Saccharomyces cerevisiae Promoter Engineering before and during the Synthetic Biology Era
Simple Summary
Abstract
1. Introduction
2. The Structure of S. cerevisiae Promoters
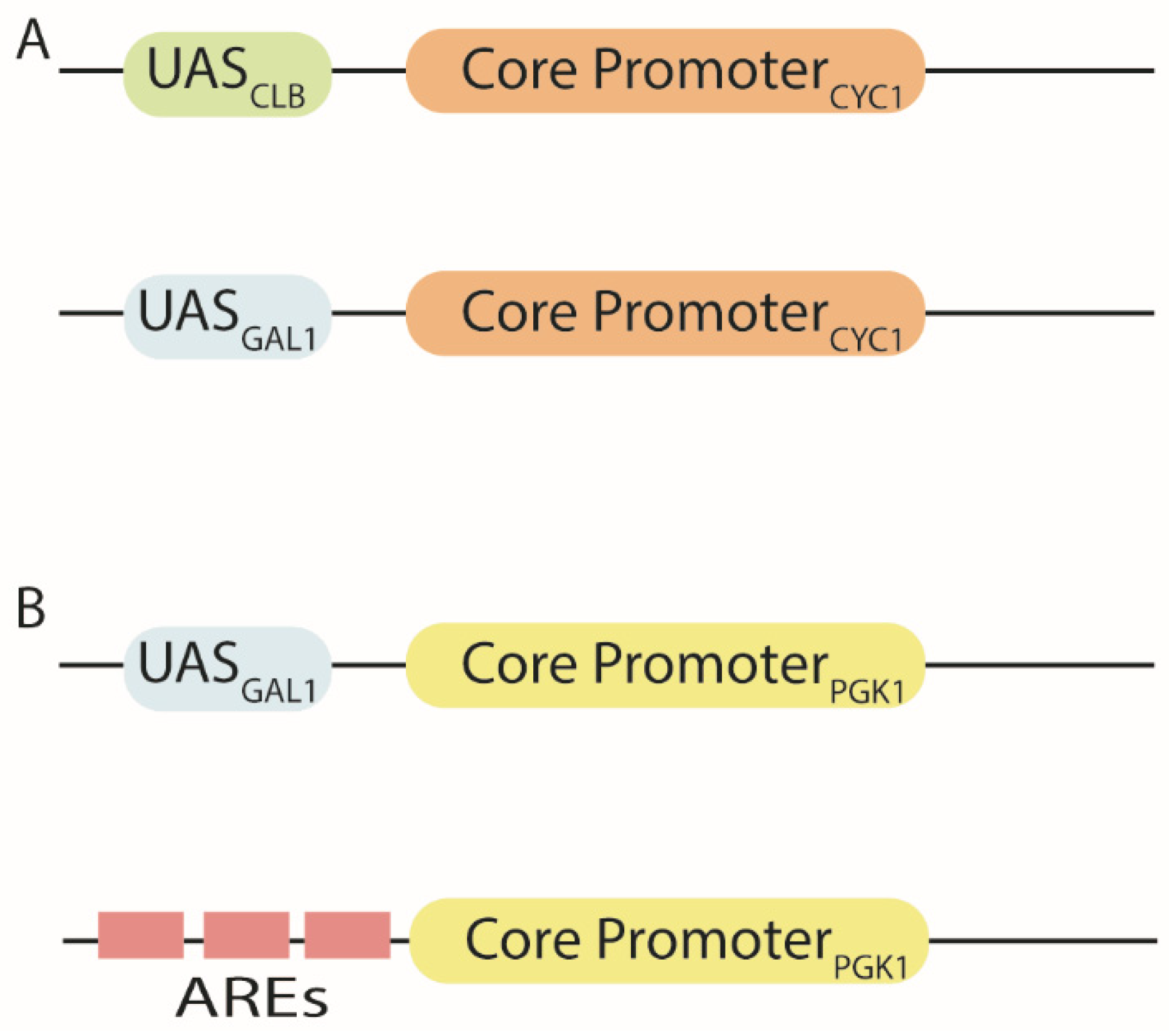
3. Hybrid Promoters
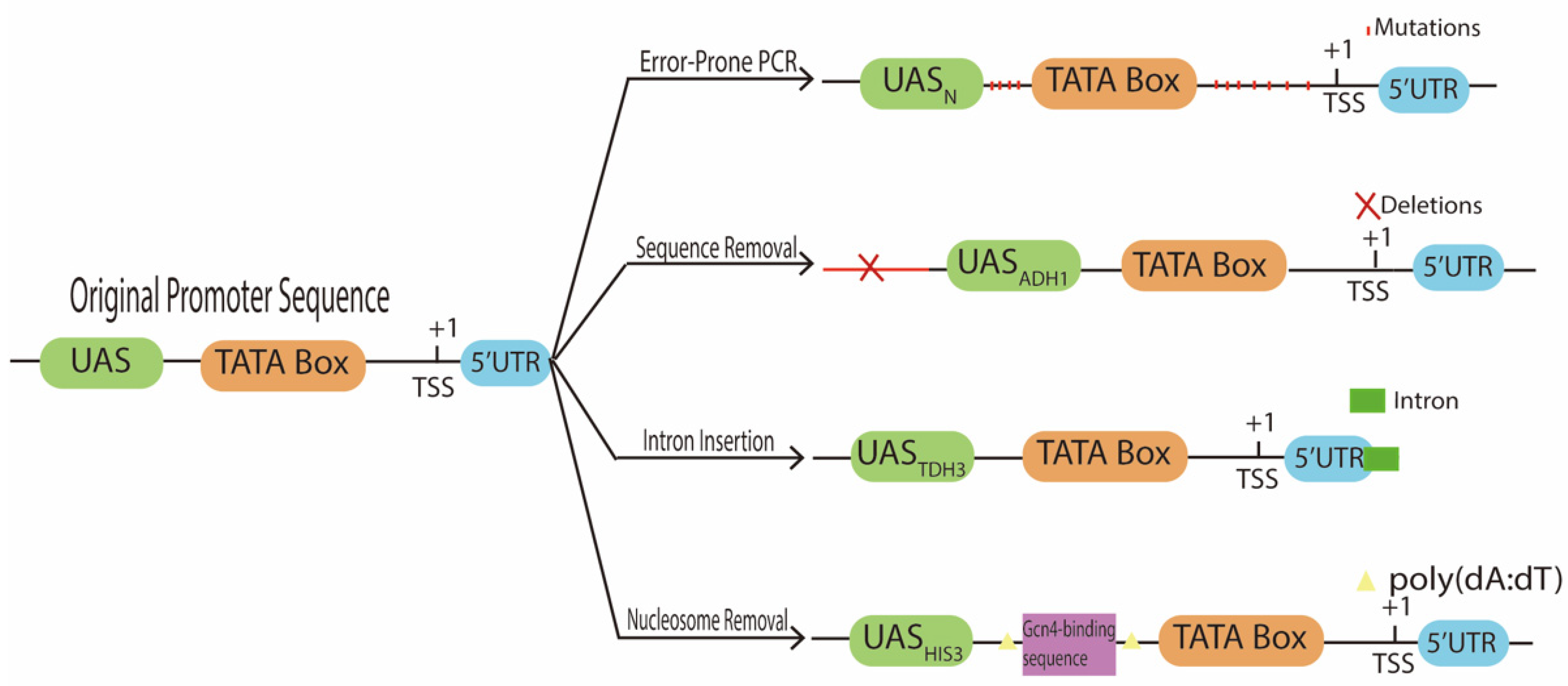
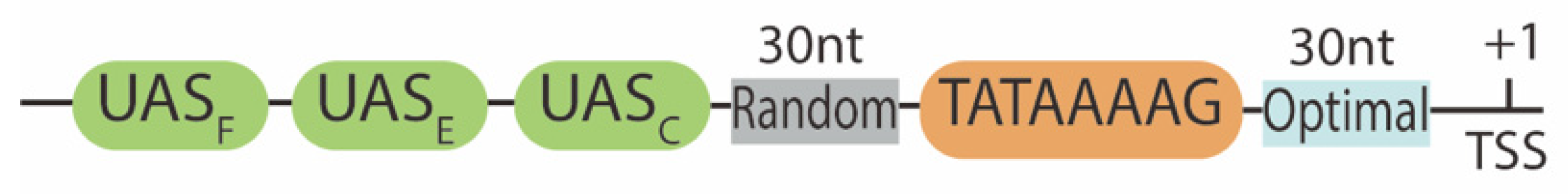
4. Promoter Sequence Modification
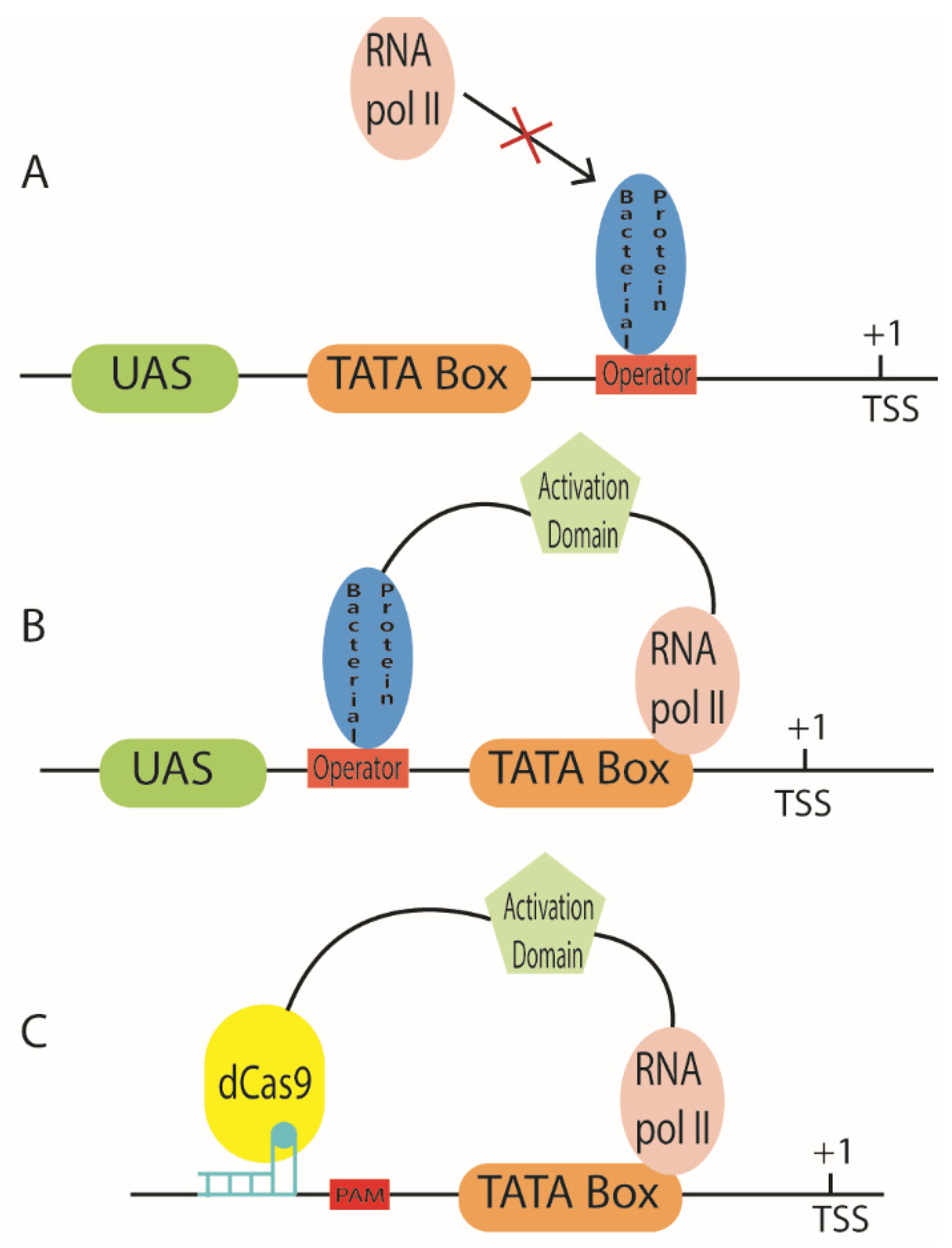
5. Synthetic Promoters Regulated by Bacterial Proteins
5.1. LexA-Regulated Promoters
5.2. TetR-Regulated Promoters
5.3. LacI-Regulated Promoters
5.4. XylR-Regulated Promoters
5.5. FadR- and FapR-Regulated Promoters
5.6. MetJ- and BenM-Regulated Promoters
6. Templates for Synthetic Transcription Factors
6.1. Zinc Finger Protein (ZFP)-Regulated Promoters
6.2. TAL Effector-Regulated Promoters
6.3. CRISPR-Cas-Regulated Promoters
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Elowitz, M.B.; Leibler, S. A synthetic oscillatory network of transcriptional regulators. Nat. Cell Biol. 2000, 403, 335–338. [Google Scholar] [CrossRef]
- Gardner, T.S.; Cantor, C.R.; Collins, J.J. Construction of a genetic toggle switch in Escherichia coli. Nat. Cell Biol. 2000, 403, 339–342. [Google Scholar] [CrossRef] [PubMed]
- Endy, D. Foundations for engineering biology. Nat. Cell Biol. 2005, 438, 449–453. [Google Scholar] [CrossRef] [PubMed]
- Pickens, L.B.; Tang, Y.; Chooi, Y.-H. Metabolic Engineering for the Production of Natural Products. Annu. Rev. Chem. Biomol. Eng. 2011, 2, 211–236. [Google Scholar] [CrossRef] [PubMed]
- Ro, D.-K.; Paradise, E.M.; Ouellet, M.; Fisher, K.J.; Newman, K.L.; Ndungu, J.M.; Ho, K.A.; Eachus, R.A.; Ham, T.S.; Kirby, J.; et al. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nat. Cell Biol. 2006, 440, 940–943. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Zhang, Y.; Nielsen, J. Synthetic Biology of Yeast. Biochem. 2019, 58, 1511–1520. [Google Scholar] [CrossRef]
- Auxillos, J.Y.; Garcia-Ruiz, E.; Jones, S.; Li, T.; Jiang, S.; Dai, J.; Cai, Y. Multiplex Genome Engineering for Optimizing Bioproduction in Saccharomyces cerevisiae. Biochem. 2019, 58, 1492–1500. [Google Scholar] [CrossRef]
- Lu, C.; Jeffries, T. Shuffling of Promoters for Multiple Genes to Optimize Xylose Fermentation in an Engineered Saccharomyces cerevisiae Strain. Appl. Environ. Microbiol. 2007, 73, 6072–6077. [Google Scholar] [CrossRef]
- Wisselink, H.W.; Toirkens, M.J.; Berriel, M.D.R.F.; Winkler, A.A.; Van Dijken, J.P.; Pronk, J.T.; Van Maris, A.J.A. Engineering of Saccharomyces cerevisiae for Efficient Anaerobic Alcoholic Fermentation of l-Arabinose. Appl. Environ. Microbiol. 2007, 73, 4881–4891. [Google Scholar] [CrossRef]
- Da Silva, N.A.; Srikrishnan, S. Introduction and expression of genes for metabolic engineering applications in Saccharomyces cerevisiae. FEMS Yeast Res. 2012, 12, 197–214. [Google Scholar] [CrossRef]
- Nevoigt, E.; Kohnke, J.; Fischer, C.R.; Alper, H.; Stahl, U.; Stephanopoulos, G. Engineering of Promoter Replacement Cassettes for Fine-Tuning of Gene Expression in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2006, 72, 5266–5273. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Shao, Z.; Zhao, H.; Nair, N.; Wen, F.; Xu, J.-H.; Zhao, H. Cloning and characterization of a panel of constitutive promoters for applications in pathway engineering in Saccharomyces cerevisiae. Biotechnol. Bioeng. 2012, 109, 2082–2092. [Google Scholar] [CrossRef]
- Redden, H.; Morse, N.; Alper, H.S. The synthetic biology toolbox for tuning gene expression in yeast. FEMS Yeast Res. 2014, 15, 1–10. [Google Scholar] [CrossRef] [PubMed]
- De Boer, C.G.; Vaishnav, E.D.; Sadeh, R.; Abeyta, E.L.; Friedman, N.; Regev, A. Deciphering eukaryotic gene-regulatory logic with 100 million random promoters. Nat. Biotechnol. 2020, 38, 56–65. [Google Scholar] [CrossRef]
- Hubmann, G.; Thevelein, J.M.; Nevoigt, E. Natural and Modified Promoters for Tailored Metabolic Engineering of the Yeast Saccharomyces cerevisiae. Methods Mol. Biol. 2014, 1152, 17–42. [Google Scholar] [CrossRef]
- Hahn, S.; Young, E.T. Transcriptional Regulation in Saccharomyces cerevisiae: Transcription Factor Regulation and Function, Mechanisms of Initiation, and Roles of Activators and Coactivators. Genet. 2011, 189, 705–736. [Google Scholar] [CrossRef]
- Forsburg, S.L.; Guarente, L. Mutational analysis of upstream activation sequence 2 of the CYC1 gene of Saccharomyces cerevisiae: A HAP2-HAP3-responsive site. Mol. Cell. Biol. 1988, 8, 647–654. [Google Scholar] [CrossRef]
- Ogden, J.E.; Stanway, C.; Kim, S.; Mellor, J.; Kingsman, A.J.; Kingsman, S.M. Efficient expression of the Saccharomyces cerevisiae PGK gene depends on an upstream activation sequence but does not require TATA sequences. Mol. Cell. Biol. 1986, 6, 4335–4343. [Google Scholar] [CrossRef]
- Swamy, K.B.S.; Cho, C.-Y.; Chiang, S.; Tsai, Z.T.-Y.; Tsai, H.-K. Impact of DNA-binding position variants on yeast gene expression. Nucleic Acids Res. 2009, 37, 6991–7001. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lubliner, S.; Keren, L.; Segal, E. Sequence features of yeast and human core promoters that are predictive of maximal promoter activity. Nucleic Acids Res. 2013, 41, 5569–5581. [Google Scholar] [CrossRef]
- Lubliner, S.; Regev, I.; Lotan-Pompan, M.; Edelheit, S.; Weinberger, A.; Segal, E. Core promoter sequence in yeast is a major determinant of expression level. Genome Res. 2015, 25, 1008–1017. [Google Scholar] [CrossRef]
- Redden, H.; Alper, H.S. The development and characterization of synthetic minimal yeast promoters. Nat. Commun. 2015, 6, 7810. [Google Scholar] [CrossRef]
- Wobbe, C.R.; Struhl, K. Yeast and human TATA-binding proteins have nearly identical DNA sequence requirements for transcription in vitro. Mol. Cell. Biol. 1990, 10, 3859–3867. [Google Scholar] [CrossRef] [PubMed]
- Mishra, A.K.; Vanathi, P.; Bhargava, P. The transcriptional activator GAL4-VP16 regulates the intra-molecular interactions of the TATA-binding protein. J. Biosci. 2003, 28, 423–436. [Google Scholar] [CrossRef]
- Mogno, I.; Vallania, F.; Mitra, R.; Cohen, B.A. TATA is a modular component of synthetic promoters. Genome Res. 2010, 20, 1391–1397. [Google Scholar] [CrossRef]
- Zhang, Z.; Dietrich, F.S. Mapping of transcription start sites in Saccharomyces cerevisiae using 5′ SAGE. Nucleic. Acids. Res. 2005, 33, 2838–2851. [Google Scholar] [CrossRef]
- Hahn, S.; Hoar, E.T.; Guarente, L. Each of three “TATA elements” specifies a subset of the transcription initiation sites at the CYC-1 promoter of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 1985, 82, 8562–8566. [Google Scholar] [CrossRef]
- Guarente, L.; Lalonde, B.; Gifford, P.; Alani, E. Distinctly regulated tandem upstream activation sites mediate catabolite repression of the CYC1 gene of S. cerevisiae. Cell 1984, 36, 503–511. [Google Scholar] [CrossRef]
- Guarente, L.; Ptashne, M. Fusion of Escherichia coli lacZ to the cytochrome c gene of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 1981, 78, 2199–2203. [Google Scholar] [CrossRef] [PubMed]
- Martens, C.; Krett, B.; Laybourn, P.J. RNA polymerase II and TBP occupy the repressed CYC1 promoter. Mol. Microbiol. 2001, 40, 1009–1019. [Google Scholar] [CrossRef] [PubMed]
- Guarente, L.; Yocum, R.R.; Gifford, P. A GAL10-CYC1 hybrid yeast promoter identifies the GAL4 regulatory region as an upstream site. Proc. Natl. Acad. Sci. USA 1982, 79, 7410–7414. [Google Scholar] [CrossRef]
- Blazeck, J.; Liu, L.; Redden, H.; Alper, H. Tuning Gene Expression in Yarrowia lipolytica by a Hybrid Promoter Approach. Appl. Environ. Microbiol. 2011, 77, 7905–7914. [Google Scholar] [CrossRef]
- Guarente, L. Yeast promoters and lacZ fusions designed to study expression of cloned genes in yeast. Methods Enzymol. 1983, 101, 181–191. [Google Scholar]
- Da Silva, N.A.; Bailey, J.E. Influence of plasmid origin and promoter strength in fermentations of recombinant yeast. Biotechnol. Bioeng. 1991, 37, 318–324. [Google Scholar] [CrossRef]
- Hadiji-Abbes, N.; Borchani-Chabchoub, I.; Triki, H.; Ellouz, R.; Gargouri, A.; Mokdad-Gargouri, R. Expression of HBsAg and preS2-S protein in different yeast based system: A comparative analysis. Protein Expr. Purif. 2009, 66, 131–137. [Google Scholar] [CrossRef] [PubMed]
- Sharon, E.; Kalma, Y.; Sharp, A.; Raveh-Sadka, T.; Levo, M.; Zeevi, D.; Keren, L.; Yakhini, Z.; Weinberger, A.; Segal, E. Inferring gene regulatory logic from high-throughput measurements of thousands of systematically designed promoters. Nat. Biotechnol. 2012, 30, 521–530. [Google Scholar] [CrossRef]
- Rajkumar, A.S.; Dénervaud, N.; Maerkl, S.J. Mapping the fine structure of a eukaryotic promoter input-output function. Nat. Genet. 2013, 45, 1207–1215. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Marchisio, M.A. Novel S. cerevisiae Hybrid Synthetic Promoters Based on Foreign Core Promoter Sequences. Int. J. Mol. Sci. 2021, 22, 5704. [Google Scholar] [CrossRef] [PubMed]
- Blazeck, J.; Garg, R.; Reed, B.; Alper, H.S. Controlling promoter strength and regulation in Saccharomyces cerevisiae using synthetic hybrid promoters. Biotechnol. Bioeng. 2012, 109, 2884–2895. [Google Scholar] [CrossRef]
- Bitter, G.A.; Egan, K.M. Expression of interferon-gamma from hybrid yeast GPD promoters containing upstream regulatory sequences from the GAL1-GAL10 intergenic region. Gene 1988, 69, 193–207. [Google Scholar] [CrossRef]
- Purvis, I.J.; Chotai, D.; Dykes, C.W.; Lubahn, D.B.; French, F.S.; Wilson, E.M.; Hobden, A.N. An androgen-inducible expression system for Saccharomyces cerevisiae. Gene 1991, 106, 35–42. [Google Scholar] [CrossRef]
- Iraqui, I.; Vissers, S.; Andreé, B.; Urrestarazu, A. Transcriptional Induction by Aromatic Amino Acids in Saccharomyces cerevisiae. Mol. Cell. Biol. 1999, 19, 3360–3371. [Google Scholar] [CrossRef]
- Kim, S.; Lee, K.; Bae, S.-J.; Hahn, J.-S. Promoters inducible by aromatic amino acids and gamma-aminobutyrate (GABA) for metabolic engineering applications in Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2015, 99, 2705–2714. [Google Scholar] [CrossRef]
- Leavitt, J.M.; Tong, A.; Tong, J.; Pattie, J.; Alper, H.S. Coordinated transcription factor and promoter engineering to establish strong expression elements in Saccharomyces cerevisiae. Biotechnol. J. 2016, 11, 866–876. [Google Scholar] [CrossRef]
- Brandman, O.; Stewart-Ornstein, J.; Wong, D.; Larson, A.; Williams, C.C.; Li, G.-W.; Zhou, S.; King, D.; Shen, P.S.; Weibezahn, J.; et al. A Ribosome-Bound Quality Control Complex Triggers Degradation of Nascent Peptides and Signals Translation Stress. Cell 2012, 151, 1042–1054. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Sonnenschein, N.; Pihl, T.; Pedersen, K.R.; Jensen, M.K.; Keasling, J.D. Engineering an NADPH/NADP+Redox Biosensor in Yeast. ACS Synth. Biol. 2016, 5, 1546–1556. [Google Scholar] [CrossRef] [PubMed]
- Rajkumar, A.S.; Liu, G.; Bergenholm, D.; Arsovska, D.; Kristensen, M.; Nielsen, J.; Jensen, M.K.; Keasling, J. Engineering of synthetic, stress-responsive yeast promoters. Nucleic Acids Res. 2016, 44, e136. [Google Scholar] [CrossRef]
- Rajkumar, A.S.; Özdemir, E.; Lis, A.V.; Schneider, K.; Qin, J.; Jensen, M.K.; Keasling, J.D. Engineered Reversal of Function in Glycolytic Yeast Promoters. ACS Synth. Biol. 2019, 8, 1462–1468. [Google Scholar] [CrossRef] [PubMed]
- Ruohonen, L.; Penttilä, M.; Keränen, S. Optimization ofBacillus α-amylase production bySaccharomyces cerevisiae. Yeast 1991, 7, 337–346. [Google Scholar] [CrossRef] [PubMed]
- Ruohonen, L.; Aalto, M.K.; Keranen, S. Modifications to the ADH1 promoter of Saccharomyces cerevisiae for efficient production of heterologous proteins. J. Biotechnol. 1995, 39, 193–203. [Google Scholar] [CrossRef]
- Hoshida, H.; Kondo, M.; Kobayashi, T.; Yarimizu, T.; Akada, R. 5’-UTR introns enhance protein expression in the yeast Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2016, 101, 241–251. [Google Scholar] [CrossRef]
- Raveh-Sadka, T.; Levo, M.; Shabi, U.; Shany, B.; Keren, L.; Lotan-Pompan, M.; Zeevi, D.; Sharon, E.; Weinberger, A.; Segal, E. Manipulating nucleosome disfavoring sequences allows fine-tune regulation of gene expression in yeast. Nat. Genet. 2012, 44, 743–750. [Google Scholar] [CrossRef] [PubMed]
- Alper, H.; Fischer, C.; Nevoigt, E.; Stephanopoulos, G. Tuning genetic control through promoter engineering. Proc. Natl. Acad. Sci. USA 2005, 102, 12678–12683. [Google Scholar] [CrossRef]
- Du, J.; Yuan, Y.; Si, T.; Lian, J.; Zhao, H. Customized optimization of metabolic pathways by combinatorial transcriptional engineering. Nucleic Acids Res. 2012, 40, e142. [Google Scholar] [CrossRef]
- Ingolia, N.T.; Murray, A.W. Positive-Feedback Loops as a Flexible Biological Module. Curr. Biol. 2007, 17, 668–677. [Google Scholar] [CrossRef]
- Williams, T.; Averesch, N.; Winter, G.; Plan, M.; Vickers, C.; Nielsen, L.; Krömer, J. Quorum-sensing linked RNA interference for dynamic metabolic pathway control in Saccharomyces cerevisiae. Metab. Eng. 2015, 29, 124–134. [Google Scholar] [CrossRef] [PubMed]
- Curran, K.A.; Crook, N.C.; Karim, A.S.; Gupta, A.R.; Wagman, A.M.; Alper, H.S. Design of synthetic yeast promoters via tuning of nucleosome architecture. Nat. Commun. 2014, 5, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Xi, L.; Fondufe-Mittendorf, Y.; Xia, L.; Flatow, J.; Widom, J.; Wang, J.-P. Predicting nucleosome positioning using a duration Hidden Markov Model. BMC Bioinform. 2010, 11, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Myburgh, M.W.; Rose, S.H.; Viljoen-Bloom, M. Evaluating and engineering Saccharomyces cerevisiae promoters for increased amylase expression and bioethanol production from raw starch. FEMS Yeast Res. 2020, 20. [Google Scholar] [CrossRef]
- Shi, S.; Choi, Y.W.; Zhao, H.; Tan, M.H.; Ang, E.L. Discovery and engineering of a 1-butanol biosensor in Saccharomyces cerevisiae. Bioresour. Technol. 2017, 245, 1343–1351. [Google Scholar] [CrossRef] [PubMed]
- Decoene, T.; De Maeseneire, S.L.; De Mey, M. Modulating transcription through development of semi-synthetic yeast core promoters. PLoS ONE 2019, 14, e0224476. [Google Scholar] [CrossRef]
- Song, W.; Li, J.; Liang, Q.; Marchisio, M.A. Can terminators be used as insulators into yeast synthetic gene circuits? J. Biol. Eng. 2016, 10, 19. [Google Scholar] [CrossRef]
- Guo, Z.; Sherman, F. Signals sufficient for 3′-end formation of yeast mRNA. Mol. Cell. Biol. 1996, 16, 2772–2776. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, G.; Lamantea, E.; Donati, A.; Filosto, M.; Briem, E.; Carrara, F.; Parini, R.; Simonati, A.; Santer, R.; Zeviani, M. Infantile hepatocerebral syndromes associated with mutations in the mitochondrial DNA polymerase-gammaA. Brain 2005, 128 Pt 4, 723–731. [Google Scholar] [CrossRef]
- Brambilla, A.; Mainieri, D.; Carbone, M.L.A. A simple signal element mediates transcription termination and mRNA 3′ end formation in the DEG1 gene of Saccharomyces cerevisiae. Mol. Genet. Genom. 1997, 254, 681–688. [Google Scholar] [CrossRef] [PubMed]
- Wertman, K.F.; Mount, D.W. Nucleotide sequence binding specificity of the LexA repressor of Escherichia coli K-12. J. Bacteriol. 1985, 163, 376–384. [Google Scholar] [CrossRef] [PubMed]
- Brent, R.; Ptashne, M. Mechanism of action of the lexA gene product. Proc. Natl. Acad. Sci. USA 1981, 78, 4204–4208. [Google Scholar] [CrossRef]
- Brent, R.; Ptashne, M. A bacterial repressor protein or a yeast transcriptional terminator can block upstream activation of a yeast gene. Nat. Cell Biol. 1984, 312, 612–615. [Google Scholar] [CrossRef]
- Brent, R.; Ptashne, M. A eukaryotic transcriptional activator bearing the DNA specificity of a prokaryotic repressor. Cell 1985, 43, 729–736. [Google Scholar] [CrossRef]
- Mizuno, T.; Wurtzel, E.T.; Inouye, M. Osmoregulation of gene expression. II. DNA sequence of the envZ gene of the ompB operon of Escherichia coli and characterization of its gene product. J. Biol. Chem. 1982, 257, 13692–13698. [Google Scholar] [CrossRef]
- Ruden, D.M.; Ma, J.; Li, Y.; Wood, K.; Ptashne, M. Generating yeast transcriptional activators containing no yeast protein sequences. Nat. Cell Biol. 1991, 350, 250–252. [Google Scholar] [CrossRef]
- Ma, J.; Ptashne, M. A new class of yeast transcriptional activators. Cell 1987, 51, 113–119. [Google Scholar] [CrossRef]
- Keleher, C.A.; Redd, M.J.; Schultz, J.; Carlson, M.; Johnson, A.D. Ssn6-Tup1 is a general repressor of transcription in yeast. Cell 1992, 68, 709–719. [Google Scholar] [CrossRef]
- Louvion, J.-F.; Havaux-Copf, B.; Picard, D. Fusion of GAL4-VP16 to a steroid-binding domain provides a tool for gratuitous induction of galactose-responsive genes in yeast. Gene 1993, 131, 129–134. [Google Scholar] [CrossRef]
- Triezenberg, S.J.; Kingsbury, R.C.; McKnight, S.L. Functional dissection of VP16, the trans-activator of herpes simplex virus immediate early gene expression. Genes Dev. 1988, 2, 718–729. [Google Scholar] [CrossRef] [PubMed]
- McIsaac, R.S.; Silverman, S.J.; McClean, M.N.; Gibney, P.A.; Macinskas, J.; Hickman, M.; Petti, A.A.; Botstein, D. Fast-acting and nearly gratuitous induction of gene expression and protein depletion inSaccharomyces cerevisiae. Mol. Biol. Cell 2011, 22, 4447–4459. [Google Scholar] [CrossRef]
- Gericke, A.; Hühnerfuss, H. IR reflection absorption spectroscopy: A versatile tool for studying interfacial enzymatic processes. Chem. Phys. Lipids 1994, 74, 205–210. [Google Scholar] [CrossRef]
- Ajo-Franklin, C.M.; Drubin, D.A.; Eskin, J.A.; Gee, E.P.; Landgraf, D.; Phillips, I.; Silver, P.A. Rational design of memory in eukaryotic cells. Genes Dev. 2007, 21, 2271–2276. [Google Scholar] [CrossRef]
- Ottoz, D.S.; Rudolf, F.; Stelling, J. Inducible, tightly regulated and growth condition-independent transcription factor in Saccharomyces cerevisiae. Nucleic Acids Res. 2014, 42, e130. [Google Scholar] [CrossRef]
- Dossani, Z.Y.; Apel, A.R.; Szmidt-Middleton, H.; Hillson, N.J.; Deutsch, S.; Keasling, J.D.; Mukhopadhyay, A. A combinatorial approach to synthetic transcription factor-promoter combinations for yeast strain engineering. Yeast 2017, 35, 273–280. [Google Scholar] [CrossRef]
- Rantasalo, A.; Czeizler, E.; Virtanen, R.; Rousu, J.; Lähdesmäki, H.; Penttilä, M.; Jäntti, J.; Mojzita, D. Synthetic Transcription Amplifier System for Orthogonal Control of Gene Expression in Saccharomyces cerevisiae. PLoS ONE 2016, 11, e0148320. [Google Scholar] [CrossRef] [PubMed]
- Rantasalo, A.; Kuivanen, J.; Penttilä, M.; Jäntti, J.; Mojzita, D. Synthetic Toolkit for Complex Genetic Circuit Engineering in Saccharomyces cerevisiae. ACS Synth. Biol. 2018, 7, 1573–1587. [Google Scholar] [CrossRef] [PubMed]
- Labow, M.A.; Baim, S.B.; Shenk, T.; Levine, A.J. Conversion of the lac repressor into an allosterically regulated transcriptional activator for mammalian cells. Mol. Cell. Biol. 1990, 10, 3343–3356. [Google Scholar] [CrossRef]
- Rantasalo, A.; Landowski, C.P.; Kuivanen, J.; Korppoo, A.; Reuter, L.; Koivistoinen, O.; Valkonen, M.; Penttilä, M.; Jäntti, J.; Mojzita, D. A universal gene expression system for fungi. Nucleic Acids Res. 2018, 46, e111. [Google Scholar] [CrossRef] [PubMed]
- Hillen, W.; Berens, C. Mechanisms Underlying Expression of TN10 Encoded Tetracycline Resistance. Annu. Rev. Microbiol. 1994, 48, 345–369. [Google Scholar] [CrossRef]
- Gossen, M.; Bujard, H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. USA 1992, 89, 5547–5551. [Google Scholar] [CrossRef] [PubMed]
- Gossen, M.; Freundlieb, S.; Bender, G.; Muller, G.; Hillen, W.; Bujard, H. Transcriptional activation by tetracyclines in mammalian cells. Science 1995, 268, 1766–1769. [Google Scholar] [CrossRef]
- Garí, E.; Piedrafita, L.; Aldea, M.; Herrero, E. A Set of Vectors with a Tetracycline-Regulatable Promoter System for Modulated Gene Expression inSaccharomyces cerevisiae. Yeast 1997, 13, 837–848. [Google Scholar] [CrossRef]
- Belli, G. An activator/repressor dual system allows tight tetracycline-regulated gene expression in budding yeast. Nucleic Acids Res. 1998, 26, 942–947. [Google Scholar] [CrossRef]
- Cuperus, J.T.; Lo, R.S.; Shumaker, L.; Proctor, J.; Fields, S. A tetO Toolkit to Alter Expression of Genes in Saccharomyces cerevisiae. ACS Synth. Biol. 2015, 4, 842–852. [Google Scholar] [CrossRef] [PubMed]
- Mnaimneh, S.; Davierwala, A.P.; Haynes, J.; Moffat, J.; Peng, W.-T.; Zhang, W.; Yang, X.; Pootoolal, J.; Chua, G.; Lopez, A.; et al. Exploration of Essential Gene Functions via Titratable Promoter Alleles. Cell 2004, 118, 31–44. [Google Scholar] [CrossRef]
- Murphy, K.F.; Balázsi, G.; Collins, J.J. Combinatorial promoter design for engineering noisy gene expression. Proc. Natl. Acad. Sci. USA 2007, 104, 12726–12731. [Google Scholar] [CrossRef] [PubMed]
- Ellis, T.; Wang, X.; Collins, J.J. Diversity-based, model-guided construction of synthetic gene networks with predicted functions. Nat. Biotechnol. 2009, 27, 465–471. [Google Scholar] [CrossRef]
- Blount, B.A.; Weenink, T.; Vasylechko, S.; Ellis, T. Rational Diversification of a Promoter Providing Fine-Tuned Expression and Orthogonal Regulation for Synthetic Biology. PLoS ONE 2012, 7, e33279. [Google Scholar] [CrossRef]
- Oehler, S.; Amouyal, M.; Kolkhof, P.; von Wilcken-Bergmann, B.; Müller-Hill, B. Quality and position of the three lac operators of E. coli define efficiency of repression. EMBO J. 1994, 13, 3348–3355. [Google Scholar] [CrossRef]
- Grilly, C.; Stricker, J.; Pang, W.L.; Bennett, M.R.; Hasty, J. A synthetic gene network for tuning protein degradation in Saccharomyces cerevisiae. Mol. Syst. Biol. 2007, 3, 127. [Google Scholar] [CrossRef]
- Marchisio, M.A. In silico design and in vivo implementation of yeast gene Boolean gates. J. Biol. Eng. 2014, 8, 6. [Google Scholar] [CrossRef] [PubMed]
- Mazumder, M.; McMillen, D.R. Design and characterization of a dual-mode promoter with activation and repression capability for tuning gene expression in yeast. Nucleic Acids Res. 2014, 42, 9514–9522. [Google Scholar] [CrossRef] [PubMed]
- Gnugge, R.; Dharmarajan, L.; Lang, M.; Stelling, J. An Orthogonal Permease-Inducer-Repressor Feedback Loop Shows Bistability. ACS Synth. Biol. 2016, 5, 1098–1107. [Google Scholar] [CrossRef] [PubMed]
- Bandiera, L.; Hou, Z.; Kothamachu, V.B.; Balsa-Canto, E.; Swain, P.S.; Menolascina, F. On-Line Optimal Input Design Increases the Efficiency and Accuracy of the Modelling of an Inducible Synthetic Promoter. Processes 2018, 6, 148. [Google Scholar] [CrossRef]
- Rodriguez, G.M.; Hussain, M.S.; Gambill, L.; Gao, D.; Yaguchi, A.; Blenner, M. Engineering xylose utilization in Yarrowia lipolytica by understanding its cryptic xylose pathway. Biotechnol. Biofuels 2016, 9, 1–15. [Google Scholar] [CrossRef]
- Teo, W.S.; Chang, M.W. Bacterial XylRs and synthetic promoters function as genetically encoded xylose biosensors inSaccharomyces cerevisiae. Biotechnol. J. 2015, 10, 315–322. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Li, S.; Zhao, H. Design and engineering of intracellular-metabolite-sensing/regulation gene circuits inSaccharomyces cerevisiae. Biotechnol. Bioeng. 2016, 113, 206–215. [Google Scholar] [CrossRef] [PubMed]
- Hector, R.E.; Mertens, J.A. A Synthetic Hybrid Promoter for Xylose-Regulated Control of Gene Expression in Saccharomyces Yeasts. Mol. Biotechnol. 2017, 59, 24–33. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.-P.; Shang, Y.; Zhang, P.; Liu, Y.; You, D.; Yin, B.-C.; Ye, B.-C. Engineering Prokaryotic Transcriptional Activator XylR as a Xylose-Inducible Biosensor for Transcription Activation in Yeast. ACS Synth. Biol. 2020, 9, 1022–1029. [Google Scholar] [CrossRef]
- Teo, W.S.; Hee, K.S.; Chang, M.W. Bacterial FadR and synthetic promoters function as modular fatty acid sensor- regulators inSaccharomyces cerevisiae. Eng. Life Sci. 2013, 13, 456–463. [Google Scholar] [CrossRef]
- Teo, W.S.; Chang, M.W. Development and characterization of AND-gate dynamic controllers with a modular synthetic GAL1 core promoter inSaccharomyces cerevisiae. Biotechnol. Bioeng. 2014, 111, 144–151. [Google Scholar] [CrossRef]
- Li, S.; Si, T.; Wang, M.; Zhao, H. Development of a Synthetic Malonyl-CoA Sensor in Saccharomyces cerevisiae for Intracellular Metabolite Monitoring and Genetic Screening. ACS Synth. Biol. 2015, 4, 1308–1315. [Google Scholar] [CrossRef]
- David, F.; Nielsen, J.; Siewers, V. Flux Control at the Malonyl-CoA Node through Hierarchical Dynamic Pathway Regulation in Saccharomyces cerevisiae. ACS Synth. Biol. 2016, 5, 224–233. [Google Scholar] [CrossRef]
- Dabirian, Y.; Teixeira, P.G.; Nielsen, J.; Siewers, V.; David, F. FadR-Based Biosensor-Assisted Screening for Genes Enhancing Fatty Acyl-CoA Pools in Saccharomyces cerevisiae. ACS Synth. Biol. 2019, 8, 1788–1800. [Google Scholar] [CrossRef]
- Dabirian, Y.; Li, X.; Chen, Y.; David, F.; Nielsen, J.; Siewers, V. Expanding the Dynamic Range of a Transcription Factor-Based Biosensor in Saccharomyces cerevisiae. ACS Synth. Biol. 2019, 8, 1968–1975. [Google Scholar] [CrossRef]
- Ambri, F.; D’Ambrosio, V.; Di Blasi, R.; Maury, J.; Jacobsen, S.A.B.; McCloskey, D.; Jensen, M.K.; Keasling, J.D. High-Resolution Scanning of Optimal Biosensor Reporter Promoters in Yeast. ACS Synth. Biol. 2019, 9, 218–226. [Google Scholar] [CrossRef]
- Qiu, C.; Chen, X.; Rexida, R.; Shen, Y.; Qi, Q.; Bao, X.; Hou, J. Engineering transcription factor-based biosensors for repressive regulation through transcriptional deactivation design in Saccharomyces cerevisiae. Microb. Cell Factories 2020, 19, 1–10. [Google Scholar] [CrossRef]
- Umeyama, T.; Okada, S.; Ito, T. Synthetic Gene Circuit-Mediated Monitoring of Endogenous Metabolites: Identification ofGAL11as a Novel Multicopy Enhancer ofS-Adenosylmethionine Level in Yeast. ACS Synth. Biol. 2013, 2, 425–430. [Google Scholar] [CrossRef]
- Skjoedt, M.L.; Snoek, T.; Kildegaard, K.R.; Arsovska, D.; Eichenberger, M.; Goedecke, T.J.; Rajkumar, A.S.; Zhang, J.; Kristensen, M.; Lehka, B.J.; et al. Engineering prokaryotic transcriptional activators as metabolite biosensors in yeast. Nat. Chem. Biol. 2016, 12, 951–958. [Google Scholar] [CrossRef] [PubMed]
- Mandell, J.G.; Barbas, C.F. Zinc Finger Tools: Custom DNA-binding domains for transcription factors and nucleases. Nucleic Acids Res. 2006, 34, W516–W523. [Google Scholar] [CrossRef] [PubMed]
- Khalil, A.S.; Lu, T.K.; Bashor, C.J.; Ramirez, C.L.; Pyenson, N.C.; Joung, J.K.; Collins, J.J. A Synthetic Biology Framework for Programming Eukaryotic Transcription Functions. Cell 2012, 150, 647–658. [Google Scholar] [CrossRef]
- McIsaac, R.S.; Oakes, B.L.; Wang, X.; Dummit, K.A.; Botstein, D.; Noyes, M.B. Synthetic gene expression perturbation systems with rapid, tunable, single-gene specificity in yeast. Nucleic Acids Res. 2012, 41, e57. [Google Scholar] [CrossRef] [PubMed]
- McIsaac, R.S.; Gibney, P.A.; Chandran, S.S.; Benjamin, K.R.; Botstein, D. Synthetic biology tools for programming gene expression without nutritional perturbations in Saccharomyces cerevisiae. Nucleic Acids Res. 2014, 42, e48. [Google Scholar] [CrossRef]
- Kotopka, B.J.; Smolke, C.D. Model-driven generation of artificial yeast promoters. Nat. Commun. 2020, 11, 1–13. [Google Scholar] [CrossRef]
- Bogdanove, A.J.; Voytas, D. TAL Effectors: Customizable Proteins for DNA Targeting. Sci. 2011, 333, 1843–1846. [Google Scholar] [CrossRef]
- De Lange, O.; Wolf, C.; Thiel, P.; Krüger, J.; Kleusch, C.; Kohlbacher, O.; Lahaye, T. DNA-binding proteins from marine bacteria expand the known sequence diversity of TALE-like repeats. Nucleic. Acids. Res. 2015, 43, 10065–10080. [Google Scholar] [CrossRef] [PubMed]
- Machens, F.; Balazadeh, S.; Mueller-Roeber, B.; Messerschmidt, K. Synthetic Promoters and Transcription Factors for Heterologous Protein Expression in Saccharomyces cerevisiae. Front. Bioeng. Biotechnol. 2017, 5, 63. [Google Scholar] [CrossRef]
- Ishino, Y.; Shinagawa, H.; Makino, K.; Amemura, M.; Nakata, A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J. Bacteriol. 1987, 169, 5429–5433. [Google Scholar] [CrossRef] [PubMed]
- Horvath, P.; Barrangou, R. CRISPR/Cas, the immune system of bacteria and archaea. Science 2010, 327, 167–170. [Google Scholar] [CrossRef] [PubMed]
- Marchisio, M.A.; Huang, Z. CRISPR-Cas type II-based Synthetic Biology applications in eukaryotic cells. RNA Biol. 2017, 14, 1286–1293. [Google Scholar] [CrossRef] [PubMed]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A Programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.S.; Larson, M.H.; Gilbert, L.A.; Doudna, J.A.; Weissman, J.S.; Arkin, A.P.; Lim, W.A. Repurposing CRISPR as an RNA-Guided Platform for Sequence-Specific Control of Gene Expression. Cell 2013, 152, 1173–1183. [Google Scholar] [CrossRef] [PubMed]
- Chavez, A.; Tuttle, M.; Pruitt, B.W.; Ewen-Campen, B.; Chari, R.; Ter-Ovanesyan, D.; Haque, S.J.; Cecchi, R.J.; Kowal, E.J.K.; Buchthal, J.; et al. Comparison of Cas9 activators in multiple species. Nat. Methods 2016, 13, 563–567. [Google Scholar] [CrossRef]
- Farzadfard, F.; Perli, S.D.; Lu, T.K. Tunable and Multifunctional Eukaryotic Transcription Factors Based on CRISPR/Cas. ACS Synth. Biol. 2013, 2, 604–613. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, L.A.; Larson, M.H.; Morsut, L.; Liu, Z.; Brar, G.A.; Torres, S.E.; Stern-Ginossar, N.; Brandman, O.; Whitehead, E.H.; Doudna, J.A.; et al. CRISPR-Mediated Modular RNA-Guided Regulation of Transcription in Eukaryotes. Cell 2013, 154, 442–451. [Google Scholar] [CrossRef]
- Zalatan, J.G.; Lee, M.E.; Almeida, R.; Gilbert, L.A.; Whitehead, E.H.; La Russa, M.; Tsai, J.; Weissman, J.S.; Dueber, J.E.; Qi, L.S.; et al. Engineering Complex Synthetic Transcriptional Programs with CRISPR RNA Scaffolds. Cell 2015, 160, 339–350. [Google Scholar] [CrossRef]
- Gander, M.W.; Vrana, J.D.; Voje, W.E.; Carothers, J.M.; Klavins, E. Digital logic circuits in yeast with CRISPR-dCas9 NOR gates. Nat. Commun. 2017, 8, 15459. [Google Scholar] [CrossRef]
- Gao, Y.; Zhao, Y. Self-processing of ribozyme-flanked RNAs into guide RNAs in vitro and in vivo for CRISPR-mediated genome editing. J. Integr. Plant Biol. 2014, 56, 343–349. [Google Scholar] [CrossRef]
- Alon, U. An Introduction to Systems Biology: Design Principles of Biological Circuits; Chapman & Hall/CRC Press: Boca Raton, FL, USA, 2006. [Google Scholar]
- Klipp, E.; Liebermeister, W.; Wierling, C.; Kowald, A.; Lehrach, H.; Herwig, R. Systems Biology: A Textbook; Wiley-VCH: Weinheim, Germany, 2009. [Google Scholar]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feng, X.; Marchisio, M.A. Saccharomyces cerevisiae Promoter Engineering before and during the Synthetic Biology Era. Biology 2021, 10, 504. https://doi.org/10.3390/biology10060504
Feng X, Marchisio MA. Saccharomyces cerevisiae Promoter Engineering before and during the Synthetic Biology Era. Biology. 2021; 10(6):504. https://doi.org/10.3390/biology10060504
Chicago/Turabian StyleFeng, Xiaofan, and Mario Andrea Marchisio. 2021. "Saccharomyces cerevisiae Promoter Engineering before and during the Synthetic Biology Era" Biology 10, no. 6: 504. https://doi.org/10.3390/biology10060504
APA StyleFeng, X., & Marchisio, M. A. (2021). Saccharomyces cerevisiae Promoter Engineering before and during the Synthetic Biology Era. Biology, 10(6), 504. https://doi.org/10.3390/biology10060504






