Simple Summary
Rheumatoid arthritis (RA) is a complex disease resulting from multiple genetic and environmental pathogenic factors. The genetic factors include single-nucleotide polymorphisms (SNPs), which have been reported to be associated with RA, but their specific role in the pathogenesis of RA remains unexplained. This study explains the potential role of RA risk-associated SNPs in its pathogenesis in order to provide a basis for understanding the genetic complexity of RA. Several roles of these SNPs are described in this study, and may also aid in the design of a therapeutic strategy for RA. Furthermore, novel potential therapeutic sites have also been researched, resulting in the identification of three novel therapeutic targets. The therapeutic strategies for the treatment of RA include inflammatory pathway-targeting drugs, which alleviate inflammation in joints. There is always a need for novel therapeutic targets that can play a role in alleviating inflammation in autoimmune diseases including RA. Therefore, these novel therapeutic sites are very important, and further experimental studies are required.
Abstract
Single-nucleotide polymorphisms (SNPs) are reported to be associated with many diseases, including autoimmune diseases. In rheumatoid arthritis (RA), about 152 SNPs are reported to account for ~15% of its heritability. These SNPs may result in the alteration of gene expression and may also affect the stability of mRNA, resulting in diseased protein. Therefore, in order to predict the underlying mechanism of these SNPs and identify novel therapeutic sites for the treatment of RA, several bioinformatics tools were used. The damaging effect of 23 non-synonymous SNPs on proteins using different tools suggested four SNPs, including rs2476601 in PTPN22, rs5029941 and rs2230926 in TNFAIP3, and rs34536443 in TYK2, to be the most damaging. In total, 42 of 76 RA-associated intronic SNPs were predicted to create or abolish potential splice sites. Moreover, the analysis of 11 RA-associated UTR SNPs indicated that only one SNP, rs1128334, located in 3′UTR of ETS1, caused functional pattern changes in BRD-BOX. For the identification of novel therapeutics sites to treat RA, extensive gene–gene interaction network interactive pathways were established, with the identification of 13 potential target sites for the development of RA drugs, including three novel target genes. The anticipated effect of these findings on RA pathogenesis may be further validated in both in vivo and in vitro studies.
1. Introduction
Rheumatoid arthritis (RA) is a systemic autoimmune disease with approximately 1% prevalence worldwide, and its presence carries the risk of irreparable functional disability of inflamed joints due to articular damage [1]. Rheumatoid joints exhibit an inflammatory environment that favors the activation of T cells, B cells, macrophages, osteoclasts, and synovial fibroblasts [1]. These cells maintain crosstalk through the production of cytokines, which upon activation induce the secretion of enzymes and other products that contribute to the destruction of cartilage and bone tissues [2]. To date, the etiology of RA remains obscure. However, some authors suggest that the over-reactive immune system in RA is due to both genetic and environmental factors [3,4]. It has been estimated that the inheritability of RA is around 65%, which underlines the importance of its genetics [5,6,7]. Among the genetic factors, several genes have been associated with RA susceptibility [7,8,9,10,11]. Genetic association studies based on different populations have identified more than 100 genomic loci [10,11,12] which account for approximately 15% of the variance [12,13]. However, the actual underlying genetic mechanism concerning SNPs has not been determined.
SNPs are genetic variations that account for ~0.1% differences in populations. The coding region contains about 50% SNPs, with ~25% being missense and ~25% being silent or synonymous [14,15]. Non-coding SNPs may change mRNA stability and promoter activity by creating or disrupting the miRNA sites, causing an altered gene expression with the consequent up-or down-regulation of a gene. The role of these variants in relation to RA risk needs to be explored for the proper elucidation of the biological pathways involved. Besides understanding the underlying disease mechanism, SNP analysis will help in the development of new drugs against RA.
In this study, 152 RA-associated SNPs were characterized and their functional importance with regard to the respective genes and their products was examined in detail. In addition, we investigated the gene–gene interaction patterns and suggested 13 potential and highly significant target sites for the development of RA drugs.
2. Results
2.1. SNP Retrieval
The first step in this study involved mining the literature from PubMed and Web of Science (Figure 1). We found 152 SNPs (located in 75 genes) in the literature which were reported to be associated with RA (Table S1). Of these SNPs, 76 SNPs were intronic (located in 51 genes), 40 SNPs were intergenic, 23 SNPs were missense (located in 18 genes), 11 SNPs were in the UTRs of 9 genes (6 SNPs in 3′UTR and 5 SNPs in 5′UTR), 1 SNP was synonymous, and 1 belonged to the splice site (Figure 2). Details on all the SNPs are provided in Table S1. The associations of these SNPs with the clinical characteristics of RA patients are provided in Table S2.
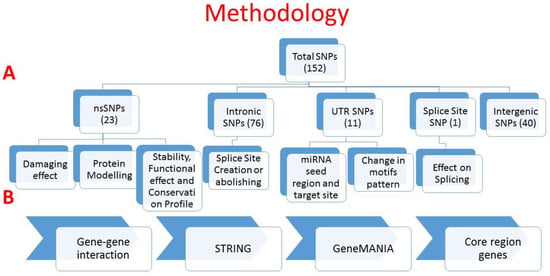
Figure 1.
Workflow of the study. (A) SNP distribution and analysis with regard to their respective effects. (B) Prediction of gene–gene interactions and core region genes. This figure was generated using Microsoft PowerPoint 2016.
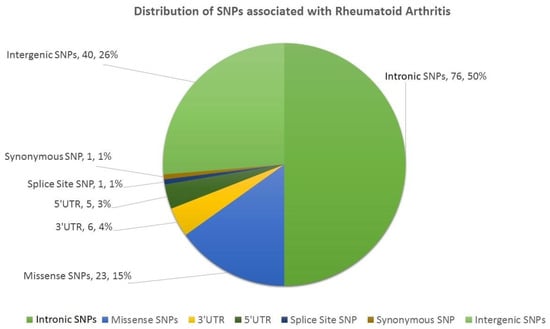
Figure 2.
Distribution of SNPs associated with RA, represented in a pie chart. This figure was generated using Microsoft Excel 2016.
2.2. Characterization of nsSNPs
The 23 nsSNPs that were retrieved from the literature and were found to be potentially associated with RA were analyzed using different tools. These nsSNPs are listed in Table 1 along with amino acid residue change and global MAFs.

Table 1.
nsSNPs along with amino acid change and global MAFs associated with RA.
2.3. Prediction of Damaging Effects of nsSNPs
The damaging effects of nsSNPs on proteins were predicted using five different in-silico tools, which included PhD-SNP, SNPs&GO, PolyPhen2, PROVEAN, and SIFT. For PhD-SNP and SNPs&GO, a threshold value of 0.5 was set and any prediction beyond this value was considered deleterious. According to these tools, all the nsSNPs were found to exhibit a neutral effect. PolyPhen2 predicted the nsSNPs to be probably damaging, possibly damaging, and benign on a scale of 0–1, with 1 being the most damaging. According to PolyPhen2, 5 out of 23 nsSNPs were predicted to be probably damaging. In the case of PROVEAN, a threshold value of −2.5 was selected and any prediction below this value was considered deleterious. Out of the total 23 nsSNPs, PROVEAN predicted four SNPs to be deleterious. In SIFT, a tolerance index (TI) of 0.05 was selected and the predictions with values less than this were considered deleterious. SIFT predicted three of the total nsSNPs to be deleterious. Finally, four nsSNPs (corresponding to three genes) which were predicted to be damaging or deleterious by at least two of the five in-silico tools were selected for further analysis (Table 2). The selected nsSNPs were cross-checked for consistency using the Ensembl genome browser (release 96), MetalR, Mutation Assessor, REVEL, and CADD. The selected nsSNPs included PTPN22 rs2476601, TNFAIP3 rs5029941 and rs2230926, and TYK2 rs34536443. Ensembl results for these four nsSNPs are listed in Table 3. These results were in accordance with our prediction results, which confirmed the reliability of our methodology. Results for all the nsSNPs are provided in Table S3.

Table 2.
SIFT, PROVEAN, PolyPhen2, SNP&GO, and PhD-SNP results for the selected nsSNPs.

Table 3.
Ensembl results for the selected four nsSNPs.
2.4. Prediction of Stability, Functional, Structural Effects, and Conservation Profile of Proteins
I-Mutant was used to predict the effects of the nsSNPs on protein stability. This tool predicted that 21 of the 23 nsSNPs would decrease protein stability, while two nsSNPs (rs2233433 and rs5029941) showed the opposite results. For the structure-based predictions, we used CUPSAT (released January 2018) (http://cupsat.tu-bs.de/, accessed on 2 February 2021) to cross-check the reliability of these predictions. The CUPSAT predicted eight nsSNPs (34.78%) to be stabilizing as compared to I-Mutant (8.70%), while 15 nsSNPs (65.22%) were predicted to be destabilizing as compared to I-Mutant (91.30%). This tool also predicted changes in energy upon amino acid substitution (Table 4). The MutPred server was used to predict different structural and functional effects, such as the creation of glycosylation and catalytic sites, altered membrane proteins, the gain of intrinsic disorder, the loss of allosteric sites, etc. Only one (rs2230926) of the 23 nsSNPs caused gain of an intrinsic disorder and loss of an allosteric site, while all the remaining nsSNPs were predicted to have no structural or functional effects on proteins. The ConSurf tool was used to predict the evolutionary conservation profile of all the amino acids of a protein. The protein FASTA sequences of each protein were submitted to ConSurf, which generated the conservation profiles of each proteins (Figure S1). Interestingly, only 2 of the 23 nsSNPs were located at buried amino acid sites and three were located at highly conserved and functional residues, while all the remaining nsSNPs were present at the exposed residues. The findings regarding the stability, functional and structural effects, and conservation profile of proteins are listed in Table 5.

Table 4.
Prediction of protein stability upon introduction of the nsSNP.

Table 5.
Results predicted by I-Mutant, MutPred, and ConSurf for the important nsSNPs.
2.5. Modeling of Proteins
The protein modeling was performed using comparative homology modeling with MODELLER v9.22. For each of the proteins, NCBI BLAST was utilized and the source database was set as the Protein Data Bank (pdb). The best-matching templates for each of the proteins were selected for homology modeling. The templates, along with the percentages of identity and coverage, are listed in Table 6. For each protein, the templates were searched and their respective pdb files were downloaded from the RCSB Protein Data Bank. Python script files were written according to the protocol by Andrej Sali Laboratory (https://salilab.org/modeller/tutorial/, accessed on 3 February 2021). For each homology model, the best models with the lowest DOPE value and highest GA341 score were selected for final modeling. The final models were viewed and studied using Chimera v1.11 (https://www.cgl.ucsf.edu/chimera/, accessed on 3 February 2021) [13]. Mutant structures were modeled using Chimera v1.11 by mutating the residue of interest. All the modeled structures along with mutated residues are given in Figure 3A–C. The RMSD values for each of the mutant proteins were calculated using TM-align for every nsSNP. Interestingly, the RMSD values for all the mutated structures were zero. To validate our designed structures, Ramachandran plot assessment was used. The RAMPAGE values for each modeled structure are listed in Table 7. All the modeled structures had outlier region residues <10%.

Table 6.
Percentage identity and coverage of the best-matching templates for query proteins.
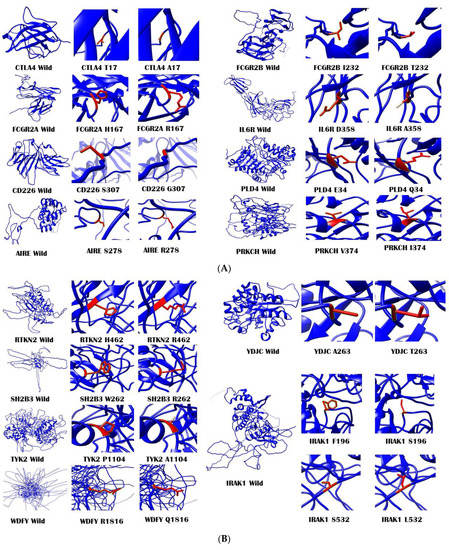
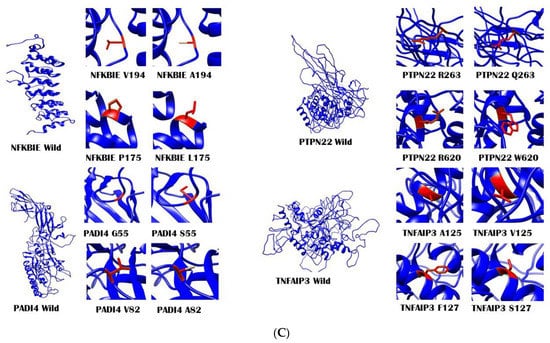
Figure 3.
Modeled structures using MODELER v9.22 for wild-type proteins along with a close-up of wild and mutated amino acid residues. (A) Modeled structures for CTLA4, FCGR2A, CD226, AIRE, FCGR2B, IL6R, PLD4, and PRKCH. (B) Modeled structures for RTKN2, YDJC, SH2B3, TYK2, WDFY4, and IRAK1. (C) Modeled structures for NFKBIE, PADI4, PTPN22, and TNFAIP3. All the protein structures were visualized, and figures were generated using Chimera v1.11 software (https://www.cgl.ucsf.edu/chimera/, accessed on 3 February 2021). The structures were then assembled and combined using Microsoft PowerPoint 2016.

Table 7.
Ramachandran plot analysis percentages of favored, allowed, and outlier residues for the modeled structures.
2.6. Characterization of Intronic SNPs
The SNPs located in the intronic regions of different genes, which were reported to be associated with RA, were compiled and subjected to characterization using ESEfinder3.0. The DNA FASTA sequences for each of the SNPs were retrieved from dbSNP database and are provided in Table S3. All the FASTA sequences, for both the wild-type and mutated proteins, were submitted, and the exon splicing enhancer sites were predicted in both the sequences separately. Of all the 76 intronic SNPs, 42 SNPs were predicted to change the functional pattern and were noted accordingly (Table 8). Of the 42 SNPs, 22 SNPs (located in 27 genes) were found to destroy potential splice sites, 16 SNPs created new splice sites, and 4 SNPs created 1 and destroyed other potential splice sites.

Table 8.
Intronic SNP effect on splicing site as predicted by ESEfinder 3.0.
2.7. Characterization of Splice Site SNPs
The splice site SNP rs2004640, located in the IRF5 gene, was characterized to investigate its potential functional effect on splicing using NetGene2 (http://www.cbs.dtu.dk/services/NetGene2/, accessed on 20 February 2021), the Alternative Splice Site Predictor (ASSP) (http://www.wangcomputing.com/assp/, accessed on 20 February 2021), ESEfinder release 3.0 (http://krainer01.cshl.edu/cgi-bin/tools/ESE3/esefinder.cgi, accessed on 20 February 2021), and Human Splicing Finder v3.1 (HSF v3.1) (http://www.umd.be/HSF3/, accessed on 20 February 2021). NetGene2 and ASSP did not predict any functional effect of this SNP on the splicing mechanism. However, ESEfinder3.0 predicted one potential splice site to be broken at 4 bp upstream of the SNP position, where the human SRSF2 protein may react. The HSF3.1 used the HSF matrices and MaxEnt algorithms to predict the creation of a new donor splice site. HSF3.1 predicted 1 potential splice site to be created, 1 enhancer SF2 motif to be broken, 3 silencer motifs (method of Sironi et al.) to be broken, 1 silencer motif to be created, and 1 silencer IIEs motif to be broken. The details of the results of ESEfinder3.0 and HSF3.1 are listed in Table 9.

Table 9.
ESEfinder3.0 and HSF3.1 prediction for SNP rs2004640 located in the IRF5 gene.
2.8. Characterization of UTR SNPs
The SNPs in the UTR region were studied using UTRScan, PolymiRTS Database, and MicroSNiPer. The DNA FASTA sequences for both the 3′UTR and 5′UTR were submitted to UTRScan, which analyzed the sequences without mutations. For this reason, the FASTA sequences for both the wild-type and mutated sequences were submitted separately (provided in Text S1), and the changes in functional patterns due to the UTR SNPs were noted. Of the 11 UTR SNPs (6 in 3′UTR and 5 in 5′UTR), only 1 SNP in the 3′UTR (rs1128334 located in ETS1 gene) was found to cause significant pattern changes, resulting in the creation of BRD-BOX. All the other SNPs did not indicate any significant changes in the expression pattern of the respective genes. The 3′UTR SNPs were further submitted to PolymiRTS database to investigate if they could disrupt or create miRNA binding site. Of the 6 SNPs in 3′UTR, 3 SNPs were predicted to create 3 miRNA binding sites and disrupt 6 miRNA binding sites. The potential effect of the UTR SNPs on destroying the possible miRNA seed region in 3′UTR was investigated using MicroSNiPer. The results showed that there were 5 SNPs in the 3′UTR which could possibly destroy 10 miRNA seed regions. The results of UTRScan, PolymiRTS database, and MicroSNiPer are listed in Table 10.

Table 10.
UTR SNPs associated with RA and their predictive regulatory role.
2.9. Gene–Gene Interactions of RA Associated Genes
STRING and GeneMANIA were used to analyze the gene–gene interactions of all the 75 genes with potential RA-associated SNPs. Both the tools were fed with gene symbols and the outcomes were recorded. The cutoff value used for both the tools was 0.1. The score ranged on a scale of 0 to 1, with 1 being the best. STRING predicted a total of 365 interactions between 60 genes (shown in Figure 4) and the details are provided in Table S4. The remaining 15 genes were not predicted to have any interaction with any of the investigated genes. A total of 18 genes were predicted to be core region genes, including IL2RB, STAT4, CTLA4, PTPN22, TYK2, BLK, GATA3, TNFAIP3, IRF4, EOMES, IL6R, IRAK1, TRAF6, CD28, CD40, TRAF1, PTPRC, and IL2RA. The GeneMANIA predictions included co-expression (71.46%), co-localization (12.75%), physical interactions (7.83%), predicted theoretical interactions (3.21%), shared protein domains (1.82%), pathways (1.66%), and genetic interactions (1.27%) (Figure 5). Additional genes which were predicted to have role in the pathogenesis of RA were IL2RB, CD3D, CD28, CD2, CD226, CD244, CCL21, CCR6, REL, BLK, IRAK1, TYK2, STAT4, TRAF1, TRAF6, PTPRC, CD247, TNFAIP3, CD83, ITK, IL6R, EOMES, ETS1, and IL2RA (Figure 6).
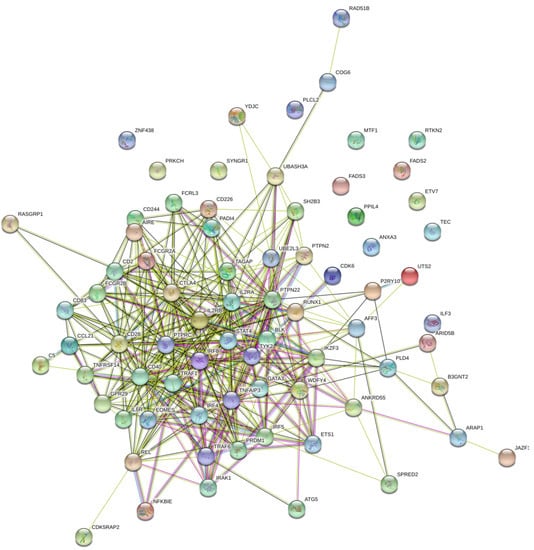
Figure 4.
Gene–gene interaction model of 75 RA-associated genes using STRING. This figure was downloaded as a high-quality image file from STRING v11.0 (https://string-db.org/, accessed on 25 February 2021). A guide, representing the different colors in this figure, is provided in Supplementary Figure S2.
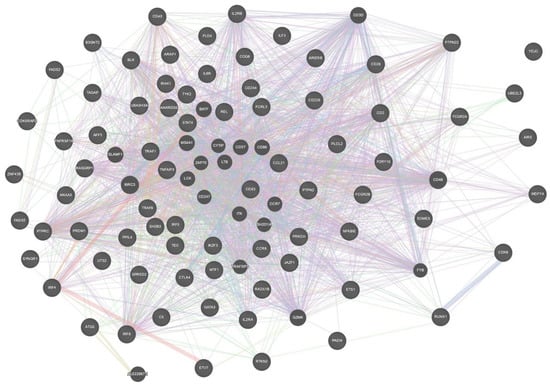
Figure 5.
Gene–gene interaction network generated by GeneMANIA for all the interaction types. This figure was downloaded as a high-quality image file from GeneMANIA v3.5.1 (https://genemania.org/, accessed on 25 February 2021).
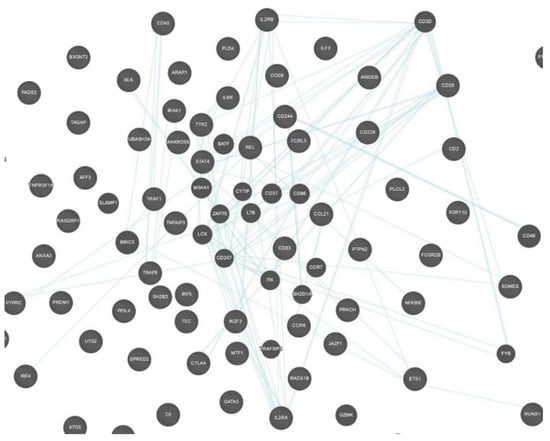
Figure 6.
Gene–gene interaction network predicted by GeneMANIA showing only co-localized genes in pathways. This figure was downloaded as a high-quality image file from GeneMANIA v3.5.1 (https://genemania.org/, accessed on 25 February 2021).
3. Discussion
SNPs hold a significant role in the pathogenesis of a disease. In-silico characterization underlines the possible functional significance of SNPs in both the coding and non-coding regions of a gene, for example through alteration of mRNA stability, promoter activity, and miRNA sites [14,15,16]. However, the actual underlying genetic mechanism concerning the SNPs has not been determined. This is similarly the case in RA-associated SNPs. Previously, some studies were carried out to identify the association of SNPs with RA [14,15,16,17], but in-silico characterizations of these SNPs are scarce.
In our study, we characterized 152 RA-associated SNPs. Of these SNPs, 76 (50%) were intronic, 40 (26.31%) were intergenic, 23 (15.13%) were missense, 6 (3.94%) were in the 3′UTR, 5 (3.28%) were in the 5′UTR, and 1 (0.67%) each belonged to the splice site and coding region, respectively. From these observations, we concluded that RA has a significant association with intronic SNPs (50%), although a large number (26.31%) of SNPs were also observed in intergenic regions. Besides, the role of SNPs in regulation of genes may be less significant (7.24%, 3′ + 5′UTRs SNPs).
nsSNPs are sometimes damaging and have a significant impact on disease pathogenesis [18]. They also contribute to altered drug responses when occurring in the active site of the drug’s target [19]. In our study, PhD-SNP and SNPs&GO predicted none of the nsSNPs to be damaging, while PolyPhen2, PROVEAN, and SIFT predicted 5 (21.74%), 4 (17.39%), and 3 (13.04%) of the nsSNPs, respectively, to be damaging (Table S3). The nsSNP TYK2 rs34536443 had the most damaging effect, with a SIFT score of 0.007, a PROVEAN score of −6.755, and a PolyPhen2 score of 1.00. The nsSNPs which were predicted to be damaging by at least two of the five tools were cross-checked for consistency using the Ensembl genome browser, MetalR, Mutation Assessor, REVEL, and CADD. The selected nsSNPs included PTPN22 rs2476601, TNFAIP3 rs5029941 and rs2230926, and TYK2 rs34536443. Notably, among these SNPs, TYK2 rs34536443 was predicted to be the most damaging, with a CADD score of 26, a REVEL score of 0.586 (0.5 threshold), a MetalR score of 0.336 (0.5 threshold), and a Mutation Assessor score of 0.36 (0.5 threshold). CADD scores of 10, 20, and 30 corresponded to 10%, 1%, and 0.1% of the most damaging SNPs, respectively. This result shows that out of 23 nsSNPs, the TYK2 rs34536443 might be the most significant in terms of functional impairment. According to I-Mutant predictions, all the nsSNPs had a deteriorating effect on protein stability, except for two SNPs (rs2233433 and rs5029941). It should be noted that the predictions of I-Mutant were based on the sequence of proteins and not on their structure. Among the predicted destabilizing nsSNPs, PTPN22 rs2476601 had the highest change in energy at −6.98 kcal/mol, followed by PADI4 rs11203366 and TNFAIP3 rs2230926, with energy change values of −5.91 kcal/mol and −4.58 kcal/mol, respectively. TYK2 rs34536443, which was found to have a significant damaging effect on protein function, was not only predicted as stabilizing but also had very high energy change of 6.73 kcal/mol. MutPred predicted only one nsSNP (TNFAIP3 rs2230926) to have functional effects. Changes were shown in eukaryotic linear motif (ELM) sites, with in a gain in intrinsic disorder and the loss of the allosteric site at position R123 in the TNFAIP3 protein. The ELM motif is a resource database which is dedicated to short linear motifs (SLiMs) [15]. These are small proteins, considered as functional modules, which play an important role in the modifications of protein sequences and protein–protein interactions (PPI) [16,17]. Many important cellular processes, such as cell signaling, protein stability, trafficking, molecular mechanism switching, and cell cycle progression are mediated by SLiMs [17,18,19,20]. Six different ELM motifs were predicted to be affected by TNFAIP3 rs2230926, including ELME000053 (GSK3 phosphorylation site), ELME000064 (CK2 phosphorylation site), ELME000106 (cyclin docking motif), ELME000146 (PCSK cleavage site), ELME000220 (FHA phosphopeptide ligands), and ELME000239 (USP7 binding motif). None of the other nsSNPs was predicted to cause any functional effect on the ELM motifs or gain or loss of any other site. ConSurf was then used to generate the conservation profiles of all the proteins, where nsSNPs were located (Figure S1). ConSurf uses solvent accessibility along with evolutionary conservation data to predict the functional and structural effect of nsSNPs that may cause human health problems [21]. Only two nsSNPs, FCGR2B rs1050501 and TNFAIP3 rs5029941, were predicted to be located at buried residues, while three nsSNPs (PTPN22 rs33996649 and rs2476601 and TYK2 rs34536443) were predicted to be located at highly exposed structural and functional residues. All the other nsSNPs were predicted to be exposed. This explains that most of the RA-associated nsSNPs (91.30%) are located at exposed residues. In order to model the protein structures and study amino acid residue changes resulting from these nsSNPs, the comparative homology modeling tool MODELER v9.22 was used. MODELLER is the most widely used tool for the comparative homology modeling of proteins. We first searched for the highest identical templates with the maximum coverage percentages. For all the proteins, the templates had >30% identity, except for PLD4, where the templates 2ZE4, 4GGJ, 2ZE9, and 1BYR had identity values of 26.52%, 26.32%, 25.97%, and 24.70%, respectively. For all the proteins, many templates were available as a result of the NCBI BLAST search. Of these, only four templates with the highest identity and maximum coverage were selected, except for YDJC, for which only one template (2I5I with 37.23%) was selected. MODELLER v9.22 predicted five structures for each of the proteins, with slight differences. The structure with the lowest discrete optimized protein energy (DOPE) value and highest GA341 score was selected. DOPE calculates the energy of proteins. It is based upon the non-interacting atoms of the modeled protein, with the radius dependent on the template [22]. GA341 scores are calculated on the basis of template and modeled structure identity percentages. Scores range from 0 to 1, where 1 shows the best-modeled structure [23]. The structures modeled with MODELLER v9.22 were then viewed and studied in Chimera v1.11, and mutant protein structures were developed. Root-mean-square deviation (RMSD) values were calculated for each of the mutant structures. RMSD values are used for the calculation of differences between the alpha-carbon backbone of wild-type and mutant protein structures [24,25]. For all the wild-type proteins, the RMSD values were 0.00, meaning that these nsSNPs may not have significant effect on any alteration in the protein’s carbon backbone. For the validation of the protein structures, Ramachandran plot analysis was carried out using RAMPAGE, which predicted all the structures to be valid. For the PLD4 protein, the templates had <30% similarities, but its modeled structure had only 4.3% outlier residues, which validated its structure.
In-silico characterization of the nsSNPs suggested that the SNPs rs33996649 and rs2476601 (PTPN22), rs5029941 and rs2230926 (TNFAIP3), and rs34536443 (TYK2) have prominent functional effects on the protein structure and function. We cross-checked these five nsSNPs with the literature and found evidence for only two nsSNPs (TNFAIP3 rs2230926 and TYK2 rs34536443). A recent study showed that TNFAIP3 rs2230926 decreased the activity of NF-κB by two-fold [26], but the exact molecular mechanism of how this decrease happened was unknown. For TYK2 rs34536443, both in-silico and in vivo studies have shown a decrease in the enzymatic activity of tyrosine kinase 2 (TYK2) due to this SNP [27,28,29]. For the two SNPs (TNFAIP3 rs2230926 and TYK2 rs34536443), our predictions are in agreement with the previous studies [26,27,28,29], while there was no literature-based evidence for the other three nsSNPs (PTPN22 rs33996649 and rs2476601 and TNFAIP3 rs5029941).
The only known functional effect of the SNPs located in the intronic regions on the gene or its product is the effect of these intronic SNPs on the splicing phenomenon. Intronic SNPs may create or abolish the interaction sites for human SR proteins. The creation of a new site for a human SR protein with higher value may result in alternative splicing, which causes the alteration of the protein. Human SR protein family members, including SRSF1, SRSF2, SRSF5, and SRSF6, have been found to have an important role in splicing mechanisms [30,31,32,33]. The binding of the U1 snRNP to the 5′ splice site and the binding of U2 snRNP to the 3′ splice site are promoted by these SR proteins, as well as the events in pre-spliceosome, and mature spliceosome are also linked with these SR proteins [34,35,36,37]. Of the 76 investigated intronic SNPs, 42 (55.26%) SNPs were predicted to cause functional changes in human SR protein binding. A total of 26 SNPs (34.21%) were predicted to demolish the SR protein binding site, while 20 SNPs (26.31%) were predicted to create a new SR protein binding site. Our results predicted 55.26% of the RA-associated intronic SNPs to cause changes in the SR protein binding sites, which may explain their functional significance in the pathogenesis of RA. However, upon a literature survey, we did not find any evidence of the characterization of any of these SNPs in-silico, in vivo, or in vitro. In vivo and in vitro studies are needed for the characterization of the functional importance of these SNPs and the association mechanism with RA and other autoimmune diseases.
The splice site SNP rs2004640 (IRF5) was investigated using different tools for its potential effect on the splicing mechanism. As it is located at the splice site, it has a prime importance in splicing of IRF5. Many studies have demonstrated that rs2004640 increases IRF5 mRNA to a level ~2-fold higher than the wild-type allele [38,39]. Clark and co-workers showed that this polymorphism decreased 1C and 1D exon usage but did not alter mRNA stability [38]. Hence, it is evident that this splice site SNP may cause alternative splicing of IRF5 mRNA and may lead to increased IRF5 production. Our predicted results are also in accordance with the literature. The creation of a potential splice site 4 bp upstream of rs2004640 was predicted by both ESEfinder3.0 and HSF3.1, with high values of 85.64 (threshold 60) and 2.9 (threshold 1.867), respectively. Both the tools predicted the same enhancer motif SRSF2 (IgM-BRCA1) to be broken, with scores of 78.92 and 2.95 by HSF3.1 and ESEfinder3.0, respectively. Therefore, our findings support the previous studies and provide an insight into the mechanism of splicing alteration, which may be caused as a result of this SNP.
Different hereditary diseases such as immune deficiency syndromes and cancer have been linked to mutations in the UTRs of the genes, which have been reported to have vital roles in the stability, translational efficiency, and localization of mRNA [40]. Both the 5′UTR and 3′UTR have key functions in mRNA stability and its expression. The processing and translation of mature mRNA can be severely affected by mutations in the UTR regions of the genes, which can lead to the changes in gene expression patterns [41]. The SNPs located in the 5′UTR are associated with the changes in mRNA stability, binding capacity to ribosomes, and translational regulation, affecting the RNA half-life. The localization, translational efficiency, polyadenylation, stability, and binding specificity of miRNA (microRNA) may be altered by SNPs in the 3′UTR, which effects the expression patterns of genes [15]. In our study, we analyzed 11 UTR SNPs, including six 3′UTR SNPs and five 5′UTR SNPs using UTRScan, the PolymiRTS database, and MicroSNiPer. UTRScan predicted only one SNP, rs1128334, located in 3′UTR of ETS1, to cause functional pattern changes in BRD-BOX. BRD-BOX is a seven-nucleotide motif, which, upon presence in the 3′UTR, controls the activation of gene. When it is lost from the UTR, it results in the hyper-activation of gene [42]. Our study suggested that the creation of BRD-BOX by rs1128334 (ETS1) would be protective against disease. Similarly, it has been shown that the presence of this SNP reduces susceptibility to autoimmune diseases [43,44]. The PolymiRTS database predicted that three 3′UTR SNPs (rs3811021 (PTPN22), rs2070197 (IRF5), and rs1128334 (ETS1)) might affect miRNAs by creating or abolishing the miRNA target sites, therefore contributing to the up- and down-regulation of genes. Similarly, MicroSNiPer predicted five 3′UTR SNPs (Table 8) which affected the miRNA target sites by changing the seed length. However, none of the 5′UTR SNPs was found to have potential role in the pathogenesis of RA, while the five 3′UTR SNPs have been proven to change the functional expression pattern of genes by various means, including destroying or creating miRNA binding sites and creating BRD-BOX.
Gene–gene interaction is an important phenomenon with a key role in the pathogenesis of multi-gene hereditary diseases. There are many genes that have been reported to have significant associations with RA, with known and unknown pathogenesis patterns. We used STRING and GeneMANIA for the prediction of different gene–gene interaction mechanisms. From STRING predictions, we found that 18 of the total 76 genes were in the core region (Figure 4), while GeneMANIA predicted type-specific interactions in which 24 genes were found to be interactive in pathways. Thirteen genes were common to both core-region genes and interactive pathway genes, including IL2RB, STAT4, TYK2, BLK, TNFAIP3, EOMES, IL6R, IRAK1, TRAF6, CD28, TRAF1, PTPRC, and IL2RA. These genes are important in the pathogenesis of RA and may be considered as potential targets for drug development. Previously, 10 genes out of these 13 were suggested for drug targeting in RA patients, as reviewed by Yamamoto and co-workers in 2015 [2]. Three genes (BLK, EOMES, and TRAF1) are predicted to be novel as potential drug targets.
The BLK gene encodes a nonreceptor tyrosine-kinase of the src family of proto-oncogenes that are typically involved in cell proliferation and differentiation. The protein has a role in B-cell receptor signaling and B-cell development. The BLK risk haplotype was found to be associated with enhanced activation of BCR-stimulated B cells with an increase in T cell–B cell collaboration, at least in part due to differential up-regulation of CD86, and with attendant effects on the isotype distribution of the switched memory B cell repertoire. This is likely to reflect a common mechanism for BLK-mediated genetic risk in autoimmune diseases associated with BLK [45]. The EOMES gene belongs to the TBR1 (T-box brain protein 1) sub-family of T-box genes that share the common DNA-binding T-box domain. The encoded protein is a transcription factor which is crucial for embryonic development of the mesoderm and the central nervous system in vertebrates. The protein may also be necessary for the differentiation of effector CD8+ T cells which are involved in defense against viral infections. The protein expression of EOMES was increased in T cells from healthy donors homozygous for the PTPN22 risk allele and correlated with a decreased number of naïve CD4+ T cells [46]. An accumulation of EOMES+CD4+ T cells was also observed in the synovial fluid of RA patients with a more pronounced production of perforin-1 in PTPN22 risk allele carriers. The protein encoded by this TRAF1 is a member of the TNF receptor (TNFR)-associated factor (TRAF) protein family. TRAF proteins associate with and mediate the signal transduction from various receptors of the TNFR superfamily. Genome-wide association studies have identified an association between SNPs in the 5′ untranslated region of the TRAF1 gene, with increased incidence and severity of rheumatoid arthritis and other rheumatic diseases. The loss of TRAF1 from chronically stimulated CD8 T cells results in desensitization of the 4-1BB signaling pathway, thereby contributing to T cell exhaustion during chronic infection. These apparently opposing roles of TRAF1 as both a positive and negative regulator of immune signaling have led to some confusion in the literature. Thus, through gene–gene interactions, we suggested 13 potential drug target sites, of which 3 were novel target genes.
All of our statements are based on evidence from the literature combined with predictive results of the in-silico tools, which used different algorithms and statistical formulas for their predictions. Our study provides a detailed insight into the mechanism of effects of different SNPs belonging to different SNP categories and associated with RA. In order to further validate the effects of these SNPs as predicted by our study, in vitro and in vivo studies are needed. Model organisms with wild-type and mutated alleles, separately, are needed to be studied for further understanding.
4. Methodology
A workflow for the complete methodology is given in Figure 1.
4.1. SNP Retrieval
RA-associated SNPs were searched in PubMed and Web of Science. The data were mined from original research articles published in indexed journals. Retrieved SNPs were divided into four categories; intronic SNPs, non-synonymous SNPs, UTR SNPs, and intergenic SNPs. Information related to the mined SNPs, such as their global minor allele frequencies (MAF), nucleotide change, amino acid residual change for nsSNPs, FASTA sequences, etc., were retrieved for each SNP from NCBI dbSNP (accessed on 2 January 2021).
4.2. Characterization of nsSNPs
The RA-associated nsSNPs were characterized as below:
4.2.1. Most Damaging Prediction
Five different in-silico tools were used to predict the effect of nsSNPs on respective proteins. These tools included: Protein Variation Effect Analyzer (PROVEAN) (http://provean.jcvi.org/seq_submit.php, accessed on 5 January 2021) [47], Sorting Intolerant from Tolerant (SIFT) (https://sift.bii.a-star.edu.sg/www/SIFT_seq_submit2.html, accessed on 5 January 2021) [48,49], SNPs&GO (http://snps.biofold.org/snps-and-go/snps-and-go.html, accessed on 5 January 2021) [50], Predictor of Human Deleterious SNP (PhD-SNP) (http://snps.biofold.org/phd-snp/phd-snp.html, accessed on 5 January 2021) [51], and Polymorphism Phenotyping 2.0 (PolyPhen 2.0) (http://genetics.bwh.harvard.edu/pph2/, accessed on 5 January 2021) [52]. The FASTA sequences of the respective proteins along with their amino acid residue changes were submitted for each nsSNP.
4.2.2. Protein Stability, Structural and Functional Effects, and Conservation Profile Prediction
To predict the effect of nsSNPs on the stability of protein, I-Mutant 2.0 (http://folding.biofold.org/i-mutant/i-mutant2.0.html, accessed on 8 January 2021) was used [53]. The predictions were made for the stability of mutated protein along with the reliability index (RI) on a scale of 0–10, where 0 and showed the minimum and maximum reliability index, respectively. The structural and functional effects of nsSNPs on protein were predicted using MutPred 1.2 (http://mutpred.mutdb.org/, accessed on 9 January 2021) [54]. This predicted the effect of substituted amino acids on proteins, with p values of <0.05 and <0.01 being considered as confident and very confident, respectively. The conservation profile of each protein was predicted with the help of the ConSurf tool (http://consurf.tau.ac.il/2016/, accessed on 15 January 2021) [55] using 50 different homologous sequences.
4.3. Protein Modeling
Protein modeling was done using MODELLER v9.22 [56]. The templates for each protein to be modeled were searched using NCBI BLAST, and those with higher percentage identity and coverage were finally chosen. These templates were later downloaded from the RCSB Protein Data Bank (http://www.rcsb.org/, accessed on 2 February 2021). Command files for each of the protein modeling were prepared separately. For the mutants, the protein structures were modeled using the in-built feature in Chimera v1.11 and the amino acid residual changes were made manually and individually in their respective protein structures. After protein modeling, TM-align (https://zhanglab.ccmb.med.umich.edu/TM-align/, accessed on 5 February 2021) was used to calculate the root-mean-square deviation (RMSD) values for each mutant and wild-type protein. The RMSD values are associated with the functional effect of nsSNP on protein, therefore showing their role in pathogenesis. The higher the RMSD values, the greater the effect of nsSNPs on protein. Later on, the protein structures were validated using Ramachandran plot assessment (http://mordred.bioc.cam.ac.uk/~rapper/rampage.php, accessed on 14 February 2021). This plot showed the percentages of favored and allowed residues, as well as outlier regions, where the structures with residues less than 10% in the outlier regions were considered as good structures.
4.4. Characterization of Intronic SNPs
The effect of intronic SNPs could be detected either by demolishing or creating the splice site in respective genes. For this purpose, we used ESEfinder 3.0 (http://krainer01.cshl.edu/cgi-bin/tools/ESE3/esefinder.cgi, accessed on 16 February 2021) [57]. The FASTA sequences of all the intronic SNPs were retrieved from the dbSNP database. The FASTA sequences for wild-type and mutated sequences were submitted to ESEfinder 3.0, separately, for all the SNPs. ESEfinder3.0 predicted the potential exonic splicing enhancer (ESE) sites that could react with any of the 4 human SR proteins, which are SF1, SF2, SF5, and SF6.
4.5. Characterization of Splice Site SNP
Only 1 splice site SNP rs2004640, located in the IRF5 gene, was reported to be associated with rheumatoid arthritis in Norwegian patients [58]. The characterization of this SNP was performed to investigate its possible effect in splicing. Four different tools were used for this purpose, which included NetGene2 (http://www.cbs.dtu.dk/services/NetGene2/, accessed on 20 February 2021) [59], the Alternative Splice Site Predictor (ASSP) (http://www.wangcomputing.com/assp/, accessed on 20 February 2021) [60], ESEfinder release 3.0 (http://krainer01.cshl.edu/cgi-bin/tools/ESE3/esefinder.cgi, accessed on 20 February 2021) [57], and Human Splicing Finder v3.1 (HSF v3.1) (http://www.umd.be/HSF3/, accessed on 20 February 2021) [61]. Default conditions were used in all the tools and DNA FASTA sequences were submitted to all the tools. Among these tools, HSF3.1 is the most advanced, as it not only predicts potential splice sites but also the exon splicing and silencing enhancers.
4.6. Characterization of UTR SNPs
SNPs in UTR regions have many important effects on mRNA stability and expression. To characterize 3′UTR and 5′UTR SNPs, UTRScan (http://itbtools.ba.itb.cnr.it/utrscan, accessed on 23 February 2021), the PolymiRTS Database 3.0 (http://compbio.uthsc.edu/miRSNP/, accessed on 23 February 2021) and MicroSNiPer (http://vm24141.virt.gwdg.de/services/microsniper/, accessed on 23 February 2021) were used. UTRScan used nucleotide sequences to identify the pattern motif in UTR regions. The DNA FASTA sequences with and without SNPs were submitted individually and the changes in the functional pattern were noted. The PolymiRTS database and MicroSNiPer identified the effect of 3′UTR SNPs on the miRNA seed region and the target site. A list of the SNPs IDs was submitted to both the tools and their effects were recorded.
4.7. Gene–Gene Interaction of RA Associated Genes
The interaction of all the genes selected for this study was investigated using STRING (https://string-db.org/, accessed on 25 February 2021) and GeneMANIA (https://genemania.org/, accessed on 25 February 2021) [62,63]. STRING predictions were based on co-expression, co-occurrence, gene fusion, biochemical and experimental data, while predictions by GeneMANIA were based on co-expression, similarity of protein domains, co-localization, and pathways. A gene list containing the official symbols of all the genes was uploaded and the results were analyzed to find the core region genes.
5. Conclusions
From our study, it was concluded that RA risk-associated SNPs play an important role in the pathogenesis of RA. They contribute to about 15% of RA heredity. Different types of SNPs have their own respective roles in RA. Missense SNPs are found to cause deleterious effect on proteins, thus leading to diseased protein. We analyzed all the RA-associated nsSNPs, and their role in RA pathogenesis was evaluated using several in-silico tools. Of the 23 nsSNPs, 4 nsSNPs (PTPN22 rs2476601, TNFAIP3 rs5029941 and rs2230926, and TYK2 rs34536443) were found to be deleterious, 21 nsSNPs were reported to decrease protein stability, 1 nsSNP (TNFAIP3 rs2230926) was reported to have functional importance (affecting ELM motifs and causing a loss of allosteric sites and a gain of intrinsic disorder), and 3 nsSNPs (PTPN22 rs33996649 and rs2476601 and TYK2 rs34536443) were reported to be located at highly conserved, functionally important, and exposed residues. Intronic SNPs represented 50% of the SNPs that are associated with RA, and our results showed that 42 of 76 intronic SNPs resulted in the alteration of human SR protein binding sites, which may contribute to the splicing mechanism. One splice site SNP (IRF5 rs2004640) was analyzed and found to be splice site donor. Five UTR SNPs (PTPN22 rs3811021, TAGAP rs4709267, IRF5 rs2070197 and rs10954213, and ETS1 rs1128334) were found to alter miRNA binding site. One SNP ETS1 rs1128334 was found to create BRD-BOX, which may down regulate gene expression. Besides SNP characterization, we also predicted gene–gene interactions to predict RA pathogenesis and identified core region genes that may act as potential targets for the development of RA drugs. Importantly, we found 13 core region genes, including IL2RB, STAT4, TYK2, BLK, TNFAIP3, EOMES, IL6R, IRAK1, TRAF6, CD28, TRAF1, PTPRC, and IL2RA. These core region genes could be used as potential therapeutic sites for the treatment of RA. Although our study was in detail and provided a comprehensive analysis of all the SNPs, experimental studies are needed for validation. Mouse models carrying any of these SNPs would be ideal for such experiments.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/biology10060501/s1, Figure S1: ConSurf results for all the proteins with missense RA-associated SNPs. These figures were downloaded as a PDF file from the ConSurf web server (https://consurf.tau.ac.il/, accessed on 15 January 2021). Figure S2: Guidelines for the in-depth understanding of STRING results. Table S1: List of SNPs associated with rheumatoid rrthritis. Table S2: Clinical associations of reported SNPs with RA patients.
Author Contributions
Conceptualization, M.A. (Mehran Akhtar); Data curation, M.A. (Maria Arshad), M.R., S.N.M., S.A.A.-F., H.A.E.-S., and F.J.; Formal analysis, M.A. (Mehran Akhtar), Z.-u.I., and M.A. (Muhammad Ali); Investigation, M.A. (Mehran Akhtar); Methodology, M.A. (Mehran Akhtar); Project administration, F.J.; Supervision, F.J.; Writing—original draft, M.A. (Mehran Akhtar) and Y.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Higher Education Commission of Pakistan, grant number NRPU 5203, and the APC was funded by the Researchers Supporting Project No. (RSP-2020/7) at King Saud University, Riyadh, Saudi Arabia.
Institutional Review Board Statement
This study did not include any living objects; therefore, it did not require approval from any institutional review boards.
Informed Consent Statement
This study did not include any human objects; therefore, it did not require any informed consent statements.
Data Availability Statement
All the data are presented in this manuscript, either in the main article or in the Supplementary Materials.
Acknowledgments
The authors would like to extend their sincere appreciation to the Higher Education Commission, Pakistan, and the Researchers Supporting Project No. (RSP-2020/7) at King Saud University, Riyadh, Saudi Arabia.
Conflicts of Interest
The authors declare that they have no conflict of interest.
References
- Halushka, M.; Fan, J.; Bentley, K.; Hsie, L.; Shen, N.; Weder, A.; Cooper, R.; Lipshutz, R.; Chakravarti, A. Patterns of sin-gle-nucleotide polymorphisms in candidate genes for blood-pressure homeostasis. Nat. Genet. 1999, 22, 239–247. [Google Scholar] [CrossRef]
- Yamamoto, K.; Okada, Y.; Suzuki, A.; Kochi, Y. Genetics of rheumatoid arthritis in Asia—Present and future. Nat. Rev. Rheumatol. 2015, 11, 375–379. [Google Scholar] [CrossRef] [PubMed]
- Silman, A.J. Epidemiology of rheumatoid arthritis. APMIS 1994, 102, 721–728. [Google Scholar] [CrossRef]
- Aho, K.; Koskenvuo, M.; Tuominen, J.; Kaprio, J. Occurrence of rheumatoid arthritis in a nationwide series of twins. J. Rheumatol. 1986, 13, 899–902. [Google Scholar] [PubMed]
- Silman, A.; Macgregor, A.; Thomson, W.; Holligan, S.; Carthy, D.; Farhan, A.; Ollier, W. Twin con-cordance rates for rheumatoid arthritis: Results from a nationwide study. Rheumatology 1993, 32, 903–907. [Google Scholar] [CrossRef] [PubMed]
- MacGregor, A.; Snieder, H.; Rigby, A.; Koskenvuo, M.; Kaprio, J.; Aho, K.; Silman, A. Characterizing the quantitative genetic contribution to rheumatoid arthritis using data from twins. Arthritis Rheum. 2000, 43, 30–37. [Google Scholar] [CrossRef]
- Van der Woude, D.; Houwing-Duistermaat, J.; Toes, R.; Huizinga, T.; Thomson, W.; Worthington, J.; van der Helm-van Mil, A.; de Vries, R. Quantitative heritability of anti-citrullinated protein antibody-positive and anti-citrullinated protein anti-body-negative rheumatoid arthritis. Arthritis Rheum. 2009, 60, 916–923. [Google Scholar] [CrossRef]
- Terao, C.; Ikari, K.; Nakayamada, S.; Takahashi, Y.; Yamada, R.; Ohmura, K.; Hashimoto, M.; Furu, M.; Ito, H.; Fujii, T.; et al. A twin study of rheumatoid arthritis in the Japanese population. Mod. Rheumatol. 2016, 26, 685–689. [Google Scholar] [CrossRef]
- Okada, Y.; Wu, D.; Trynka, G.; Raj, T.; Terao, C.; Ikari, K.; Kochi, Y.; Ohmura, K.; Suzuki, A.; Yoshida, S.; et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature 2013, 506, 376–381. [Google Scholar] [CrossRef]
- Lenz, T.L.; Deutsch, A.J.; Han, B.; Hu, X.; Okada, Y.; Eyre, S.; Knapp, M.; Zhernakova, A.; Huizinga, T.W.; Abecasis, G.R.; et al. Widespread non-additive and interaction effects within HLA loci modulate the risk of autoimmune diseases. Nat. Genet. 2015, 47, 1085–1090. [Google Scholar] [CrossRef]
- Stahl, E.A.; Diabetes Genetics Replication and Meta-analysis Consortium; Wegmann, D.; Trynka, G.; Gutierrez-Achury, J.; Do, R.; Voight, B.F.; Kraft, P.; Chen, R.; Kallberg, H.J.; et al. Bayesian inference analyses of the polygenic architecture of rheumatoid arthritis. Nat. Genet. 2012, 44, 483–489. [Google Scholar] [CrossRef]
- Kim, K.; Bang, S.-Y.; Lee, H.-S.; Bae, S.-Y.B.H.-S.L.S.-C. Update on the genetic architecture of rheumatoid arthritis. Nat. Rev. Rheumatol. 2017, 13, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.; Goddard, T.; Huang, C.; Couch, G.; Greenblatt, D.; Meng, E.; Ferrin, T. UCSF Chimera? A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Akhtar, M.; Jamal, T.; Jamal, H.; Din, J.U.; Jamal, M.; Arif, M.; Arshad, M.; Jalil, F. Identification of most damaging nsSNPs in human CCR6 gene: In silico analyses. Int. J. Immunogenet. 2019, 46, 459–471. [Google Scholar] [CrossRef]
- Akhtar, M.; Khan, S.; Ali, Y.; Haider, S.; Din, J.U.; Islam, Z.-U.; Jalil, F. Association study of CCR6 rs3093024 with Rheumatoid Arthritis in a Pakistani cohort. Saudi J. Biol. Sci. 2020, 27, 3354–3358. [Google Scholar] [CrossRef] [PubMed]
- Davey, N.E.; Van Roey, K.; Weatheritt, R.J.; Toedt, G.; Uyar, B.; Altenberg, B.; Budd, A.; Diella, F.; Dinkel, H.; Gibson, T.J. Attributes of short linear motifs. Mol. Biosyst. 2012, 8, 268–281. [Google Scholar] [CrossRef]
- Van Roey, K.; Uyar, B.; Weatheritt, R.J.; Dinkel, H.; Seiler, M.; Budd, A.; Gibson, T.J.; Davey, N.E. Short linear motifs: Ubiquitous and functionally diverse protein interaction modules directing cell regulation. Chem. Rev. 2014, 114, 6733–6778. [Google Scholar] [CrossRef] [PubMed]
- Diella, F. Understanding eukaryotic linear motifs and their role in cell signaling and regulation. Front. Biosci. 2008, 13, 603. [Google Scholar] [CrossRef]
- Van Roey, K.; Gibson, T.J.; Davey, N.E. Motif switches: Decision-making in cell regulation. Curr. Opin. Struct. Biol. 2012, 22, 378–385. [Google Scholar] [CrossRef]
- Van Roey, K.; Dinkel, H.; Weatheritt, R.J.; Gibson, T.J.; Davey, N.E. The switches. ELM resource: A compendium of conditional regulatory interaction interfaces. Sci. Signal. 2013, 6, rs7. [Google Scholar] [CrossRef]
- Ramensky, V.; Bork, P.; Sunyaev, S. Human non-synonymous SNPs: Server and survey. Nucleic Acids Res. 2002, 30, 3894–3900. [Google Scholar] [CrossRef]
- Shen, M.-Y.; Sali, A. Statistical potential for assessment and prediction of protein structures. Protein Sci. 2006, 15, 2507–2524. [Google Scholar] [CrossRef] [PubMed]
- John, B. Comparative protein structure modeling by iterative alignment, model building and model assessment. Nucleic Acids Res. 2003, 31, 3982–3992. [Google Scholar] [CrossRef]
- Carugo, O.; Pongor, S. A normalized root-mean-square distance for comparing protein three-dimensional structures. Protein Sci. 2001, 10, 1470–1473. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Skolnick, J. TM-align: A protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005, 33, 2302–2309. [Google Scholar] [CrossRef]
- Zhang, B.; Nakamura, B.N.; Perlman, A.; Alipour, O.; Abbasi, S.Q.; Sohn, P.; Gulati, A.; Moore, G.; Hwang, C.; Sheibani, S.; et al. Identification of functional missense single-nucleotide polymorphisms in TNFAIP3 in a predominantly Hispanic population. J. Clin. Transl. Sci. 2018, 2, 350–355. [Google Scholar] [CrossRef]
- Couturier, N.; Bucciarelli, F.; Nurtdinov, R.N.; Debouverie, M.; Lebrun-Frenay, C.; Defer, G.; Moreau, T.; Confavreux, C.; Vukusic, S.; Cournu-Rebeix, I.; et al. Tyrosine kinase 2 variant influences T lymphocyte polarization and multiple sclerosis susceptibility. Brain 2011, 134, 693–703. [Google Scholar] [CrossRef]
- Dendrou, C.; Cortes, A.; Shipman, L.; Evans, H.; Attfield, K.; Jostins, L. Resolving TYK2 locus genotype-to-phenotype differ-ences in autoimmunity. Sci. Transl. Med. 2016, 8, 363ra149. [Google Scholar] [CrossRef]
- Lesgidou, N.; Eliopoulos, E.; Goulielmos, G.N.; Vlassi, M. Insights on the alteration of functionality of a tyrosine kinase 2 variant: A molecular dynamics study. Bioinformatics 2018, 34, i781–i786. [Google Scholar] [CrossRef]
- Fu, X.-D.; Maniatis, T. Factor required for mammalian spliceosome assembly is localized to discrete regions in the nucleus. Nat. Cell Biol. 1990, 343, 437–441. [Google Scholar] [CrossRef] [PubMed]
- Ge, H.; Zuo, P.; Manley, J.L. Primary structure of the human splicing factor ASF reveals similarities with Drosophila regulators. Cell 1991, 66, 373–382. [Google Scholar] [CrossRef]
- Krainer, A.R.; Mayeda, A.; Kozak, D.; Binns, G. Functional expression of cloned human splicing factor SF2: Homology to RNA-binding proteins, U1 70K, and Drosophila splicing regulators. Cell 1991, 66, 383–394. [Google Scholar] [CrossRef]
- Zahler, A.M.; Lane, W.S.; Stolk, J.A.; Roth, M.B. SR proteins: A conserved family of pre-mRNA splicing factors. Genes Dev. 1992, 6, 837–847. [Google Scholar] [CrossRef]
- Cho, S.; Hoang, A.; Sinha, R.; Zhong, X.-Y.; Fu, X.-D.; Krainer, A.R.; Ghosh, G. Interaction between the RNA binding domains of Ser-Arg splicing factor 1 and U1-70K snRNP protein determines early spliceosome assembly. Proc. Natl. Acad. Sci. USA 2011, 108, 8233–8238. [Google Scholar] [CrossRef]
- Fu, X.D.; Maniatis, T. The 35-kDa mammalian splicing factor SC35 mediates specific interactions between U1 and U2 small nuclear ribonucleoprotein particles at the 3′ splice site. Proc. Natl. Acad. Sci. USA 1992, 89, 1725–1729. [Google Scholar] [CrossRef]
- Kohtz, J.D.; Jamison, S.F.; Will, C.L.; Zuo, P.; Lührmann, R.; Garcia-Blanco, M.A.; Manley, J.L. Protein–protein interactions and 5′-splice-site recognition in mammalian mRNA precursors. Nat. Cell Biol. 1994, 368, 119–124. [Google Scholar] [CrossRef] [PubMed]
- Roscigno, R.F.; Garcia-Blanco, M.A. SR proteins escort the U4/U6.U5 tri-snRNP to the spliceosome. RNA 1995, 1, 692–706. [Google Scholar] [PubMed]
- Clark, D.; Lambert, J.; Till, R.; Argueta, L.; Greenhalgh, K.; Henrie, B.; Poole, B.D. Molecular Effects of Autoimmune-Risk Promoter Poly-morphisms on Expression, Exon Choice, and Translational Efficiency of Interferon Regulatory Factor. J. Interferon Cytokine Res. 2014, 34, 354–365. [Google Scholar] [CrossRef]
- Hedl, M.; Yan, J.; Abraham, C. IRF5 and IRF5 Disease-Risk Variants Increase Glycolysis and Human M1 Macrophage Polariza-tion by Regulating Proximal Signaling and Akt2 Activation. Cell Rep. 2016, 16, 2442–2455. [Google Scholar] [CrossRef] [PubMed]
- Grillo, G.; Turi, A.; Licciulli, F.; Mignone, F.; Liuni, S.; Banfi, S.; Gennarino, V.; Horner, D.; Pavesi, G.; Picardi, E.; et al. UTRdb and UTRsite (RELEASE 2010): A collection of sequences and regulatory motifs of the untranslated regions of eukary-otic mRNAs. Nucleic Acids Res. 2009, 38, D75–D80. [Google Scholar] [CrossRef]
- Pesole, G.; Mignone, F.; Gissi, C.; Grillo, G.; Licciulli, F.; Liuni, S. Structural and functional features of eukaryotic mRNA un-translated regions. Gene 2001, 276, 73–81. [Google Scholar] [CrossRef]
- Lai, E.C.; Posakony, J.W. The Bearded box, a novel 3′ UTR sequence motif, mediates negative post-transcriptional regulation of Bearded and Enhancer of split Complex gene expression. Development 1997, 124, 4847–4856. [Google Scholar] [CrossRef]
- Shan, S.; Dang, J.; Li, J.; Yang, Z.; Zhao, H.; Xin, Q.; Liu, Q. ETS1 variants confer susceptibility to ankylosing spondylitis in Han Chinese. Arthritis Res. Ther. 2014, 16, R87. [Google Scholar] [CrossRef]
- Yang, W.; Shen, N.; Ye, D.; Liu, Q.; Zhang, Y.; Qian, X.; Asian Lupus Genetics Consortium. Genome-Wide Association Study in Asian Populations Identifies Vari-ants in ETS1 and WDFY4 Associated with Systemic Lupus Erythematosus. PLoS Genet. 2010, 6, e1000841. [Google Scholar] [CrossRef]
- Simpfendorfer, K.; Armstead, B.; Shih, A.; Li, W.; Curran, M.; Manjarrez-Orduño, N.; Lee, A.; Diamond, B.; Gregersen, P. Autoimmune Disease-Associated Haplotypes of BLK Exhibit Lowered Thresholds for B Cell Activation and Expansion of Ig Class-Switched B Cells. Arthritis Rheumatol. 2015, 67, 2866–2876. [Google Scholar] [CrossRef]
- Chemin, K.; Ramsköld, D.; Diaz-Gallo, L.; Herrath, J.; Houtman, M.; Tandre, K.; Rönnblom, L.; Catrina, A.; Malmström, V. EOMES-positive CD4+T cells are increased inPTPN22(1858T) risk allele carriers. Eur. J. Immunol. 2018, 48, 655–669. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.; Sims, G.E.; Murphy, S.; Miller, J.R.; Chan, A.P. Predicting the Functional Effect of Amino Acid Substitutions and Indels. PLoS ONE 2012, 7, e46688. [Google Scholar] [CrossRef]
- Ng, P.C.; Henikoff, S. Predicting the Effects of Amino Acid Substitutions on Protein Function. Annu. Rev. Genom. Hum. Genet. 2006, 7, 61–80. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Henikoff, S.; Ng, P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009, 4, 1073–1081. [Google Scholar] [CrossRef] [PubMed]
- Capriotti, E.; Calabrese, R.; Fariselli, P.; Martelli, P.L.; Altman, R.B.; Casadio, R. WS-SNPs&GO: A web server for predicting the dele-terious effect of human protein variants using functional annotation. BMC Genom. 2013, 3, S6. [Google Scholar]
- Capriotti, E.; Calabrese, R.; Casadio, R. Predicting the insurgence of human genetic diseases associated to single point pro-teinmutations with support vector machines and evolutionary information. Bioinformatics 2006, 22, 2729–2734. [Google Scholar] [CrossRef] [PubMed]
- Adzhubei, I.A.; Schmidt, S.; Peshkin, L.; Ramensky, V.E.; Gerasimova, A.; Bork, P.; Sunyaev, S.R. A method and server for predicting damag-ing missense mutations. Nat. Methods 2010, 7, 248–249. [Google Scholar] [CrossRef]
- Capriotti, E.; Fariselli, P.; Casadio, R. I-Mutant2.0: Predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Res. 2005, 33, W306–W310. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Krishnan, V.G.; Mort, M.E.; Xin, F.; Kamati, K.K.; Cooper, D.N.; Mooney, S.D.; Radivojac, P. Automated inference of molecular mechanisms of disease from amino acid substitutions. Bioinformatics 2009, 25, 2744–2750. [Google Scholar] [CrossRef]
- Berezin, C.; Glaser, F.; Rosenberg, J.; Paz, I.; Pupko, T.; Fariselli, P.; Ben-Tal, N. ConSeq: The Identification of Functionallyand Structurally Important Residues in Protein Sequences. Bioinformatics 2004, 20, 1322–1324. [Google Scholar] [CrossRef] [PubMed]
- Šali, A.; Blundell, T.L. Comparative Protein Modelling by Satisfaction of Spatial Restraints. J. Mol. Biol. 1993, 234, 779–815. [Google Scholar] [CrossRef]
- Smith, P.J.; Zhang, C.; Wang, J.; Chew, S.L.; Zhang, M.Q.; Krainer, A.R. An increased specificity score matrix for the prediction of SF2/ASF-specific exonic splicing enhancers. Hum. Mol. Genet. 2006, 15, 2490–2508. [Google Scholar] [CrossRef]
- Nordang, G.B.N.; Viken, M.K.; Amundsen, S.S.; Sanchez, E.S.; Flatø, B.; Førre Øystein, T.; Martin, J.; Kvien, T.K.; Lie, B.A. Interferon regulatory factor 5 gene polymorphism confers risk to several rheumatic diseases and correlates with expression of alternative thymic transcripts. Rheumatology 2011, 51, 619–626. [Google Scholar] [CrossRef] [PubMed]
- Hebsgaard, S.M.; Korning, P.G.; Tolstrup, N.; Engelbrecht, J.; Rouze, P.; Brunak, S. Splice site prediction in Arabidopsis thaliana DNA by combining local and global sequence information. Nucleic Acids Res. 1996, 24, 3439–3452. [Google Scholar] [CrossRef]
- Wang, M.; Marín, A. Characterization and Prediction of Alternative Splice Sites. Gene 2006, 366, 219–227. [Google Scholar] [CrossRef] [PubMed]
- Desmet, F.O.; Hamroun, D.; Lalande, M.; Collod-Beroud, G.; Claustres, M.; Beroud, C. Human Splicing Finder: An online bioinfor-matics tool to predict splicing signals. Nucleic Acid Res. 2009, 37, e67. [Google Scholar] [CrossRef] [PubMed]
- Warde-Farley, D.; Donaldson, S.L.; Comes, O.; Zuberi, K.; Badrawi, R.; Chao, P.; Franz, M.; Grouios, C.; Kazi, F.; Lopes, C.T.; et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010, 38, W214–W220. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).