedlings were treated with distilled water [
62]. The next day, the seedlings were inoculated with 3 g of wheat seeds colonized with
R. solani [
36,
63]. Plants were kept in the greenhouse in a high humidity chamber for 12 h before and 12 h after inoculation to maintain high humidity and facilitate infection. Ten pots with one seedling per pot were used in each treatment. Disease severity was scored 15 days later using a previously reported scale [
26]. The disease index (DI) was scored as described previously [
64]; briefly, a score of 0 indicated no visible necrotic lesions, 1 indicated root necrosis up to 2.5 mm in length, 2 indicated necrosis 2.5–5.0 mm in length, 3 indicated necrosis larger than 5.0 mm, 4 indicated lesions covering the crown and shoots, and 5 indicated seedling damping-off.
2.11.2. Preparation of Inoculum of Bacterial Pathogens and Bacterial Antagonists
C. m. subsp.
michiganensis and
X. e. pv.
perforans were selected to evaluate the biocontrol activity of FG106 on tomato plants in vivo. Inocula of both pathogens and the biocontrol agent (FG106) were prepared from bacterial cells grown for 48 h on nutrient agar (NA) (Sigma-Aldrich, Taufkirchen, Germany). Single colonies were transferred into LB broth and subsequently incubated at 28 °C for 48 h in a rotary shaker at 150 revolutions/min (rpm). The bacterial cultures were then centrifuged for 15 min at 7500 rpm, and the pellets were resuspended in sterile water, after which their density was adjusted to 2 × 10
8 cfu/mL (OD600 = 0.1), and they were used in pathogen challenge experiments with 4–5 true-leaved tomato seedlings [
65,
66,
67].
2.11.3. Plant Material and Inoculation of Bacterial Endophytes
Three-week-old seedlings of the hybrid tomato cultivar ‘SIR ELYAN F
1′ were obtained from a local nursery [
65]. Fifteen seedlings were tested for each treatment. The trials were designed to evaluate biocontrol effect of FG106 on the formation of bacterial cankers
C. m. subsp.
michiganensis and bacterial spots
X. e. pv.
perforans. Plants were maintained in a growth chamber at 68–80% RH and 25 °C, with 16/8 h of light and darkness daily. Experiments were conducted in duplicate. The bacterial inoculations were performed by soil drenching with 20 mL of the appropriate bacterial suspensions. Tomato seedlings were harvested after 30 days. Negative controls consisted of seedlings drenched with tap water instead of the pathogen suspensions.
2.11.4. Plant Competition with Bacterial Pathogens
Inoculation of
C. m. subsp.
michiganensis on the plants was performed as described previously [
65]. Briefly, after 7 days of treatment with the putative BCA or water (negative control), 20 mL of
C. m. subsp.
michiganensis solution was poured onto the soil near the stem crown. To facilitate bacterial penetration, the roots were damaged by cutting with a scalpel at three points located 2 cm from the stem crown. After 4 weeks, bacterial canker symptoms were assigned a disease index score on a scale ranging from 0 to 5, where 0 means no symptoms, 1 refers to loss of turgor and chlorosis, 2 is wilt and/or cankers > 0.5 cm in diameter in 1 or 2 leaves, 3 indicates wilt or cankers > 0.5 cm in diameter in 3 or more leaves, 4 represents fully withered plants, and 5 denotes dead plants [
66]. Area under disease progress curves (AUDPCs) were generated based on weekly monitoring data for 4 weeks post-incubation [
65].
X. e. pv.
perforans was sprayed onto the abaxial and adaxial leaf surfaces of tomato seedlings using hand-trigger sprayers 3 days after treating the soil with either FG106 or water (negative control). The RH was increased to promote bacterial penetration, and the inoculated plants were pre-incubated and post-incubated for 24 h under polyethylene sheets. Ten days after pathogen inoculation, 10 tomato leaflets per plant were sampled randomly. Spots and lesions on the leaflets were counted and the leaflet area determined, and disease severity was evaluated as number of lesions/cm
2. The leaflet area was quantified by image processing and analysis using the ImageJ software package [
68,
69]. The percentage reduction in disease severity caused by the presence of FG106 was calculated relative to that seen in negative controls as described previously [
68].
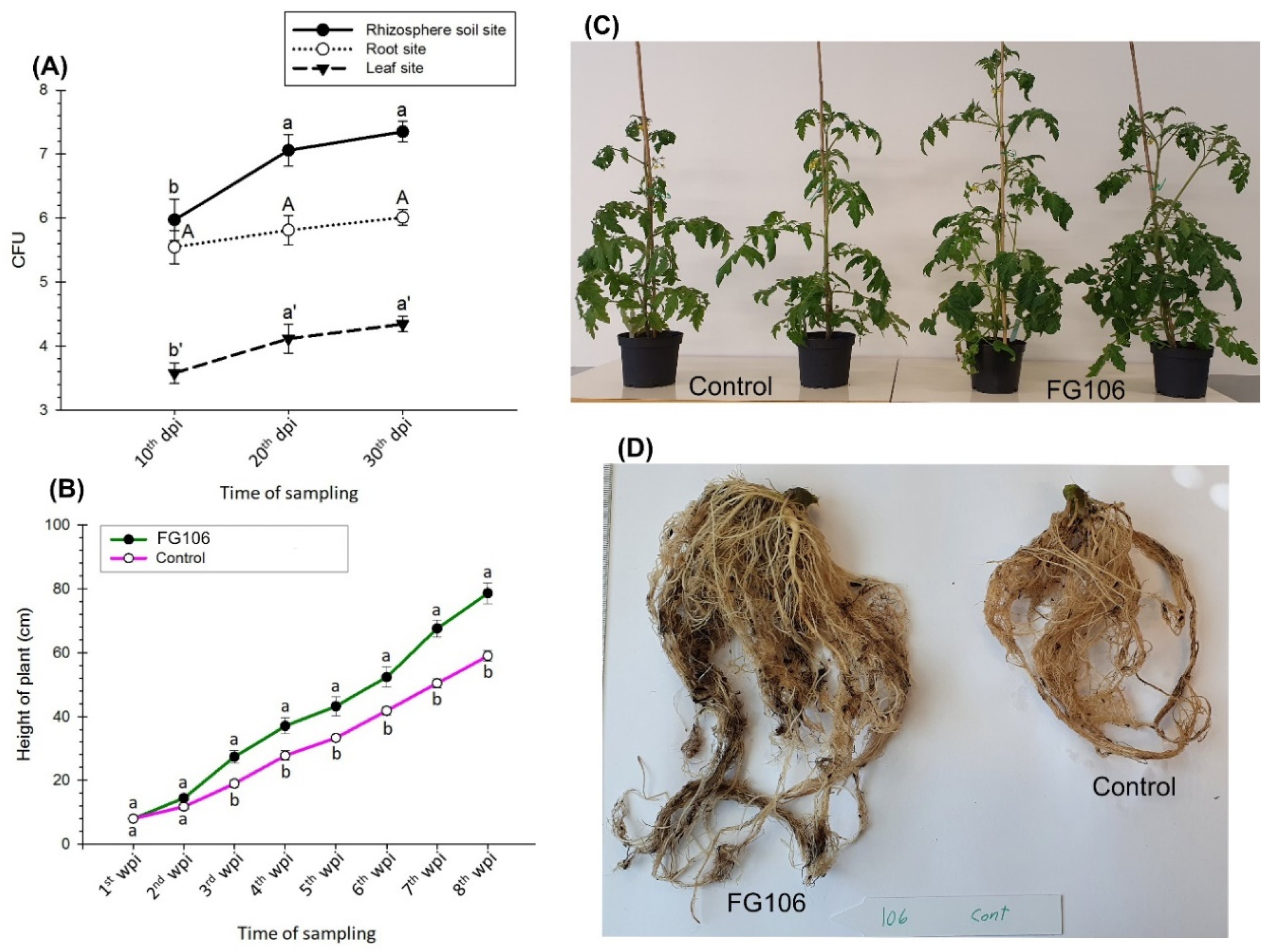
2.12. Colonization of Tomato Seedlings by Strain FG106
Tomato plants (Money Maker) were planted in a seedling tray and then transferred to pots after 4 weeks. During the transfer, the roots were immersed in a suspension of FG106 with a concentration of 2 × 108 cfu/mL, while negative controls were immersed in water. All seedlings were kept at 25–28 °C and 60% humidity with a 14/10 h light/dark cycle at a light intensity of approximately 500 μmol m−2 s−1.
The number of colony forming units per gram of sample was determined using TSA media supplemented with chloramphenicol (Sigma-Aldrich, Taufkirchen, Germany) with a final concentration of 100 µg mL
−1. To evaluate the presence of bacteria in the leaves and roots, the leaf and root surfaces were surface sterilized for 5 min with 3% sodium hypochlorite, followed by three washes with sterile water. Water from the final wash was used as a negative control. Plant tissue was homogenized aseptically with a pestle and mortar, and bacterial CFUs were enumerated by dilution and plate counting as described previously [
70]. Homogenized samples (1 g per replicate) of the rhizosphere, root, and leaves were added to 9 mL of 1× PBS solution, separately. Samples were then vortexed for 1 min and serially diluted at 1:10 until a 10
9 dilution was reached. Aliquots of 100 µL were plated from each dilution onto agar plates and then incubated at 28 °C for 48 h. Untreated plants maintained under similar conditions were also subjected to this procedure to detect naturally occurring bacteria capable of growing on the TSA medium [
71].
Bacterial populations in the rhizosphere, roots, and leaves of the plants were assessed at 15, 30, and 45 days post-inoculation (dpi) to investigate the survival of the bacteria in the plant and rhizosphere. The height, chlorophyll content, number of leaves, and fresh and dry weight of the seedlings were also recorded. Six replicates were analyzed per treatment [
70,
72,
73].
2.13. Genome Sequencing and Assembly
The P. aeruginosa strain FG106 was grown in 20 mL of LB medium and incubated for 18 h at 28 °C. After this period, total genomic DNA was extracted using the Nanobind CBB Big DNA Kit (Circulomics Kit, Baltimore, MD, USA) according to the manufacturer’s recommendations. DNA quality and quantity were checked by agarose gel electrophoresis and using a NanoDrop instrument (NanoDrop Technologies, South San Francisco, CA, USA). Library construction was performed using the Illumina TruSeq PCR-free kit (Thermo Scientific, Waltham, MA, USA)) with an insert size of 670 bp. The FG106 genome was sequenced at the SciLifeLab, Sweden, on a MiSeq instrument (MSC 2.5.0.5/RTA 1.18.54) with a 2 × 300 bp reads setup using ‘Version3’ chemistry. The SPAdes version 3.14.1 software package was used for sequence assembly and quality assessment. The whole genome sequencing raw data were submitted to the BioProject: ID PRJNA767521. The genome assembly data have been deposited at DDBJ/ENA/GenBank under the accession: JAJNEF010000000.
2.14. Predicted 16S rRNA Gene
The 16S rRNA gene sequence was predicted based on the constructed primary genome assembly using RNAmmer 1.2 4, which predicts 5s/8s, 16s/18s, and 23s/28s ribosomal RNA in full genome sequences [
74].
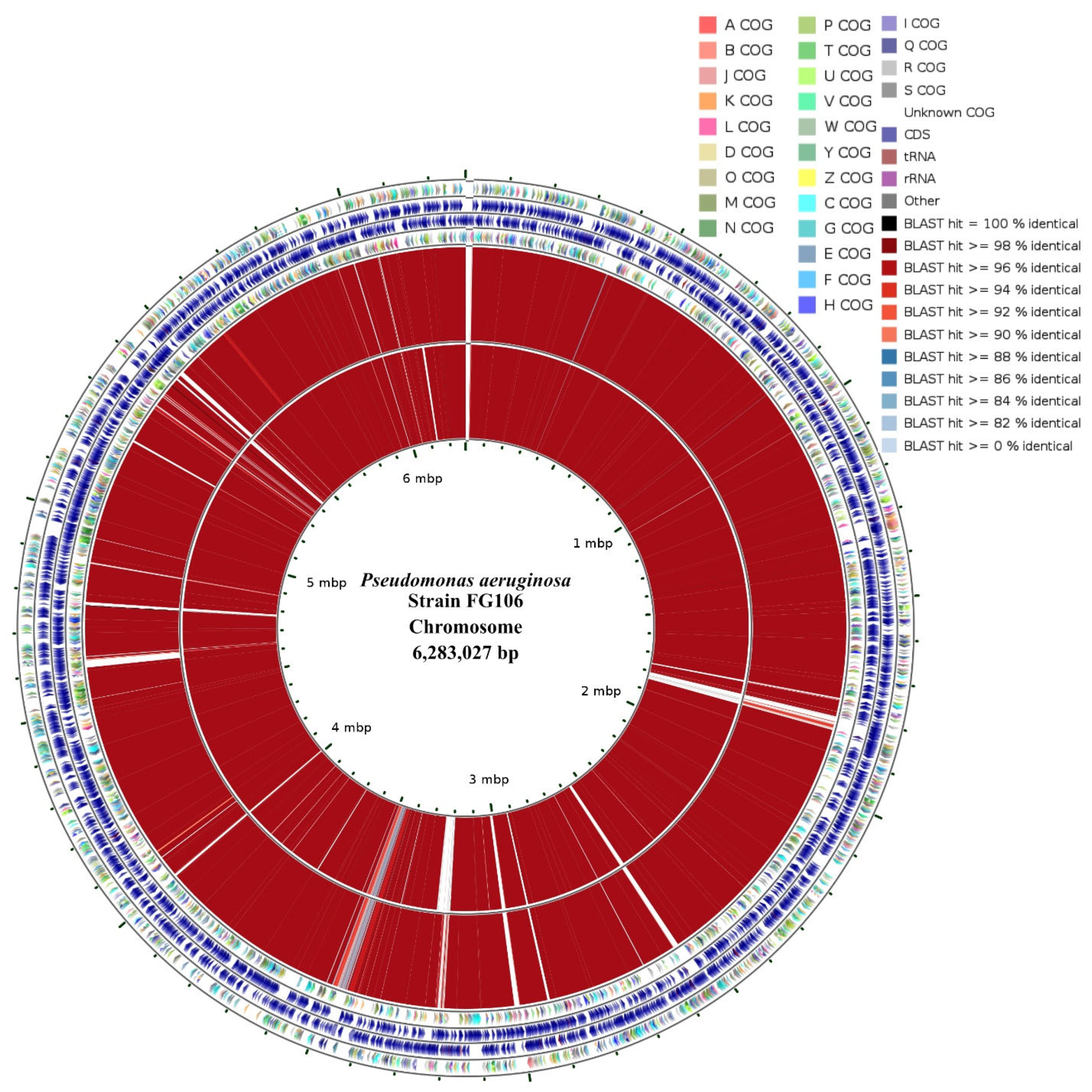
2.15. Draft Assembly Circular Map
A circular map of genome features was constructed using the CGView tool [
75]. Final draft assembly was performed using the CGView Server. The CGView Comparison Tool (CCT) package [
76] was used to perform clusters of orthologous groups (COG) classifications and generate a circular plot showing DNA-vs.-DNA mappings for the FG106 strain and two reference strains: P.aeru-DSM.50071 (NZ_CP012001) and P.aeru-M18 (NC_017548).
2.16. Gene Detection and Analysis of Coding Genes from FG106
The virulence factor database (VFDB) [
77] was searched to retrieve bacterial VF categories for detected genes, and additional factors were added from the literature. Comparative mapping was performed by BLAST searching using eight complete and closely related genomes of
P. aeruginosa strains, namely, P.aeru-LESB58, P.aeru-PA7, P.aeru-PAO1, P.aeru-UCBPP-PA14, P.aeru-DSM.50071, P.aeru-M18, P.aeru-PAO581, and P.aeru-L10.
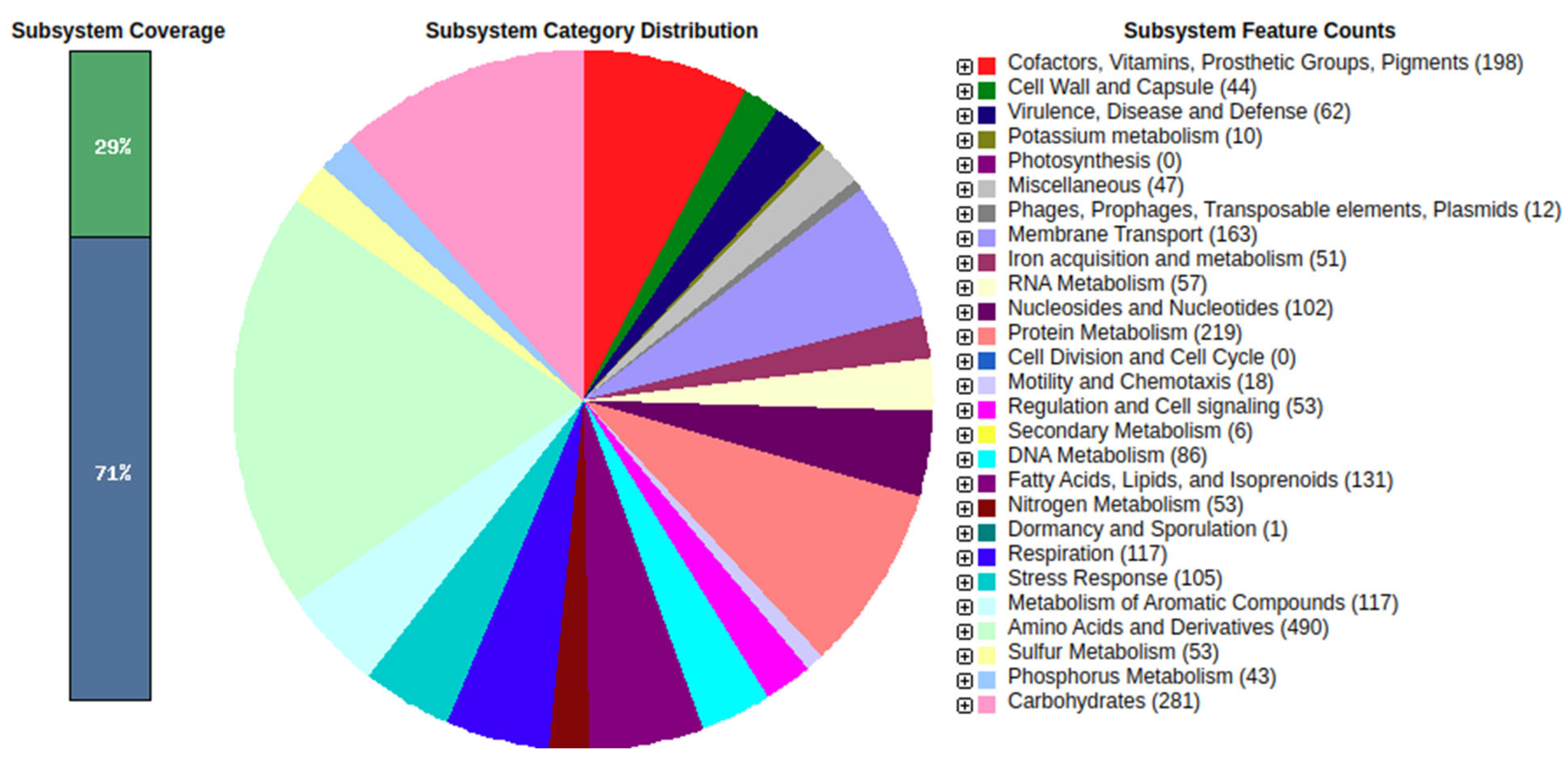
2.17. Draft Genome Gene Prediction, Annotation and Functional Characterization
Gene prediction and annotation based on the draft genome were performed using the RAST annotation server (Rapid Annotation using Subsystem Technology (
http://rast.nmpdr.org (accessed on 16 November 2021)) with the following annotation parameters: genetic code = 11, E value cut-off for selection, and pinneMetricd CDSs = 1 × 10
−20.
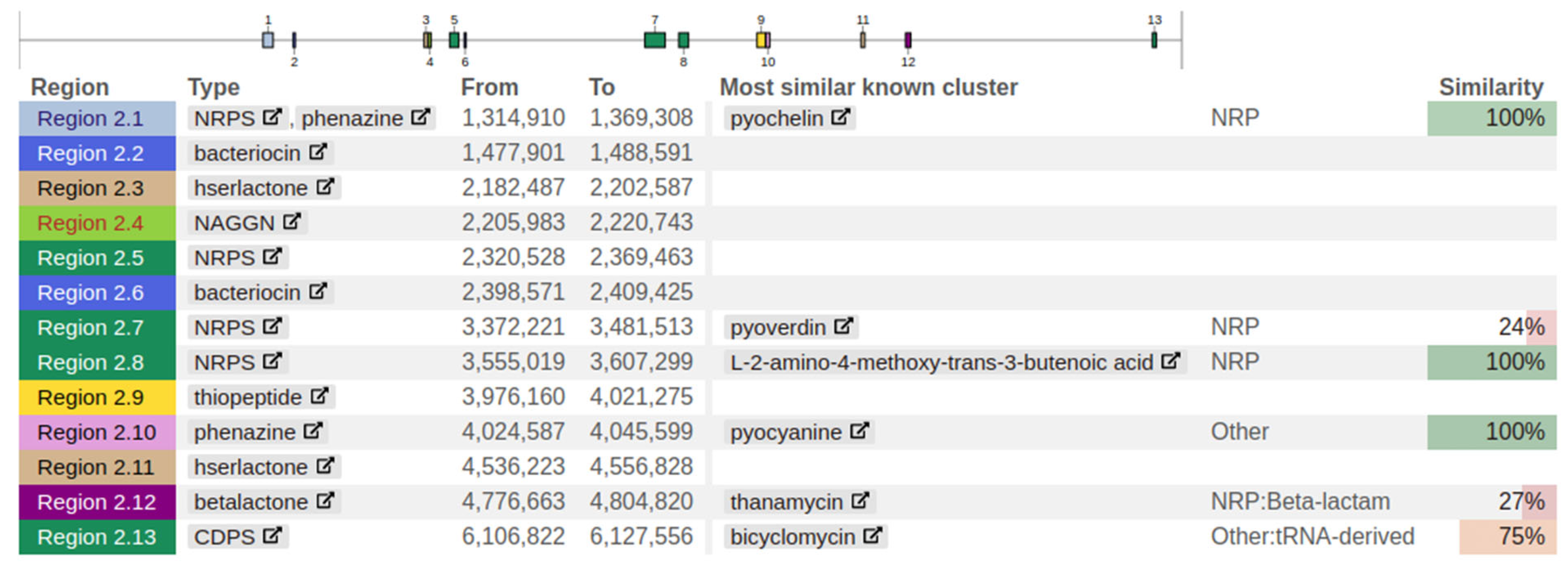
2.18. Secondary Metabolite Analysis
Secondary metabolite analysis was performed using antiSMASH to identify gene clusters encoding enzymes synthesizing secondary metabolites belonging to all known broad chemical classes.
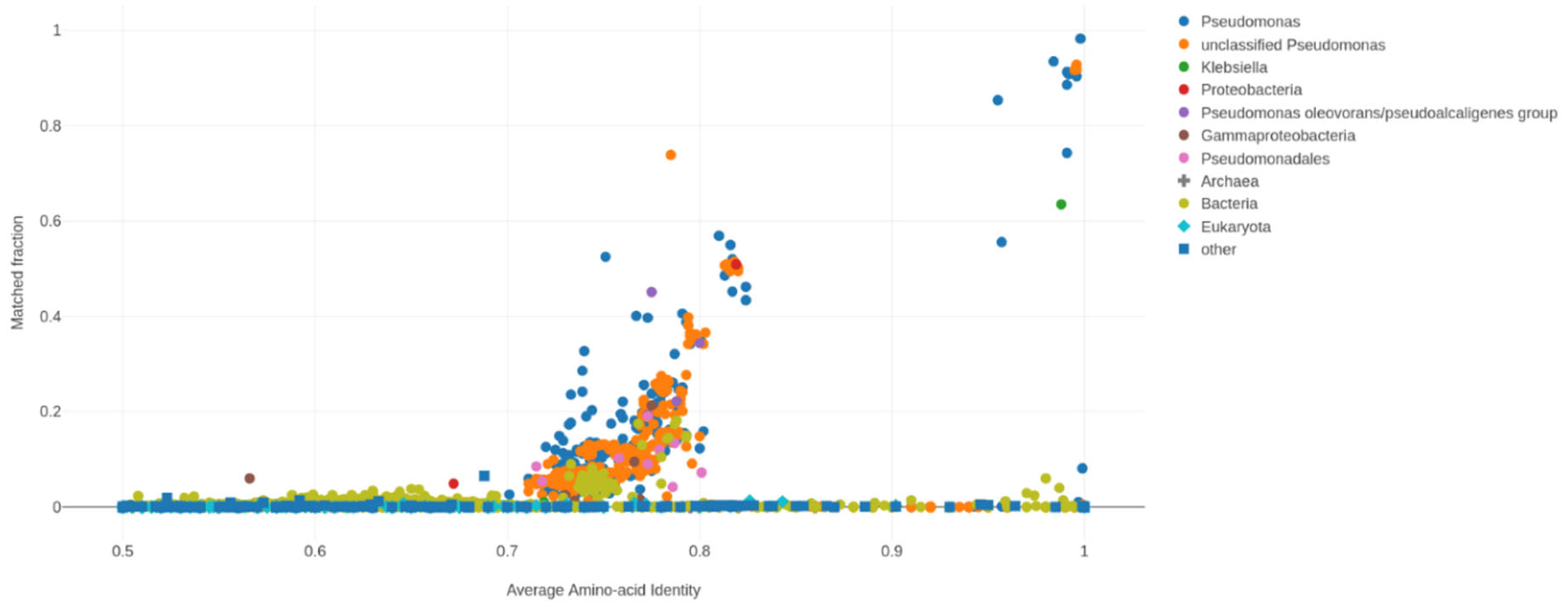
2.19. Comparative Genomics and Phylogenomics
The average nucleotide identity (ANI) value is widely used to compare prokaryotic genome sequences and to classify and identify bacteria [
78]. ANI was calculated using the nearest reference genome (
P. aeruginosa PAO1) to the sample’s assembled genome (FG106) using Chulabs’s online Average Nucleotide Identity (ANI) calculator. The average amino acid identity (AAI) is a very robust measure of genetic and evolutionary relatedness between two strains that correlates strongly with DNA–DNA association values (the classical tool for species delineation in prokaryotes) and the genome’s mutation rate. For the phylogeny analysis based on the complete genome, we used iTOL (
https://itol.embl.de (accessed on 16 November 2021)) with Dendroscope with the whole genomes of 37
P. aeruginosa strains (including the genome of strain FG106 and 36 other genomes), which led to constructing a phylogenetic tree based on maximum parsimony. This analysis grouped strain FG106 in a clade with five other
P. aeruginosa strains (RTE4, RP73, SCV20265, C3719, DK2).
2.20. Statistical Analysis
Data from the experiments on the antagonistic effects of FG106, the tomato growth experiments, chlorophyll content measurements, and the bioassays were analyzed using SAS statistical software version 9.2 (SAS Institute, 2009). One-way ANOVA was used to compare parameters with at least three independent groups. Tukey’s test with a significance threshold of p < 0.05 was used to compare differences between paired means. For parameters with two independent groups, differences between means were evaluated using the t-test.
4. Discussion
A strain of Pseudomonas spp. designated FG106 was isolated from the rhizosphere of healthy tomato plants. Phenotypic and phylogenetic analyses revealed that this strain belongs to P. aeruginosa. In particular, P. aeruginosa has many interesting features that can be agriculturally valuable, mainly due to its robustness, ability to compete with pathogens, and capacity to produce secondary metabolites that inhibit pathogen growth. Accordingly, its strains are potential biocontrol agents.
The new strain’s ability to control the pathogenicity of several pathogens was evaluated in vitro and in vivo, revealing that strain FG106 can solubilize phosphate and thus promote lateral root development and mineral absorption by the host plant [
85]. It also produced siderophores, which increase plant tissue growth and root length while also increasing pathogen resistance by solubilizing otherwise insoluble iron [
86]. Siderophore production by FG106 also contributes to its ability to compete effectively with pathogens for scarce iron [
87], enabling FG106 to efficiently occupy an ecological niche in the rhizosphere. Another important bioactive compound produced by this strain is IAA, which increases root length. Research demonstrated that
Pseudomonas strains produce higher levels of IAA than other beneficial bacteria [
88]. The impact of this compound on the host plant is due to its status as a key phytohormone that regulates plant growth, nutrient uptake, and activation of defense responses [
89]. It also plays important roles in the formation of root tissues; changes in auxin levels correlate with changes in root growth [
90]. Another important quality of FG106 is its ability to produce ammonia, which facilitates the absorption of nitrate and ammonium by the host plant. Furthermore, it can produce hydrogen cyanide (HCN) and form biofilms, enabling it to effectively limit the growth of phytopathogens. Finally, FG106 secretes active protease and lipase enzymes into the culture medium, which further inhibited the growth of the tested phytopathogens. Ahmadzadeh and Sharifi-Tehrani [
91] have shown that these enzymes contribute to the degradation of pathogens’ cell walls. Similarly, Zaiha et al. [
92] showed that the production of proteases, chitinases, and 1-3-gluconases by beneficial strains contributes to plant resistance to the pathogen
R. solani.
The inhibitory effect of strain FG106 differed between the tested pathogens in vitro. Its antagonistic activity was highest against
C. m. subsp.
michiganensis and
P. infestans, followed by
A. alternata,
P. colocasiae,
X. e. pv.
perforans,
B. cinerea and
R. solani. Conversely, Chandra et al. [
14] found that
P.
aeruginosa was most successful at inhibiting
A. alternata and that it limited the pathogen’s growth by inhibiting mycelial growth, radial growth, and spore germination.
Inoculation with the strain FG106 had a significant effect on germination of tomato seeds under greenhouse conditions. Previous research reports indicated that other strains of this bacterial species, such as CQ-4, stimulate seed growth and plant germination on tomato [
93]. We also found that the biomass of FG106 treatment in the rhizosphere was higher than in the roots and leaves. Treatment of plants with this strain led to increases in seedling height and in the fresh and dry weight of shoots and roots. Furthermore, chlorophyll levels increased significantly in seedlings treated with FG106, probably due to an increase in their ability to absorb micronutrients. Iron is an essential micronutrient for plants that is involved in several processes including chlorophyll synthesis, maintenance of chloroplast structure, and photosynthesis. All of these processes increase plant growth and phytopathogen resistance. The capacity of FG106 to control pathogens was also tested in vivo, revealing that it successfully controlled
P. infestans in detached leaf assays and reduced the appearance and spread of disease symptoms. Although gray mold colonized the leaf surface, the size of the disease spots was smaller than in positive control plants not treated with FG106.
We sequenced the genome of P. aeruginosa strain FG106, which was found to be 62,081,727 bp in length and to contain 5941 protein-coding genes along with 63 RNA-only genes. Comparative analysis revealed that the closest hit for strain FG106 is P. aeruginosa strain DSM 50,071; these two strains exhibited 99.93% sequence identity in the 16S rRNA gene. A variety of genes associated with anti-pathogen activity including putative TTSS and T6SS genes as well as genes apparently involved in siderophore biosynthesis were identified. In addition, the genome contains gene collections associated with the production of volatile compounds, adhesion proteins, and enzymes including proteases and ACC deaminase. These results indicate that FG106 can produce cell wall-degrading enzymes and plant growth factors.
Further functional research and comparative genomic analyses will be necessary to fully evaluate the potential of biocontrol strategies based on endophytism. However, our results are consistent with other studies showing that
P. aeruginosa strains can improve the growth of inoculated plants and protect against various pathogens [
94,
95].
5. Conclusions
Our study revealed that P. aeruginosa strain FG106, a Pseudomonadales belonging to the Gammaproteobacteria that was originally isolated from the roots of healthy nursery tomato plants, produces a wide range of biologically active metabolites and is a potentially valuable biocontrol agent. Our findings suggest that biological control using this strain could facilitate the integrated and sustainable management of several fungal and bacterial pathogens on tomato, potato, taro, and strawberry crops. To develop a sustainable and effective pathogen control method, we believe that multiple control strategies including fertilization, resistant cultivars, and the use of biocontrol agents will have to be applied in combination. Future research should, therefore, investigate the ability of this bacterial strain to control other plant pathogens and the mechanisms underpinning this control.