The Regulatory Mechanism of Feeding a Diet High in Rice Grain on the Growth and microRNA Expression Profiles of the Spleen, Taking Goats as an Artiodactyl Model
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Design and Animal Management
2.2. Sample Collection
2.3. LPS Determination
2.4. RNA Isolation, Small RNA Library Construction, and Sequencing
2.5. Bioinformatics Analysis of Solexa Sequencing Data
2.6. Quantitative PCR
2.7. Statistical Analysis
3. Results
3.1. Visceral Organ Development and LPS Concentration
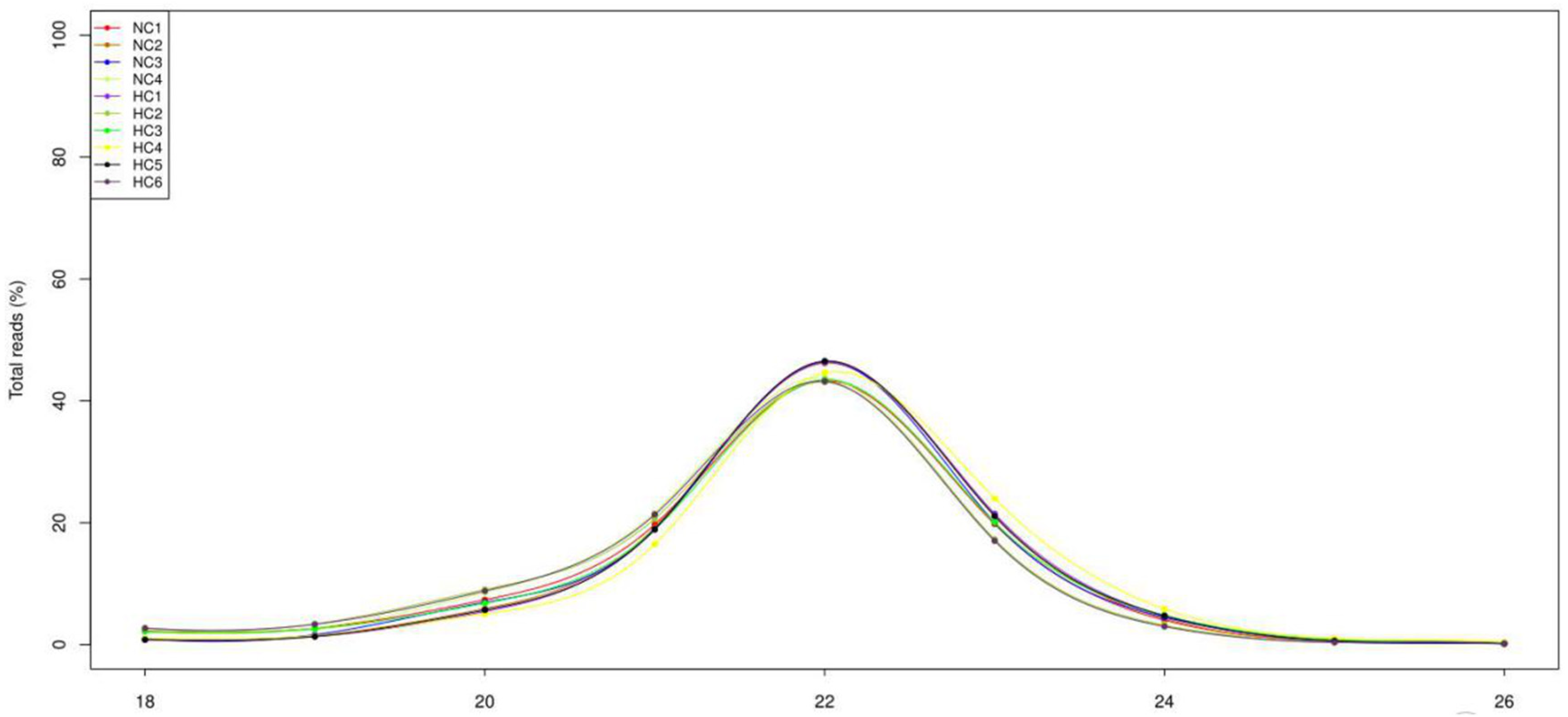
3.2. Overview of the Deep-Sequencing Data
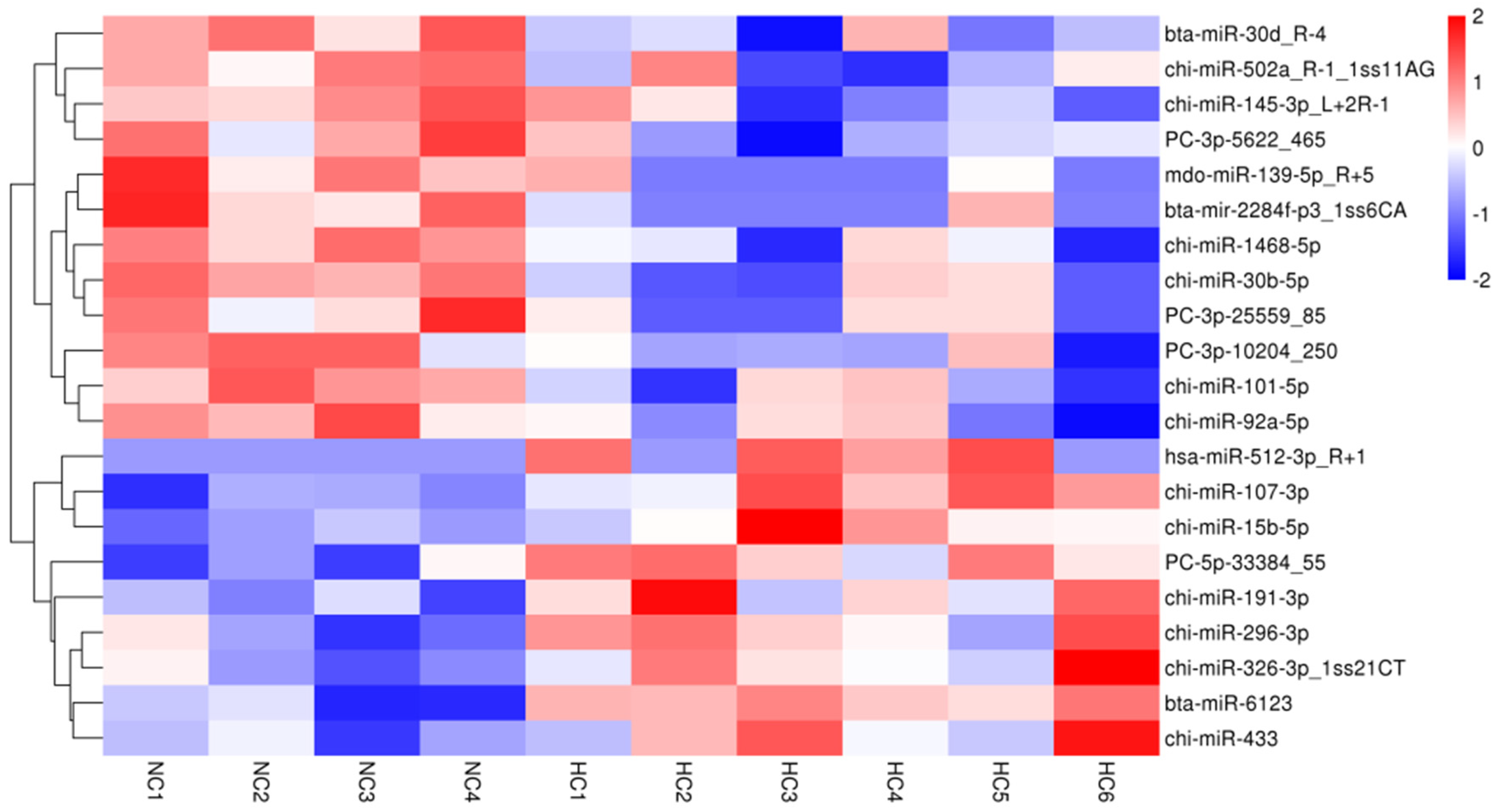
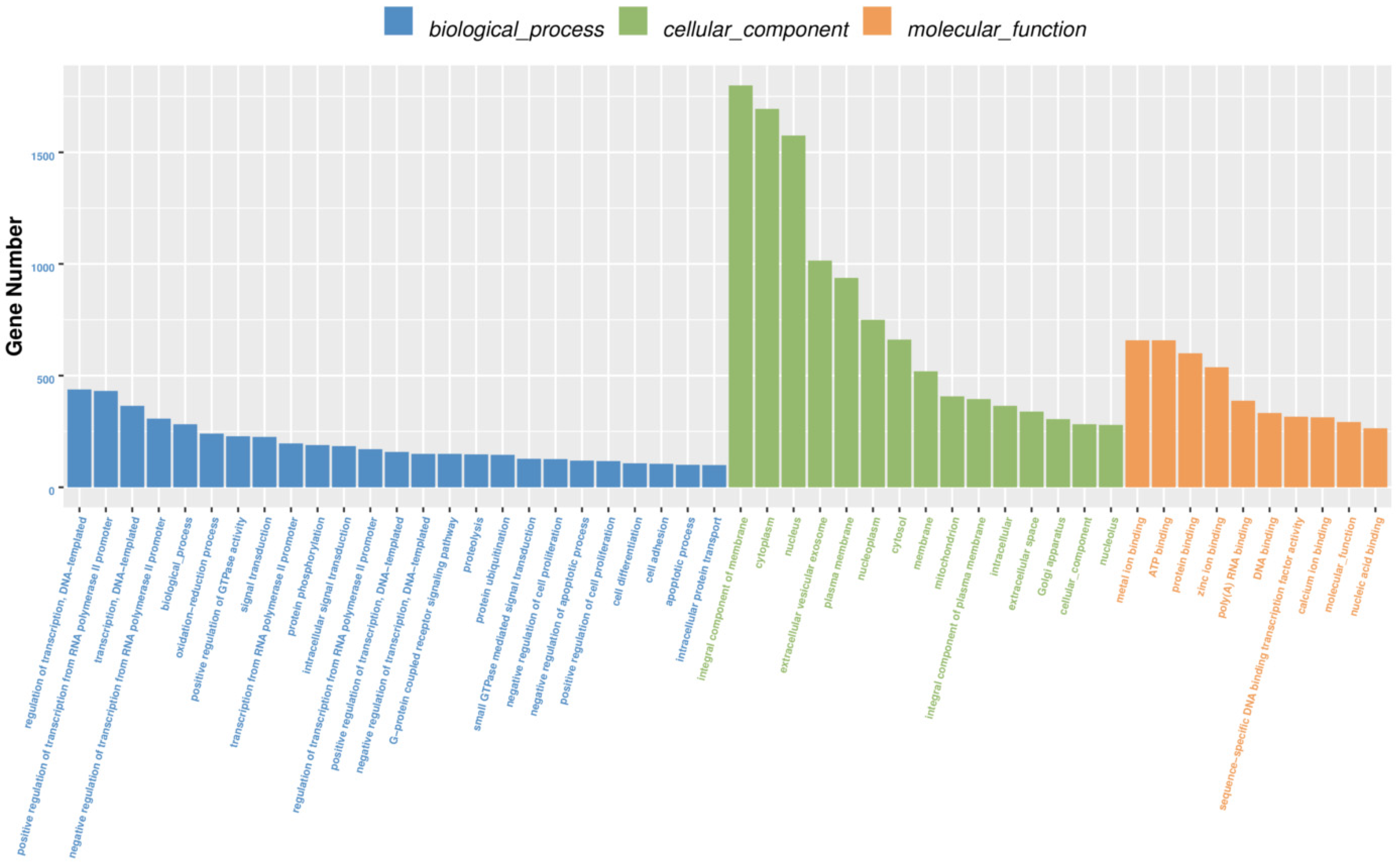
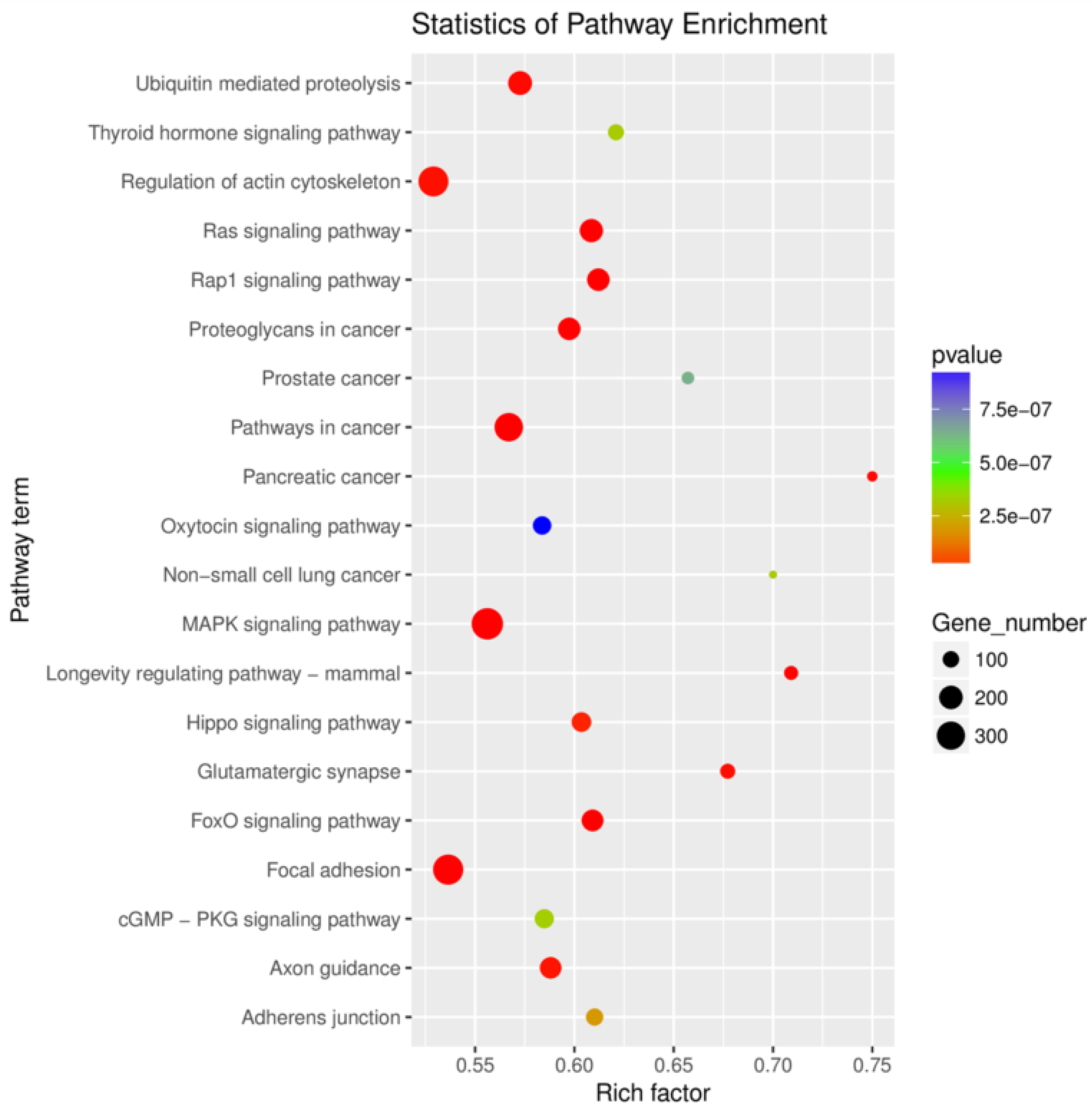
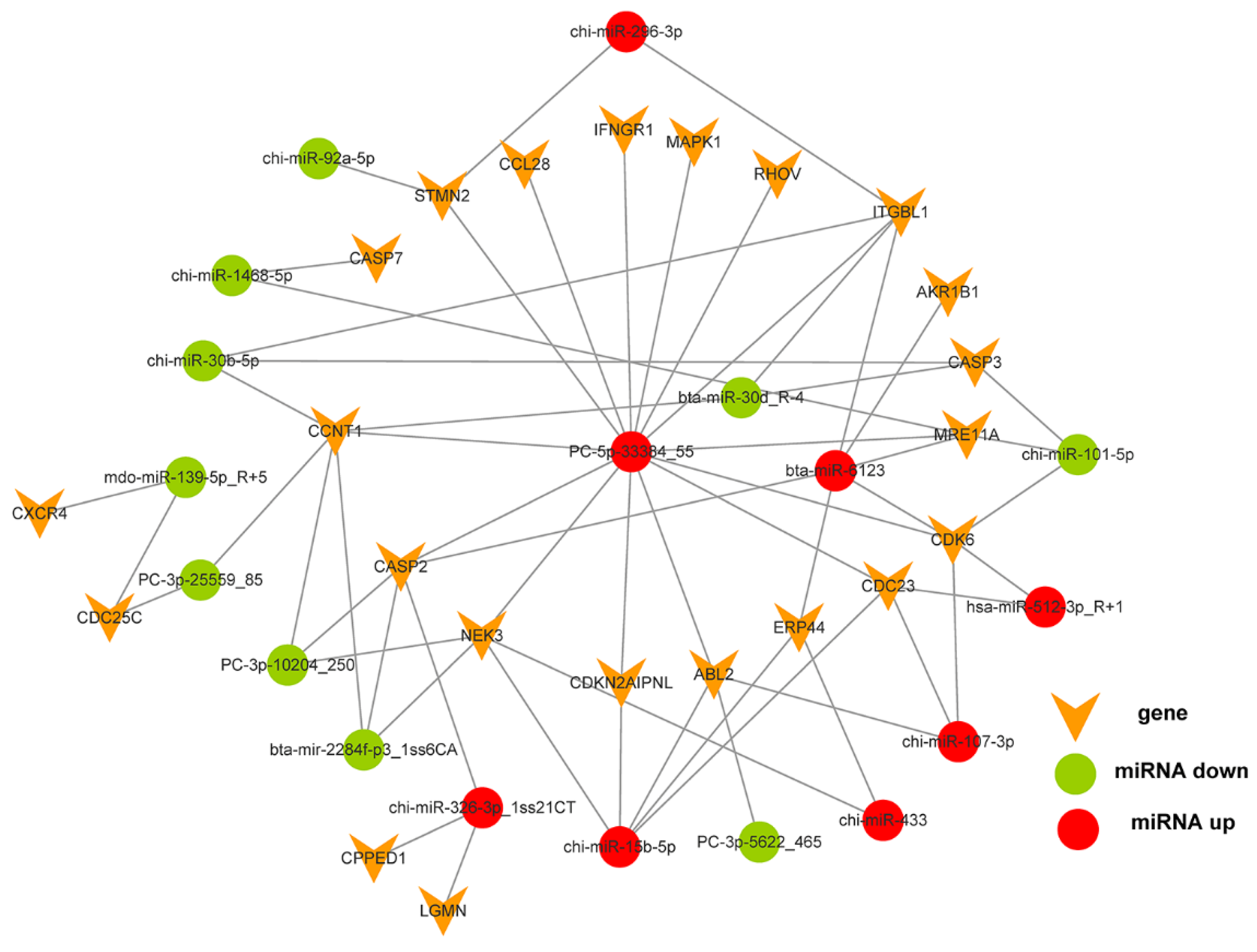
3.3. Differentially Expressed microRNAs, Gene Function, Pathway and Interactive Network
3.4. Targeted Gene Expression
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mebius, R.E.; Kraal, G. Structure and function of the spleen. Nat. Rev. Immunol. 2005, 5, 606–616. [Google Scholar] [CrossRef]
- Bronte, V.; Pittet, M.J. The spleen in local and systemic regulation of immunity. Immunity 2013, 39, 806–818. [Google Scholar] [CrossRef] [Green Version]
- Pereira, J.P.; Kelly, L.M.; Cyster, J.G. Finding the right niche: B-cell migration in the early phases of T-dependent antibody responses. Int. Immunol. 2010, 22, 413–419. [Google Scholar] [CrossRef] [PubMed]
- Mohr, A.M.; Mott, J.L. Overview of microRNA biology. Semin. Liver Dis. 2015, 35, 3–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patil, V.S.; Zhou, R.; Rana, T.M. Gene regulation by non-coding RNAs. Crit. Rev. Biochem. Mol. Biol. 2014, 49, 16–32. [Google Scholar] [CrossRef]
- Raisch, J.; Darfeuille-Michaud, A.; Hang Thi Thu, N. Role of microRNAs in the immune system, inflammation and cancer. World J. Gastroenterol. 2013, 19, 2985–2996. [Google Scholar] [CrossRef]
- He, J.-J.; Ma, J.; Wang, J.-L.; Xu, M.-J.; Zhu, X.-Q. Analysis of miRNA expression profiling in mouse spleen affected by acute Toxoplasma gondii infection. Infect. Genet. Evol. 2016, 37, 137–142. [Google Scholar] [CrossRef] [PubMed]
- Lei, L.; Hu, Y.; Guo, M.; Lu, J.; Zheng, W.; Xu, H.; Chen, C.; Xu, L. Change on composition of immune cells in spleen of miRNA-126 knockdown mice. Chin. J. Immunol. 2016, 32, 460–464. [Google Scholar]
- Zheng, W.; Zhao, J.; Zhu, S.; Xu, H.; Lei, L.; Lu, J.; Jia, L.; Chu, F.; Wang, H.; Xu, L. The effect of miRNA-7 knock down on the function of T lymphocyte of spleen in vitro. Curr. Immunol. 2016, 36, 358–363. [Google Scholar]
- Maldonado-Aviles, J.G.; Guarnieri, D.J.; Zhu, X.; DiLeone, R.J. Down-regulation of miRNAs in the brain and development of diet-induced obesity. Int. J. Dev. Neurosci. 2018, 64, 2–7. [Google Scholar] [CrossRef]
- Yamada, K.; Takizawa, S.; Ohgaku, Y.; Asami, T.; Furuya, K.; Yamamoto, K.; Takahashi, F.; Hamajima, C.; Inaba, C.; Endo, K.; et al. MicroRNA 16-5p is upregulated in calorie-restricted mice and modulates inflammatory cytokines of macrophages. Gene 2020, 725, 144191. [Google Scholar] [CrossRef] [PubMed]
- Huo, W.; Zhu, W.; Mao, S. Effects of feeding increasing proportions of corn grain on concentration of lipopolysaccharide in the rumen fluid and the subsequent alterations in immune responses in goats. Asian-Australas. J. Anim. Sci. 2013, 26, 1437–1445. [Google Scholar] [CrossRef] [Green Version]
- Khafipour, E.; Krause, D.O.; Plaizier, J.C. A grain-based subacute ruminal acidosis challenge causes translocation of lipopolysaccharide and triggers inflammation. J. Dairy Sci. 2009, 92, 1060–1070. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Xu, T.; Zhu, W.; Mao, S. High-grain feeding alters caecal bacterial microbiota composition and fermentation and results in caecal mucosal injury in goats. Br. J. Nutr. 2014, 112, 416–427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tao, S.; Duanmu, Y.; Dong, H.; Tian, J.; Ni, Y.; Zhao, R. A high-concentrate diet induced colonic epithelial barrier disruption is associated with the activating of cell apoptosis in lactating goats. BMC Vet. Res. 2014, 10, 235. [Google Scholar] [CrossRef] [Green Version]
- Chang, G.; Zhang, K.; Xu, T.; Jin, D.; Seyfert, H.-M.; Shen, X.; Zhuang, S. Feeding a high-grain diet reduces the percentage of LPS clearance and enhances immune gene expression in goat liver. BMC Vet. Res. 2015, 11, 67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, K.J.; Zheng, M.L.; Ren, A.; Zhou, C.S.; Yan, Q.X.; Tan, Z.L.; Zhang, P.H.; Yi, K.L. Effects of high rice diet on growth performance, nutrients apparent digestibility, nitrogen metabolism, blood parameters and rumen fermentation in growing goats. Kafkas Univ. Vet. Fak. Derg. 2019, 25, 749–755. [Google Scholar]
- Jiao, J.Z.; Zhang, X.L.; Wang, M.; Zhou, C.S.; Yan, Q.X.; Tan, Z.L. Linkages between Epithelial microbiota and host transcriptome in the ileum during high-grain challenges: Implications for gut homeostasis in goats. J. Agric. Food Chem. 2019, 67, 551–561. [Google Scholar] [CrossRef]
- Zhang, L.; Liang, R.; Zhang, G.; Zhai, Z.; Deng, Y.; Li, J.; Cui, S. Analysis of the microRNA expression profiles in feline kidney cell line infected with feline panleukopenia virus. Infect Genet. Evol. 2019, 75, 103945. [Google Scholar] [CrossRef]
- Li, C.; Xiong, T.; Zhou, M.; Wan, L.; Xi, S.; Liu, Q.; Chen, Y.; Mao, H.; Liu, S.; Chen, B. Characterization of microRNAs during Embryonic Skeletal Muscle Development in the Shan Ma Duck. Animals 2020, 10, 1417. [Google Scholar] [CrossRef]
- Li, X.; Shahid, M.Q.; Wu, J.; Wang, L.; Liu, X.; Lu, Y. Comparative small RNA analysis of pollen development in autotetraploid and diploid rice. Int. J. Mol. Sci. 2016, 17, 499. [Google Scholar] [CrossRef] [PubMed]
- Yan, Q.; Xu, J.; Wu, X.; Su, D.; Tan, Z. Stage-specific feed intake restriction differentially regulates placental traits and proteome of goats. Br. J. Nutr. 2018, 119, 1119–1132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, Q.; Tong, H.; Tang, S.; Tan, Z.; Han, X.; Zhou, C. L-theanine administration modulates the absorption of dietary nutrients and expression of transporters and receptors in the intestinal mucosa of rats. Biomed Res. Int. 2017, 2017, 9747256. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Liu, J.-H.; Xu, T.-T.; Zhum, W.-Y.; Mao, S.-Y. A high-grain diet alters the omasal epithelial structure and expression of tight junction proteins in a goat model. Vet. J. 2014, 201, 95–100. [Google Scholar] [CrossRef]
- Emmanuel, D.G.V.; Madsen, K.L.; Churchill, T.A.; Dunn, S.M.; Ametaj, B.N. Acidosis and lipopolysaccharide from Escherichia coli B: 055 cause hyperpermeability of rumen and colon tissues. J. Dairy Sci. 2007, 90, 5552–5557. [Google Scholar] [CrossRef] [Green Version]
- Escate, R.; Padro, T.; Badimon, L. Epigenetic regulation through miR-139-5p during human monocyte-to macrophage differentiation. Eur. J. Clin. Investig. 2018, 48, 124. [Google Scholar]
- Kasravi, F.B.; Brecht, W.J.; Weisgraber, K.H.; Harris, H.W. Induction of cytokine tolerance requires internalization of chylomicron-bound LIPS into hepatocytes. J. Surg. Res. 2003, 115, 303–309. [Google Scholar] [CrossRef]
- He, L.; Thomson, J.M.; Hemann, M.T.; Hernando-Monge, E.; Mu, D.; Goodson, S.; Powers, S.; Cordon-Cardo, C.; Lowe, S.W.; Hannon, G.J.; et al. A microRNA polycistron as a potential human oncogene. Nature 2005, 435, 828–833. [Google Scholar] [CrossRef]
- Hennessy, E.J.; Sheedy, F.J.; Santamaria, D.; Barbacid, M.; O’Neill, L.A.J. Toll-like Receptor-4 (TLR4) down-regulates microRNA-107, increasing macrophage adhesion via cyclin-dependent kinase 6. J. Biol. Chem. 2011, 286, 25531–25539. [Google Scholar] [CrossRef] [Green Version]
- Ahonen, M.A.; Haridas, P.A.N.; Mysore, R.; Wabitsch, M.; Fischer-Posovszky, P.; Olkkonen, V.M. MiR-107 inhibits CDK6 expression, differentiation, and lipid storage in human adipocytes. Mol. Cell. Endocrinol. 2019, 479, 110–116. [Google Scholar] [CrossRef] [Green Version]
- Tan, H.Y.; Zhao, L.; Song, R.P.; Liu, Y.L.; Wang, L.M. The long noncoding RNA SNHG1 promotes nucleus pulposus cell proliferation through regulating miR-326 and CCND1. Am. J. Physiol. Cell Physiol. 2018, 315, C21–C27. [Google Scholar] [CrossRef]
- Jiang, H.; Liang, M.; Jiang, Y.Q.; Zhang, T.; Mo, K.X.; Su, S.W.; Wang, A.P.; Zhu, Y.Y.; Huang, G.Q.; Zhou, R.J. The lncRNA TDRG1 promotes cell proliferation, migration and invasion by targeting miR-326 to regulate MAPK1 expression in cervical cancer. Cancer Cell Int. 2019, 19, 152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, C.; Liu, C.; Kang, J.; Zhao, G.; Ye, Z.; Huang, S.; Li, Z.; Wu, Z.; Pei, G. MicroRNA miR-326 regulates T-H-17 differentiation and is associated with the pathogenesis of multiple sclerosis. Nat. Immunol. 2009, 10, U1252–U1254. [Google Scholar] [CrossRef] [PubMed]
- Kurashina, R.; Kikuchi, K.; Iwaki, J.; Yoshitake, H.; Takeshita, T.; Takizawa, T. Placenta- specific miRNA (miR-512-3p) targets PPP3R1 encoding the calcineurin B regulatory subunit in BeWo cells. J. Obstet. Gynaecol. Res. 2014, 40, 650–660. [Google Scholar] [CrossRef]
- Li, J.; Lei, H.; Xu, Y.; Tao, Z.-Z. miR-512-5p suppresses tumor growth by targeting hTERT in telomerase positive head and neck squamous cell carcinoma in vitro and in vivo. PLoS ONE 2015, 10, e0135265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, Y.; Wang, C.; Zhang, D.; Chu, X.; Zhang, Y.; Li, J. miR-15b-5p ameliorated high glucose-induced podocyte injury through repressing apoptosis, oxidative stress, and inflammatory responses by targeting Sema3A. J. Cell. Physiol. 2019, 234, 20869–20878. [Google Scholar] [CrossRef]
- Tsai, Y.-C.; Kuo, M.-C.; Hung, W.-W.; Wu, L.-Y.; Wu, P.-H.; Chang, W.-A.; Kuo, P.-L.; Hsu, Y.-L. High glucose induces mesangial cell apoptosis through miR-15b-5p and promotes diabetic nephropathy by extracellular vesicle delivery. Mol. Ther. J. Am. Soc. Gene Ther. 2020, 28, 963–974. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Chen, R.; Zhang, Y. miR-296-3p targets APEX1 to suppress cell migration and invasion of non-small-cell lung cancer. Oncol. Lett. 2019, 18, 2612–2618. [Google Scholar] [CrossRef] [Green Version]
- Kakizaki, T.; Hatakeyama, H.; Nakamaru, Y.; Takagi, D.; Mizumachi, T.; Sakashita, T.; Kano, S.; Homma, A.; Fukuda, S. Role of microRNA-296-3p in the malignant transformation of sinonasal inverted papilloma. Oncol. Lett. 2017, 14, 987–992. [Google Scholar] [CrossRef] [Green Version]
- Volinia, S.; Calin, G.A.; Liu, C.G.; Ambs, S.; Cimmino, A.; Petrocca, F.; Visone, R.; Iorio, M.; Roldo, C.; Ferracin, M.; et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. USA 2006, 103, 2257–2261. [Google Scholar] [CrossRef] [Green Version]
- Tian, X.; Zhang, Z. miR-191/DAB2 axis regulates the tumorigenicity of estrogen receptor-positive breast cancer. IUBMB Life 2018, 70, 71–80. [Google Scholar] [CrossRef] [Green Version]
- Nagpal, N.; Kulshreshtha, R. rniR-191: An emerging player in disease biology. Front. Genet. 2014, 5, 99. [Google Scholar] [CrossRef] [Green Version]
- Di Leva, G.; Piovan, C.; Gasparini, P.; Ngankeu, A.; Taccioli, C.; Briskin, D.; Cheung, D.G.; Bolon, B.; Anderlucci, L.; Alder, H.; et al. Estrogen Mediated-Activation of miR-191/425 Cluster modulates tumorigenicity of breast cancer cells depending on estrogen receptor status. PLoS Genet. 2013, 9, e1003311. [Google Scholar] [CrossRef]
- Zhang, T.; Jiang, K.; Zhu, X.; Zhao, G.; Wu, H.; Deng, G.; Qiu, C. miR-433 inhibits breast cancer cell growth via the MAPK signaling pathway by targeting Rap1a. Int. J. Biol. Sci. 2018, 14, 622–632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, D.; Jian, X.; Xu, P.; Zhu, R.; Wang, Y. Linc01234 promotes cell proliferation and metastasis in oral squamous cell carcinoma via miR-433/PAK4 axis. BMC Cancer 2020, 20, 107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.C.; Ma, Y.; Meng, P.S.; Han, J.L.; Yu, H.Y.; Bi, L.J. miR-433 inhibits oral squamous cell carcinoma (OSCC) cell growth and metastasis by targeting HDAC6. Oral Oncol. 2015, 51, 674–682. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.Y.; Guo, D.D.; Liu, B.; Yin, X.W.; Wei, H.X.; Tang, K.; Bi, H.S. Regulatory role of rno-miR-30b-5p in IL-10 and toll-like receptor 4 expressions of t lymphocytes in experimental autoimmune uveitis in vitro. Mediat. Inflamm. 2018, 2018, 2574067. [Google Scholar] [CrossRef] [Green Version]
- Tian, L.; Ji, X.F.; Yang, L.; Chang, N.; Duan, X.H.; Yang, L.; Li, L.Y. Mir-30b-5p participates in NLRP3 inflammasome expression of bone marrow-derived monocytes/macrophages in mouse chronic liver injury. Hepatology 2018, 68, 62A–63A. [Google Scholar]
- Zhou, T.; Chen, Y.L. The functional mechanisms of miR-30b-5p in acute lung injury in children. Med. Sci. Monit. 2019, 25, 40–51. [Google Scholar] [CrossRef]
- Zhao, F.Y.; Qu, Y.; Zhu, J.H.; Zhang, L.; Huang, L.; Liu, H.T.; Li, S.P.; Mu, D.Z. miR-30d-5p plays an important role in autophagy and apoptosis in developing rat brains after hypoxic-ischemic injury. J. Neuropathol. Exp. Neurol. 2017, 76, 709–719. [Google Scholar] [CrossRef] [Green Version]
- Slee, E.A.; Adrain, C.; Martin, S.J. Executioner caspase-3,-6, and-7 perform distinct, non-redundant roles during the demolition phase of apoptosis. J. Biol. Chem. 2001, 276, 7320–7326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liou, L.Y.; Herrmann, C.H.; Rice, A.P. Human immunodeficiency virus type 1 infection induces cyclin T1 expression in macrophages. J. Virol. 2004, 78, 8114–8119. [Google Scholar] [CrossRef] [Green Version]
- Liou, L.-Y.; Haaland, R.E.; Herrmann, C.H.; Rice, A.P. Cyclin T1 but not cyclin T2a is induced by a post-transcriptional mechanism in PAMP-activated monocyte-derived macrophages. J. Leukoc. Biol. 2006, 79, 388–396. [Google Scholar] [CrossRef]
- Jiang, K.; Zhi, T.L.; Xu, W.H.; Xu, X.P.; Wu, W.N.; Yu, T.F.; Nie, E.; Zhou, X.; Bao, Z.Y.; Jin, X.; et al. MicroRNA-1468-5p inhibits glioma cell proliferation and induces cell cycle arrest by targeting RRM1. Am. J. Cancer Res. 2017, 7, 784–800. [Google Scholar]
- Saini, R.V.; Wilson, C.; Finn, M.W.; Wang, T.; Krensky, A.M.; Clayberger, C. Granulysin delivered by cytotoxic cells damages endoplasmic reticulum and activates caspase-7 in target cells. J. Immunol. 2011, 186, 3497–3504. [Google Scholar] [CrossRef] [Green Version]
- Lamkanfi, M.; Moreira, L.O.; Makena, P.; Spierings, D.C.J.; Boyd, K.; Murray, P.J.; Green, D.R.; Kanneganti, T.-D. Caspase-7 deficiency protects from endotoxin-induced lymphocyte apoptosis and improves survival. Blood 2009, 113, 2742–2745. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.X.; Zhou, S.N.; Zhang, L.M.; Zhang, J.; Wang, J.S.; Huang, C. Microrna-502a (mir-502a) post-transcriptionally regulates erbb4 and inhibits cell proliferation in esophageal squamous cell carcinoma. Ann. Oncol. 2012, 23, 32. [Google Scholar] [CrossRef]
- Huang, Y.; Du, K.L.; Guo, P.Y.; Zhao, R.M.; Wang, B.; Zhao, X.L.; Zhang, C.Q. IL-16 regulates macrophage polarization as a target gene of mir-145-3p. Mol. Immunol. 2019, 107, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Zou, F.Y.; Mao, R.R.; Yang, L.Y.; Lin, S.C.; Lei, K.C.; Zheng, Y.H.; Ding, Y.; Zhang, P.; Cai, G.X.; Liang, X.; et al. Targeted deletion of miR-139-5p activates MAPK, NF-κB and STAT3 signaling and promotes intestinal inflammation and colorectal cancer. FEBS J. 2016, 283, 1438–1452. [Google Scholar] [CrossRef] [PubMed]
- Jin, W.W.; Ibeagha-Awemu, E.M.; Liang, G.X.; Beaudoin, F.; Zhao, X.; Guan, L.L. Transcriptome microRNA profiling of bovine mammary epithelial cells challenged with Escherichia coli or Staphylococcus aureus bacteria reveals pathogen directed microRNA expression profiles. BMC Genom. 2014, 15, 181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, C.; Wu, H.; Wang, Y.; Zhu, S.; Liu, J.; Fang, X.; Chen, H. Circular RNA of cattle casein genes are highly expressed in bovine mammary gland. J. Dairy Sci. 2016, 99, 4750–4760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, D.; Chen, T.; Xie, M.; Li, M.; Zeng, B.; Sun, R.; Zhu, Y.; Ye, D.; Wu, J.; Sun, J.; et al. Oral administration of bovine and porcine milk exosome alter miRNAs profiles in piglet serum. Sci. Rep. 2020, 10, 6983. [Google Scholar] [CrossRef]
- Wang, C.Q.; Xu, C.; Fu, X.L.; Jiang, Y.Y. Schisandrin B suppresses liver fibrosis in rats by targeting miR-101-5p through the TGF-beta signaling pathway. Artif. Cells Nanomed. Biotechnol. 2020, 48, 473–478. [Google Scholar]
- Lei, Y.; Wang, Q.L.; Shen, L.; Tao, Y.Y.; Liu, C.H. MicroRNA-101 suppresses liver fibrosis by downregulating PI3K/Akt/mTOR signaling pathway. Clin. Res. Hepatol. Gastroenterol. 2019, 43, 575–584. [Google Scholar] [CrossRef]
- Nunez-Hernandez, F.; Perez, L.J.; Munoz, M.; Vera, G.; Accensi, F.; Sanchez, A.; Rodriguez, F.; Nunez, J.I. Differential expression of porcine microRNAs in African swine fever virus infected pigs: A proof-of-concept study. Virol. J. 2017, 14, 198. [Google Scholar] [CrossRef]
- Kohram, F.; Fallah, P.; Shamsara, M.; Bolandi, Z.; Rassoulzadegan, M.; Soleimani, M.; Ghanbarian, H. Cell type-dependent functions of microRNA-92a. J. Cell. Biochem. 2018, 119, 5798–5804. [Google Scholar] [CrossRef] [PubMed]
- Valente, G.; Ozmen, L.; Novelli, F.; Geuna, M.; Palestro, G.; Forni, G.; Garotta, G. Distribution of interferon-gamma receptor in human tissues. Eur. J. Immunol. 1992, 22, 2403–2412. [Google Scholar] [CrossRef] [PubMed]
- Novelli, F.; Bernabei, P.; Ozmen, L.; Rigamonti, L.; Allione, A.; Pestka, S.; Garotta, G.; Forni, G. Switching on of the proliferation or apoptosis of activated human T lymphocytes by IFN-gamma is correlated with the differential expression of the alpha- and beta-chains of its receptor. J. Immunol. 1996, 157, 1935–1943. [Google Scholar]
- Li, Q.; Wang, Z.; Jin, X.; Xu, Y.; Guo, X.; Chen, W.; Dong, W. Expression characteristic of IFNGR in stellate ganglion of female goats. Chin. J. Immunol. 2014, 30, 604–608. [Google Scholar]
- Li, Q.; Wang, Z.; Jin, X.; Xu, Y.; Chen, W.; Huang, S.; Sun, M. Expression of IFNGR in the cranial cervical ganglion of goat. Acat Agric. Boreali-Occident. Sin. 2015, 24, 7–12. [Google Scholar]
- Li, Q.; Wang, Z.; Jin, X.; Xu, Y.; Guo, X.; Dong, W.; Liu, W. Expression of IFNGR in the celiac superior mesenteric ganglion of goats. Sci. Agric. Sin. 2015, 48, 959–965. [Google Scholar]
- Ma, T.; Jiang, H.; Gao, Y.; Zhao, Y.; Dai, L.; Xiong, Q.; Xu, Y.; Zhao, Z.; Zhang, J. Microarray analysis of differentially expressed microRNAs in non-regressed and regressed bovine corpus luteum tissue; microRNA-378 may suppress luteal cell apoptosis by targeting the interferon gamma receptor 1 gene. J. Appl. Genet. 2011, 52, 481–486. [Google Scholar] [CrossRef] [PubMed]
| Item | NC | HC |
|---|---|---|
| Ingredient composition (%) | ||
| Rice straw | 45.0 | 10.0 |
| Rice with shell | 33.2 | 54.3 |
| Soybean meal | 9.60 | 15.7 |
| Wheat bran | 6.00 | 9.80 |
| Fat powder | 3.20 | 5.20 |
| Calcium carbonate | 0.50 | 0.80 |
| Calcium bicarbonate | 1.10 | 1.80 |
| Sodium chloride | 0.60 | 1.00 |
| Premix a | 1.00 | 1.40 |
| Nutrient levels b, % of DM | ||
| Crude protein | 13.5 | 17.6 |
| Crude ash | 9.34 | 9.12 |
| Crude fat | 12.0 | 11.9 |
| Neutral detergent fiber | 49.8 | 38.4 |
| Acid detergent fiber | 36.5 | 9.51 |
| NFC c | 5.74 | 12.5 |
| DMI d, g/d | 572 | 602 |
| Item | NC | HC | p-Values |
|---|---|---|---|
| Heart, g | 64.5 ± 3.80 | 77.5 ± 6.12 | 0.002 |
| Liver, g | 293 ± 50.9 | 343 ± 57.4 | 0.142 |
| Spleen, g | 20.5 ± 3.30 | 27.0 ± 4.75 | 0.020 |
| Lung, g | 215 ± 36.1 | 260 ± 43.1 | 0.078 |
| Kidney, g | 54.7 ± 4.61 | 62.1 ± 4.18 | 0.016 |
| Heart OI, % BW | 0.383 ± 0.034 | 0.426 ± 0.012 | 0.028 |
| Liver OI, % BW | 1.74 ± 0.37 | 1.67 ± 0.038 | 0.652 |
| Spleen OI, % BW | 0.112 ± 0.007 | 0.145 ±0.026 | 0.023 |
| Lung OI, % BW | 1.29 ± 0.31 | 1.47 ± 0.089 | 0.256 |
| Kidney OI, % BW | 0.326 ± 0.038 | 0.333 ± 0.018 | 0.667 |
| ADG a, g/d | 67.1± 19.2 | 125 ± 31.9 | 0.006 |
| Item | NC | HC | p-Values |
|---|---|---|---|
| Blood a, EU/mL | 1.84 ± 0.115 | 1.77 ± 0.143 | 0.520 |
| Spleen, ng/mgprot | 180 ± 42.4 | 176 ± 38.0 | 0.889 |
| Liver, ng/mgprot | 135 ± 34.7 | 87.5 ± 6.27 | 0.015 |
| Items | NC | HC | p Values |
|---|---|---|---|
| Cell Cycle | |||
| ABL2 | 1.04 ± 0.35 | 0.88 ± 0.49 | 0.574 |
| CASP2 | 1.01 ± 0.14 | 1.25 ± 0.33 | 0.218 |
| CASP3 | 1.07 ± 0.40 | 1.14 ± 0.52 | 0.82 |
| CASP7 | 1.09 ± 0.49 | 0.38 ± 0.11 | 0.017 |
| CCNT1 | 1.01 ± 0.14 | 0.51 ± 0.25 | 0.015 |
| CDC14A | 1.04 ± 0.33 | 0.54 ± 0.31 | 0.04 |
| CDC23 | 1.02 ± 0.21 | 0.65 ± 0.10 | 0.006 |
| CDC25C | 1.06 ± 0.41 | 0.78 ± 0.15 | 0.156 |
| CDC42 | 1.10 ± 0.45 | 0.86 ± 0.24 | 0.304 |
| CDK6 | 1.02 ± 0.26 | 0.61 ± 0.22 | 0.027 |
| CDKN2AIPNL | 1.05 ± 0.38 | 0.71 ± 0.36 | 0.213 |
| ITGBL1 | 1.06 ± 0.43 | 0.75 ± 0.24 | 0.178 |
| MAPK1 | 1.04 ± 0.32 | 0.69 ± 0.33 | 0.135 |
| MRE11A | 1.08 ± 0.48 | 0.65 ± 0.24 | 0.099 |
| NEK3 | 1.02 ± 0.26 | 0.96 ± 0.32 | 0.746 |
| RHOV | 1.14 ± 0.63 | 2.32 ± 1.05 | 0.091 |
| STMN2 | 1.01 ± 0.15 | 1.00 ± 0.29 | 0.954 |
| Inflammation | |||
| CCL28 | 1.03 ± 0.31 | 0.71 ± 0.23 | 0.089 |
| CXCR4 | 1.06 ± 0.43 | 0.82 ± 0.17 | 0.246 |
| IFNGR1 | 1.03 ± 0.30 | 0.47 ± 0.24 | 0.016 |
| Other Genes | |||
| AKR1B1 | 1.01 ± 0.35 | 0.88 ± 0.49 | 0.574 |
| LGMN | 1.07 ± 0.43 | 0.95 ± 0.17 | 0.553 |
| CPPED1 | 1.03 ± 0.29 | 0.42 ± 0.18 | 0.006 |
| ERP44 | 1.01 ± 0.18 | 0.88 ± 0.28 | 0.425 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, Q.; Wang, K.; Han, X.; Tan, Z. The Regulatory Mechanism of Feeding a Diet High in Rice Grain on the Growth and microRNA Expression Profiles of the Spleen, Taking Goats as an Artiodactyl Model. Biology 2021, 10, 832. https://doi.org/10.3390/biology10090832
Yan Q, Wang K, Han X, Tan Z. The Regulatory Mechanism of Feeding a Diet High in Rice Grain on the Growth and microRNA Expression Profiles of the Spleen, Taking Goats as an Artiodactyl Model. Biology. 2021; 10(9):832. https://doi.org/10.3390/biology10090832
Chicago/Turabian StyleYan, Qiongxian, Kaijun Wang, Xuefeng Han, and Zhiliang Tan. 2021. "The Regulatory Mechanism of Feeding a Diet High in Rice Grain on the Growth and microRNA Expression Profiles of the Spleen, Taking Goats as an Artiodactyl Model" Biology 10, no. 9: 832. https://doi.org/10.3390/biology10090832
APA StyleYan, Q., Wang, K., Han, X., & Tan, Z. (2021). The Regulatory Mechanism of Feeding a Diet High in Rice Grain on the Growth and microRNA Expression Profiles of the Spleen, Taking Goats as an Artiodactyl Model. Biology, 10(9), 832. https://doi.org/10.3390/biology10090832






