Ablation of Lrp4 in Schwann Cells Promotes Peripheral Nerve Regeneration in Mice
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Generation and Genotyping of Mouse Lines
2.2. Reagents
2.3. Immunofluorescence
2.4. Western Blot Analysis
2.5. BrdU Staining
2.6. TUNEL Staining
2.7. NMJ Electrophysiology
2.8. Denervation Experiment
2.9. Biotin Tracing
2.10. Quantitative Real-Time PCR (qRT-PCR)
2.11. Electron Microscopy
2.12. Statistical Analysis
3. Results
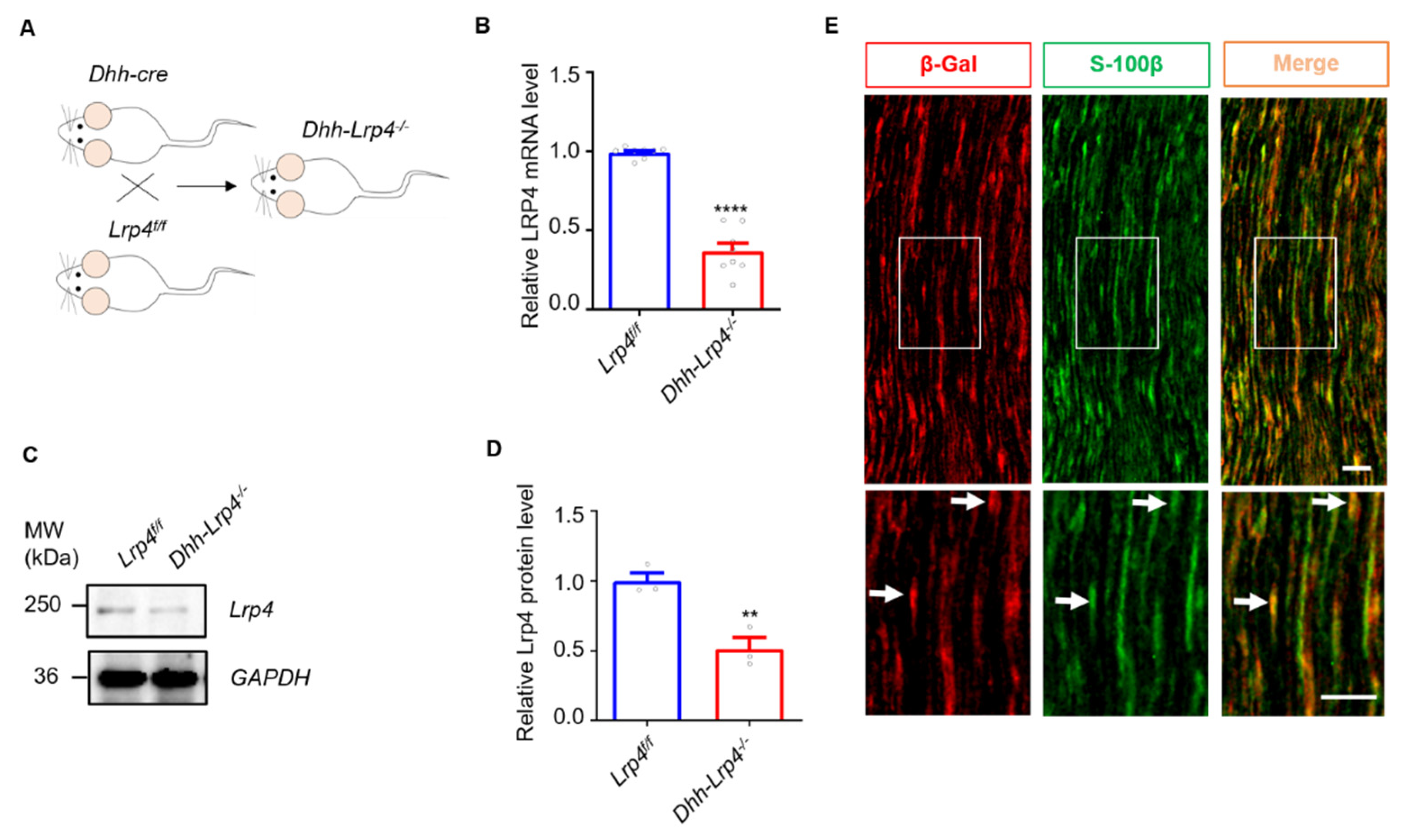
3.1. Lrp4 Expression in SCs
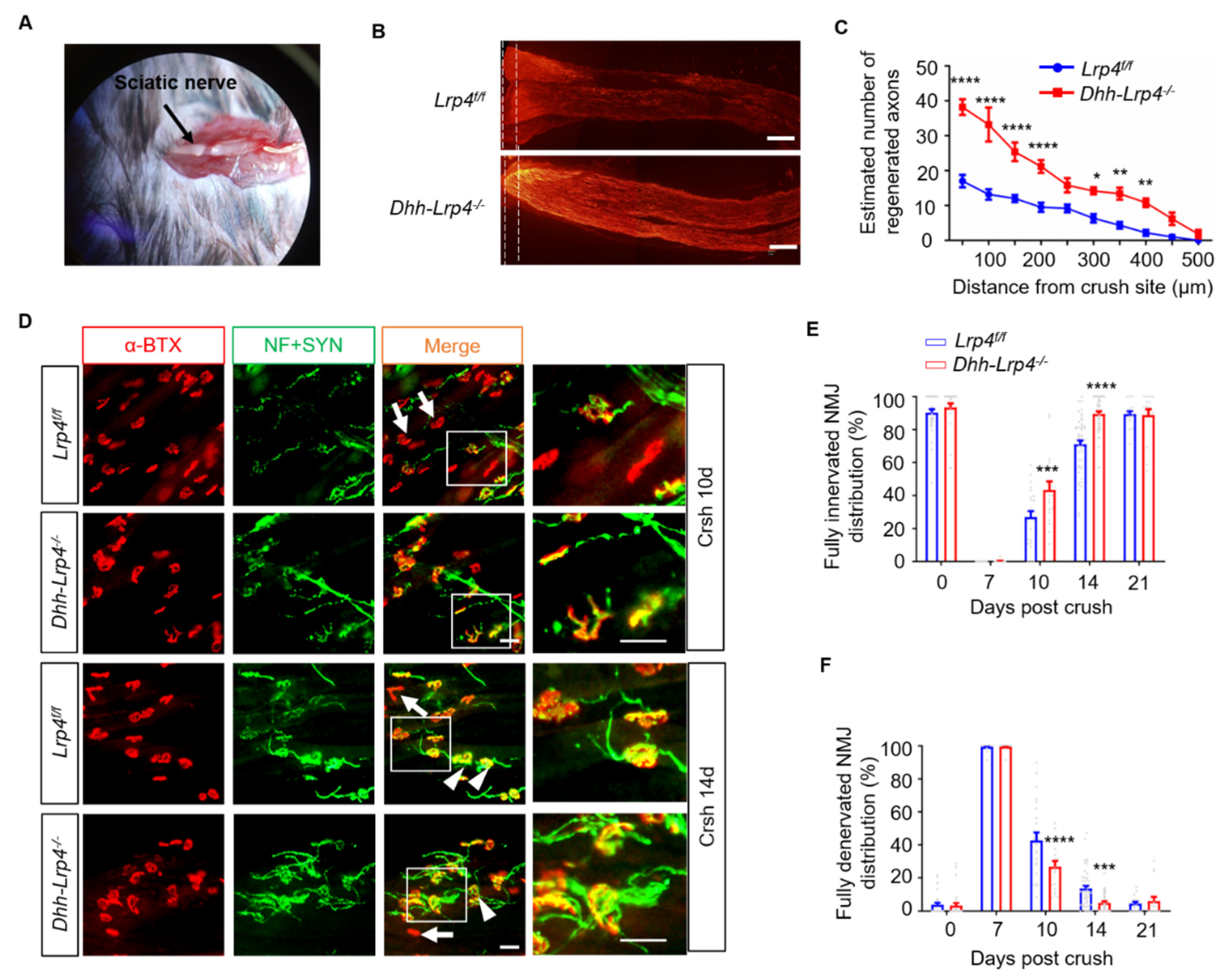
3.2. Nerve Regeneration Promotion in the Lrp4 cKO Mice
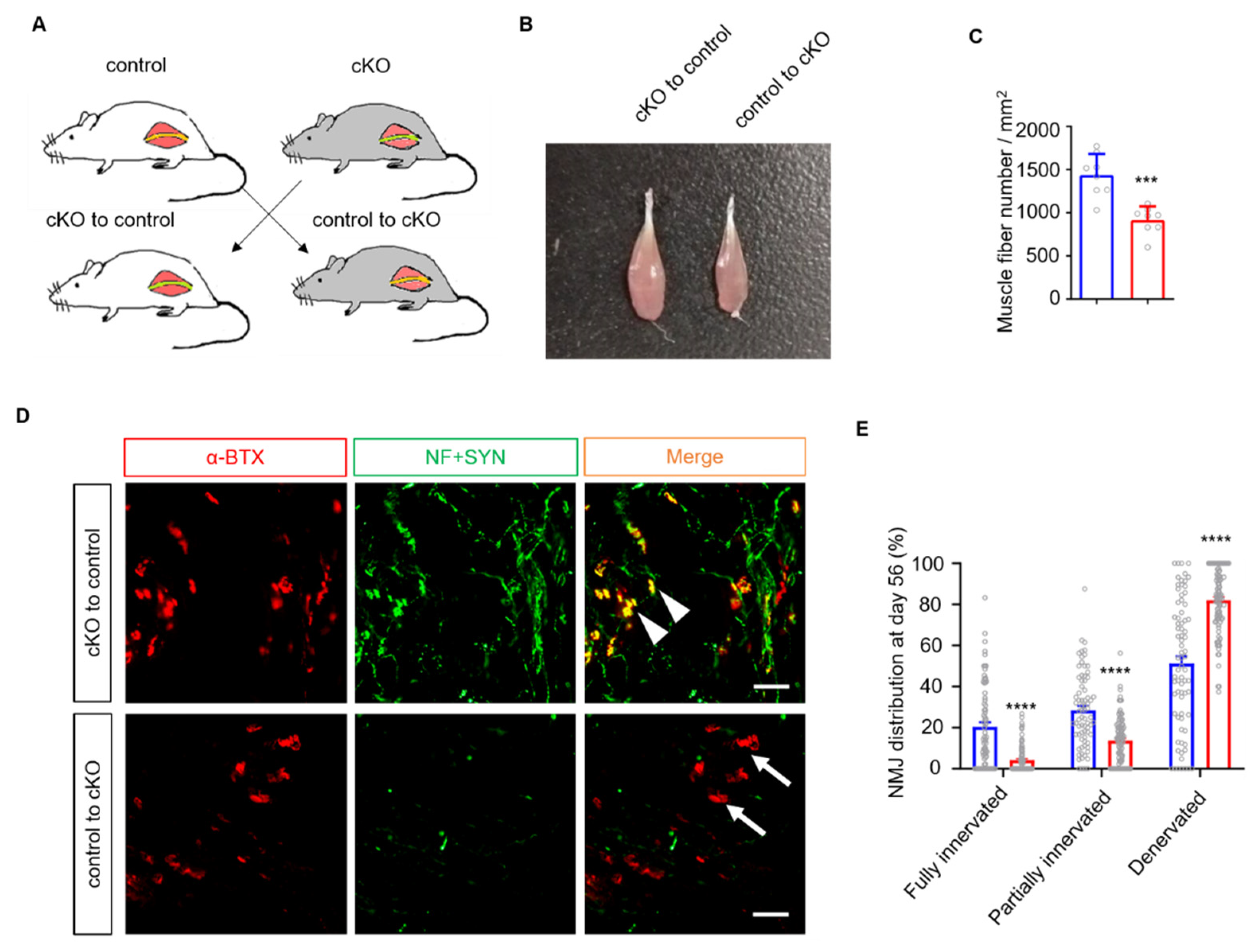
3.3. The mSCs Rather Than tSCs Are Involved in Accelerating Nerve Regeneration in Lrp4 cKO Mice
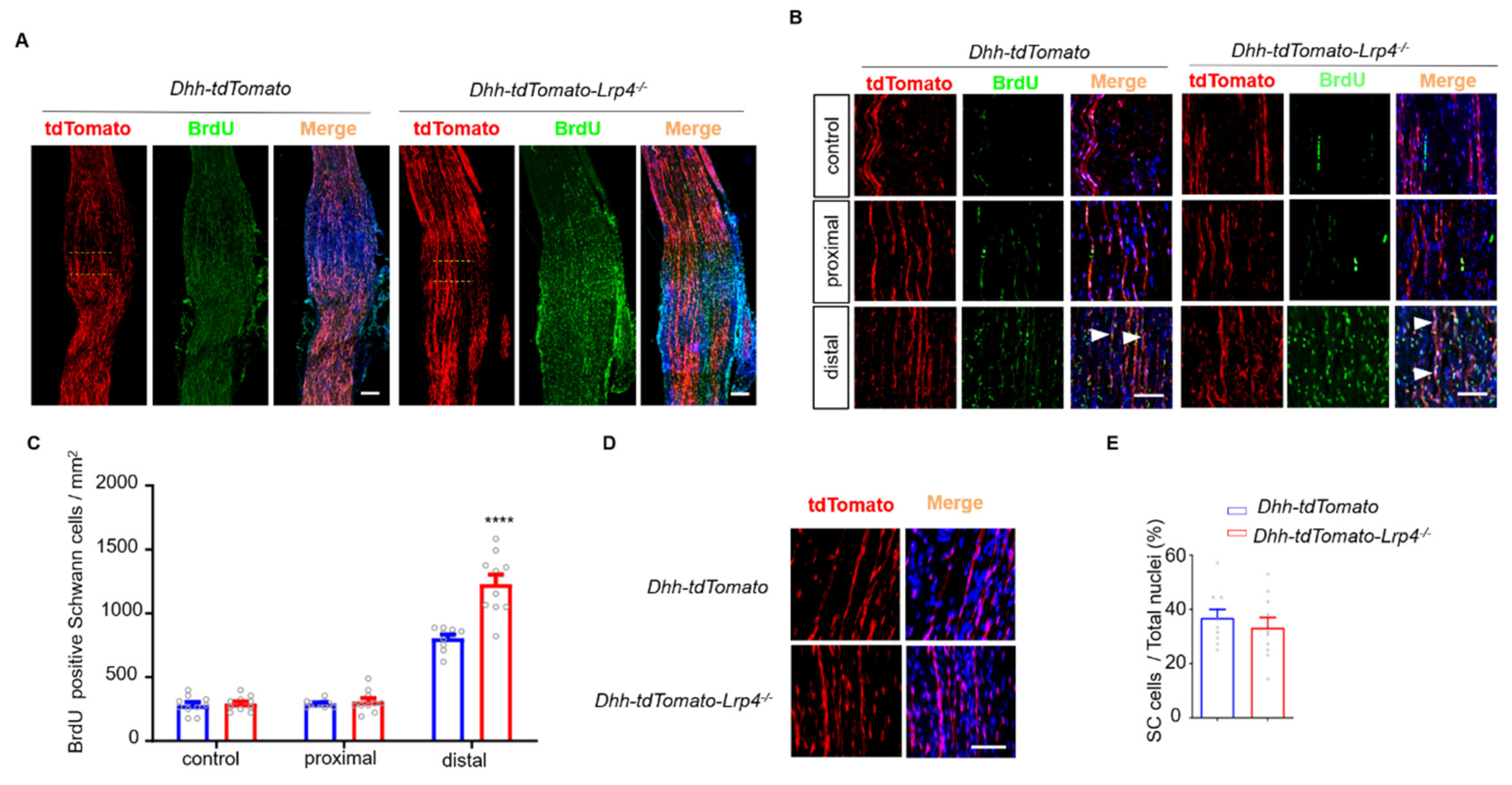
3.4. Enhanced SC Proliferation in Lrp4 cKO Mice after Nerve Damage
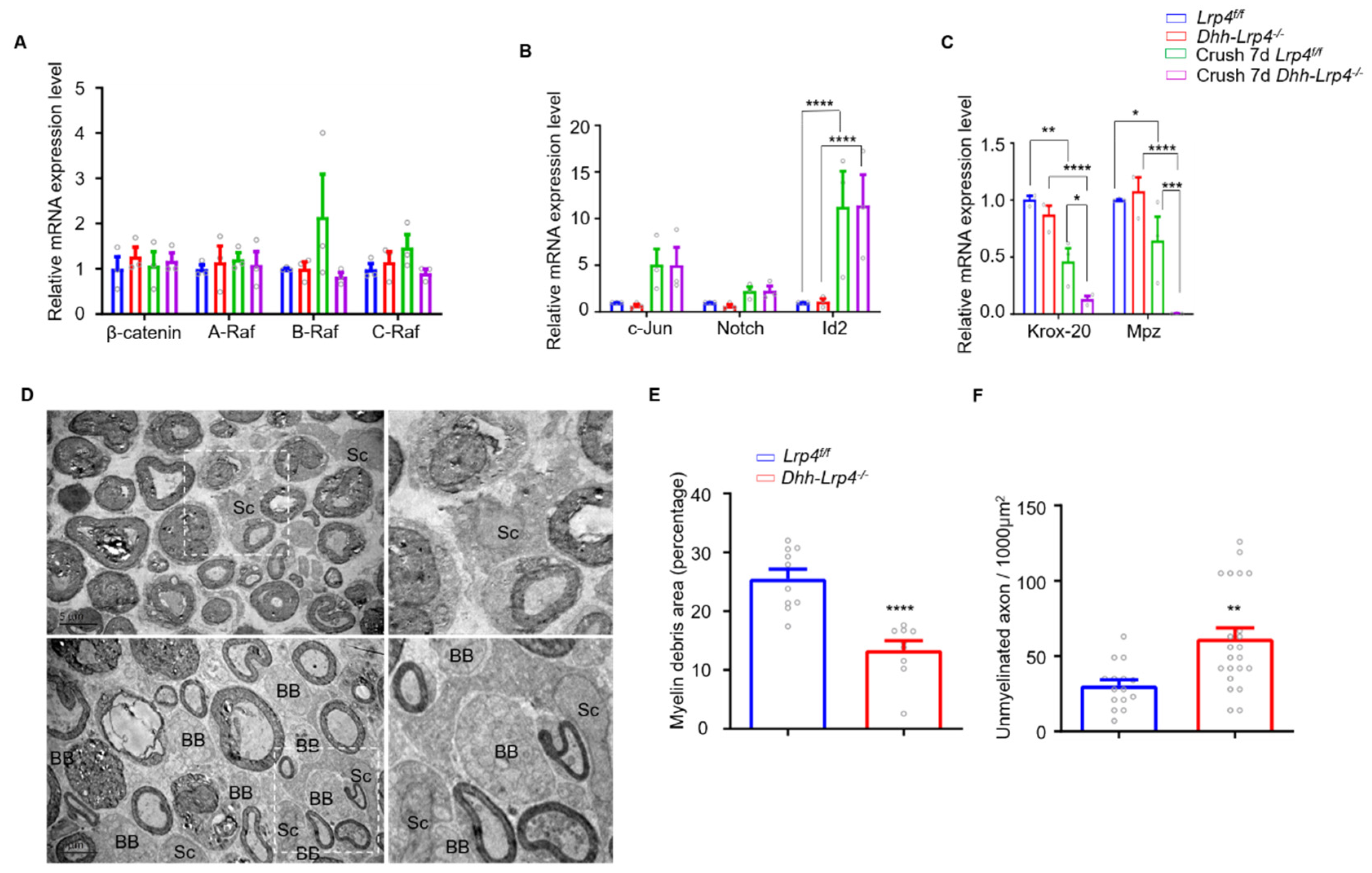
3.5. Lrp4 Deficiency Promotes Demyelination and Downregulates Krox-20 after Nerve Injury
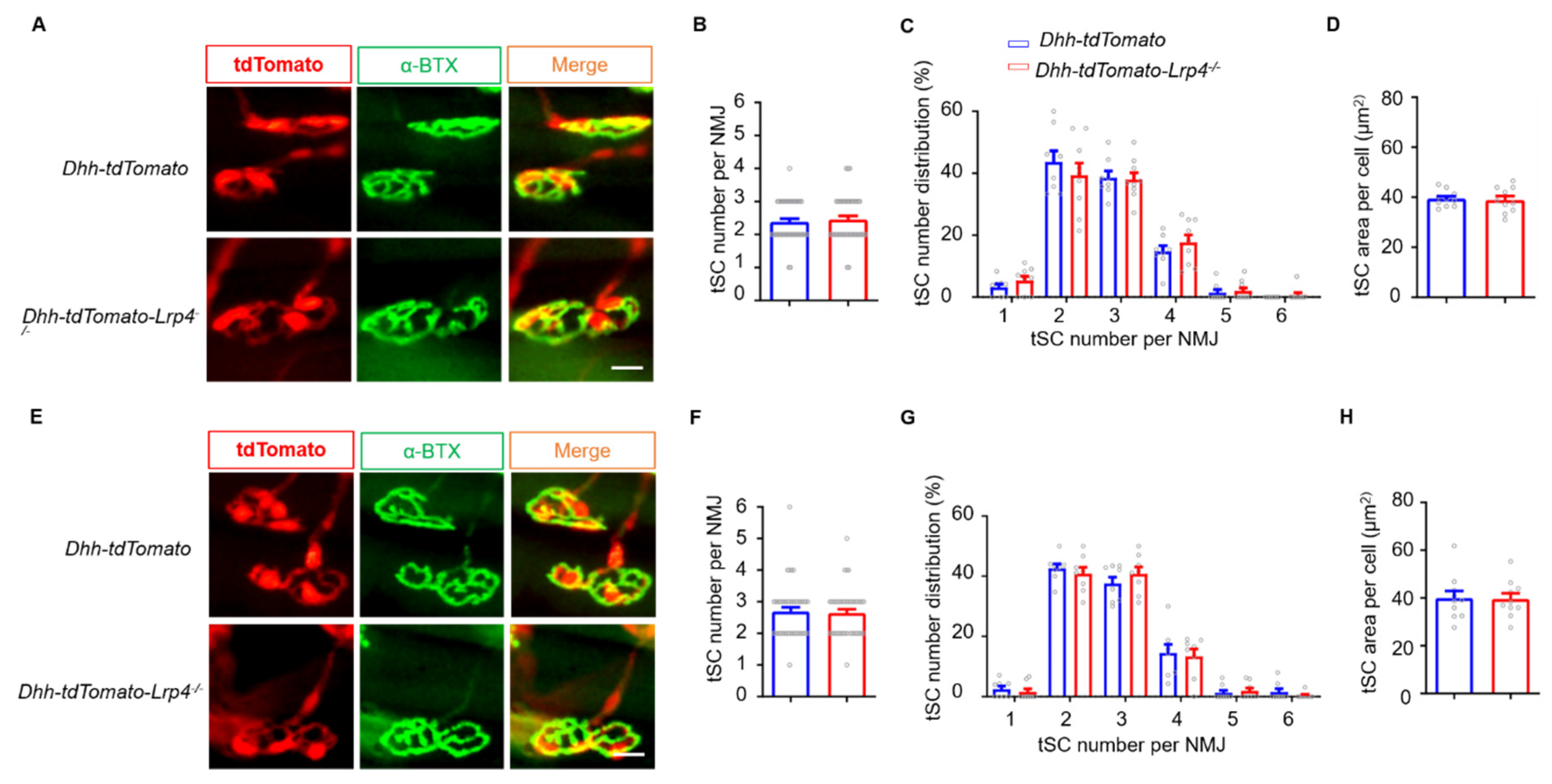
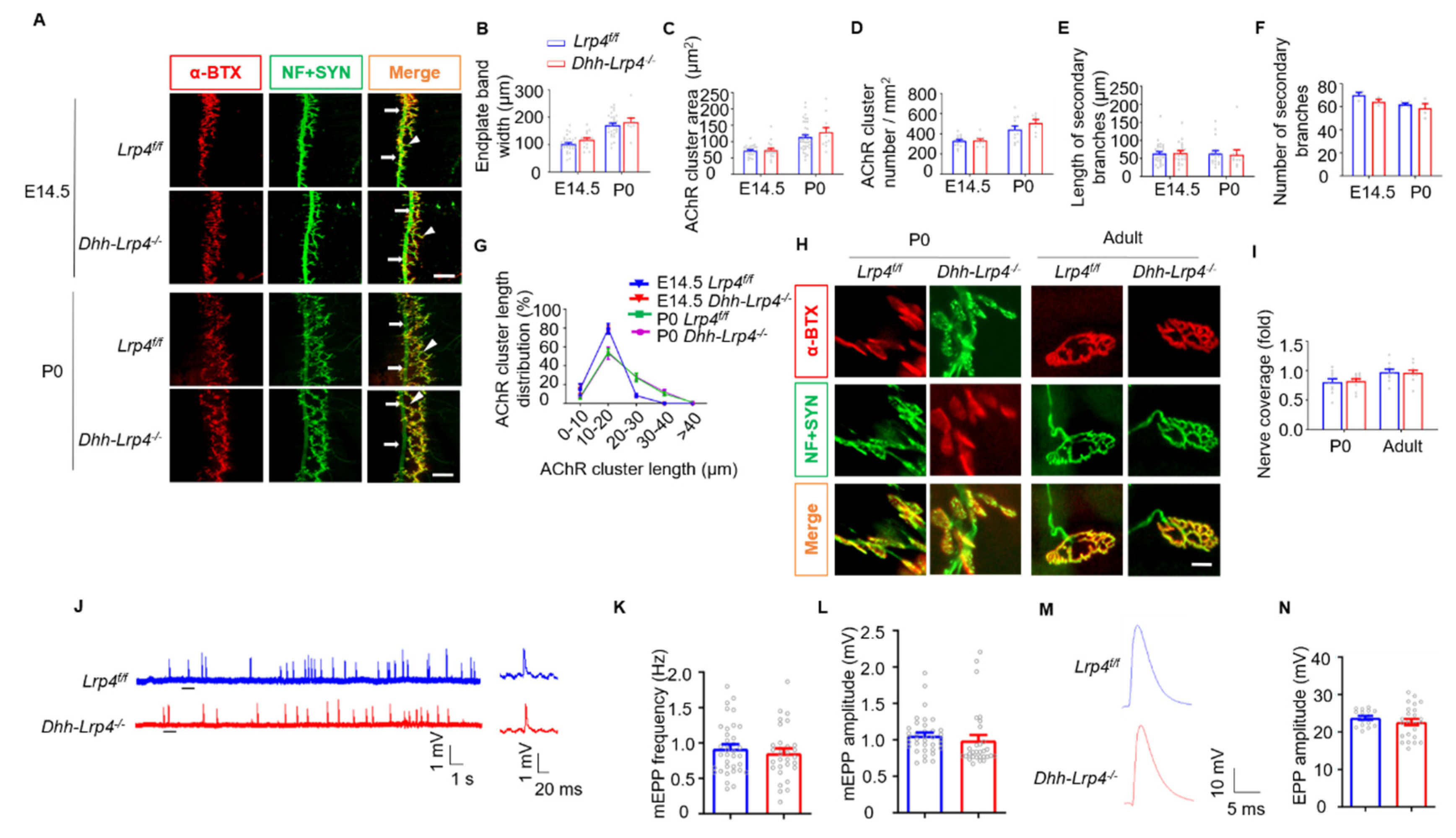
3.6. Normal NMJ Formation and Transmission in SC Lrp4 cKO Mice
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Ethics Statement
References
- Zhu, S.; Ge, J.; Wang, Y.; Qi, F.; Ma, T.; Wang, M.; Yang, Y.; Liu, Z.; Huang, J.; Luo, Z. A synthetic oxygen carrier-olfactory ensheathing cell composition system for the promotion of sciatic nerve regeneration. Biomaterial 2014, 35, 1450–1461. [Google Scholar] [CrossRef] [PubMed]
- Min, Q.; Parkinson, D.B.; Dun, X. Migrating Schwann cells direct axon regeneration within the peripheral nerve bridge. Glia 2021, 69, 235–254. [Google Scholar] [CrossRef] [PubMed]
- Morris, J.K.; Lin, W.; Hauser, C.; Marchuk, Y.; Getman, D.; Lee, K.-F. Rescue of the Cardiac Defect in ErbB2 Mutant Mice Reveals Essential Roles of ErbB2 in Peripheral Nervous System Development. Neuron 1999, 23, 273–283. [Google Scholar] [CrossRef]
- Jessen, K.R.; Mirsky, R. The Success and Failure of the Schwann Cell Response to Nerve Injury. Front. Cell. Neurosci. 2019, 13, 33. [Google Scholar] [CrossRef]
- Darabid, H.; Arbour, D.; Robitaille, R. Glial Cells Decipher Synaptic Competition at the Mammalian Neuromuscular Junction. J. Neurosci. 2013, 33, 1297–1313. [Google Scholar] [CrossRef] [PubMed]
- Smith, I.W.; Mikesh, M.; Lee, Y.I.; Thompson, W.J. Terminal Schwann cells participate in the competition underlying neuromuscular synapse elimination. J. Neurosci. 2013, 33, 17724–17736. [Google Scholar] [CrossRef] [PubMed]
- Barik, A.; Li, L.; Sathyamurthy, A. Schwann Cells in Neuromuscular Junction Formation and Maintenance. J. Neurosci. 2016, 36, 9770–9781. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.I.; Thompson, W.J.; Harlow, M.L. Schwann cells participate in synapse elimination at the developing neuromuscular junction. Curr. Opin. Neurobiol. 2017, 47, 176–181. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Lu, Y.; Shen, C.; Patel, N.; Gan, L.; Xiong, W.C.; Mei, L. Distinct Roles of Muscle and Motoneuron LRP4 in Neuromuscular Junction Formation. Neuron 2012, 75, 94–107. [Google Scholar] [CrossRef]
- Yumoto, N.; Kim, N.; Burden, S.J. Lrp4 is a retrograde signal for presynaptic differentiation at neuromuscular synapses. Nat. Cell Biol. 2012, 489, 438–442. [Google Scholar] [CrossRef]
- Barik, A.; Lu, Y.; Sathyamurthy, A. LRP4 is critical for neuromuscular junction maintenance. J. Neurosci. 2014, 34, 13892–13905. [Google Scholar] [CrossRef]
- Leupin, O.; Piters, E.; Halleux, C.; Hu, S.; Kramer, I.; Morvan, F.; Bouwmeester, T.; Schirle, M.; Bueno-Lozano, M.; Fuentes, F.J.R.; et al. Bone Overgrowth-associated Mutations in the LRP4 Gene Impair Sclerostin Facilitator Function. J. Biol. Chem. 2011, 286, 19489–19500. [Google Scholar] [CrossRef] [PubMed]
- Bullock, W.A.; Hoggatt, A.M.; Horan, D.J. Lrp4 Mediates Bone Homeostasis and Mechanotransduction through Interaction with Sclerostin In Vivo. iScience 2019, 20, 205–215. [Google Scholar] [CrossRef] [PubMed]
- Tian, J.; Shao, J.; Liu, C.; Hou, H.-Y.; Chou, C.-W.; Shboul, M.; Li, G.-Q.; El-Khateeb, M.; Samarah, O.Q.; Kou, Y.; et al. Deficiency of lrp4 in zebrafish and human LRP4 mutation induce aberrant activation of Jagged–Notch signaling in fin and limb development. Cell. Mol. Life Sci. 2018, 76, 163–178. [Google Scholar] [CrossRef] [PubMed]
- Ohazama, A.; Porntaveetus, T.; Ota, M.S.; Herz, J.; Sharpe, P.T. Lrp4: A novel modulator of extracellular signaling in craniofacial organogenesis. Am. J. Med. Genet. Part A 2010, 152A, 2974–2983. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Pawlik, B.; Elcioglu, N. LRP4 mutations alter Wnt/beta-catenin signaling and cause limb and kidney malformations in Cenani-Lenz syndrome. Am. J. Hum. Genet. 2010, 86, 696–706. [Google Scholar] [CrossRef]
- Ahn, Y.; Sims, C.; Logue, J.M.; Weatherbee, S.D.; Krumlauf, R. Lrp4 and Wise interplay controls the formation and patterning of mammary and other skin appendage placodes by modulating Wnt signaling. Development 2013, 140, 583–593. [Google Scholar] [CrossRef]
- Mosca, T.J.; Luginbuhl, D.J.; Wang, I.E. Presynaptic LRP4 promotes synapse number and function of excitatory CNS neurons. eLife 2017, 6. [Google Scholar] [CrossRef]
- Gomez, A.M.; Froemke, R.C.; Burden, S.J. Synaptic plasticity and cognitive function are disrupted in the absence of Lrp4. Elife 2014, 3, e04287. [Google Scholar] [CrossRef]
- Zhang, H.; Sathyamurthy, A.; Liu, F.; Li, L.; Zhang, L.; Dong, Z.; Cui, W.; Sun, X.; Zhao, K.; Wang, H.; et al. Agrin-Lrp4-Ror2 signaling regulates adult hippocampal neurogenesis in mice. eLife 2019, 8. [Google Scholar] [CrossRef] [PubMed]
- Tzartos, J.S.; Zisimopoulou, P.; Rentzos, M.; Karandreas, N.; Zouvelou, V.; Evangelakou, P.; Tsonis, A.; Thomaidis, T.; Lauria, G.; Andreetta, F.; et al. LRP 4 antibodies in serum and CSF from amyotrophic lateral sclerosis patients. Ann. Clin. Transl. Neurol. 2013, 1, 80–87. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Chen, W.; Tan, Z. A Role of Low-Density Lipoprotein Receptor-Related Protein 4 (LRP4) in Astrocytic Abeta Clearance. J. Neurosci. 2020, 40, 5347–5361. [Google Scholar] [CrossRef]
- Ye, X.-C.; Hu, J.-X.; Li, L.; Li, Q.; Tang, F.-L.; Lin, S.; Sun, N.; Sun, X.-D.; Cui, G.-Y.; Mei, L.; et al. Astrocytic Lrp4 (Low-Density Lipoprotein Receptor–Related Protein 4) Contributes to Ischemia-Induced Brain Injury by Regulating ATP Release and Adenosine-A2AR (Adenosine A2A Receptor) Signaling. Stroke 2018, 49, 165–174. [Google Scholar] [CrossRef] [PubMed]
- DePew, A.; Mosca, T. Conservation and Innovation: Versatile Roles for LRP4 in Nervous System Development. J. Dev. Biol. 2021, 9, 9. [Google Scholar] [CrossRef] [PubMed]
- Gribble, K.D.; Walker, L.J.; Saint-Amant, L.; Kuwada, J.Y.; Granato, M. The synaptic receptor Lrp4 promotes peripheral nerve regeneration. Nat. Commun. 2018, 9, 2389. [Google Scholar] [CrossRef]
- Remédio, L.; Gribble, K.D.; Lee, J.K.; Kim, N.; Hallock, P.T.; Delestrée, N.; Mentis, G.Z.; Froemke, R.C.; Granato, M.; Burden, S.J. Diverging roles for Lrp4 and Wnt signaling in neuromuscular synapse development during evolution. Genes Dev. 2016, 30, 1058–1069. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.-D.; Li, L.; Liu, F.; Huang, Z.-H.; Bean, J.C.; Jiao, H.-F.; Barik, A.; Kim, S.-M.; Wu, H.; Shen, C.; et al. Lrp4 in astrocytes modulates glutamatergic transmission. Nat. Neurosci. 2016, 19, 1010–1018. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Zhang, M.; Luo, B.; Jing, H.; Yu, Y.; Wang, S.; Luo, S. Lrp4 in hippocampal astrocytes serves as a negative feedback factor in seizures. Cell Biosci. 2020, 10, 135. [Google Scholar] [CrossRef]
- Liang, C.; Tao, Y.; Shen, C.; Tan, Z.; Xiong, W.-C.; Mei, L. Erbin Is Required for Myelination in Regenerated Axons after Injury. J. Neurosci. 2012, 32, 15169–15180. [Google Scholar] [CrossRef]
- Zhao, K.; Shen, C.; Lu, Y.; Huang, Z.; Li, L.; Rand, C.D.; Pan, J.; Sun, X.-D.; Tan, Z.; Wang, H.; et al. Muscle Yap Is a Regulator of Neuromuscular Junction Formation and Regeneration. J. Neurosci. 2017, 37, 3465–3477. [Google Scholar] [CrossRef]
- Kang, H.; Tian, L.; Mikesh, M.; Lichtman, J.W.; Thompson, W.J. Terminal Schwann Cells Participate in Neuromuscular Synapse Remodeling during Reinnervation following Nerve Injury. J. Neurosci. 2014, 34, 6323–6333. [Google Scholar] [CrossRef]
- Castro, R.; Taetzsch, T.; Vaughan, S.K.; Godbe, K.; Chappell, J.; Settlage, R.E.; Valdez, G. Specific labeling of synaptic schwann cells reveals unique cellular and molecular features. eLife 2020, 9. [Google Scholar] [CrossRef]
- Adilakshmi, T.; Ness-Myers, J.; Madrid-Aliste, C.; Fiser, A.; Tapinos, N. A Nuclear Variant of ErbB3 Receptor Tyrosine Kinase Regulates Ezrin Distribution and Schwann Cell Myelination. J. Neurosci. 2011, 31, 5106–5119. [Google Scholar] [CrossRef]
- Hirata, K.; Kawabuchi, M. Myelin phagocytosis by macrophages and nonmacrophages during Wallerian degeneration. Microsc. Res. Tech. 2002, 57, 541–547. [Google Scholar] [CrossRef]
- Vargas, M.E.; Watanabe, J.; Singh, S.J.; Robinson, W.H.; Barres, B.A. Endogenous antibodies promote rapid myelin clearance and effective axon regeneration after nerve injury. Proc. Natl. Acad. Sci. USA 2010, 107, 11993–11998. [Google Scholar] [CrossRef]
- Jessen, K.R.; Mirsky, R. Negative regulation of myelination: Relevance for development, injury, and demyelinating disease. Glia 2008, 56, 1552–1565. [Google Scholar] [CrossRef]
- Jessen, K.R.; Mirsky, R. Signals that determine Schwann cell identity*. J. Anat. 2002, 200, 367–376. [Google Scholar] [CrossRef]
- Chen, Z.-L.; Yu, W.-M.; Strickland, S. Peripheral Regeneration. Annu. Rev. Neurosci. 2007, 30, 209–233. [Google Scholar] [CrossRef]
- Decker, L.; Desmarquet-Trin-Dinh, C.; Taillebourg, E.; Ghislain, J.; Vallat, J.-M.; Charnay, P. Peripheral Myelin Maintenance Is a Dynamic Process Requiring Constant Krox20 Expression. J. Neurosci. 2006, 26, 9771–9779. [Google Scholar] [CrossRef]
- Liu, Y.; Sugiura, Y.; Padgett, D. Postsynaptic development of the neuromuscular junction in mice lacking the gamma-subunit of muscle nicotinic acetylcholine receptor. J. Mol. Neurosci. 2010, 40, 21–26. [Google Scholar] [CrossRef][Green Version]
- Chen, F.; Liu, Y.; Sugiura, Y.; Allen, P.D.; Gregg, R.G.; Lin, W. Neuromuscular synaptic patterning requires the function of skeletal muscle dihydropyridine receptors. Nat. Neurosci. 2011, 14, 570–577. [Google Scholar] [CrossRef]
- Zhao, K.; Shen, C.; Li, L.; Wu, H.; Xing, G.; Dong, Z.; Jing, H.; Chen, W.; Zhang, H.; Tan, Z.; et al. Sarcoglycan Alpha Mitigates Neuromuscular Junction Decline in Aged Mice by Stabilizing LRP4. J. Neurosci. 2018, 38, 8860–8873. [Google Scholar] [CrossRef]
- Lin, M.; Xiong, W.-C.; Mei, L. Neuromuscular Junction Formation, Aging, and Disorders. Annu. Rev. Physiol. 2018, 80, 159–188. [Google Scholar]
- Abdesselem, H.; Shypitsyna, A.; Solis, G.P. No Nogo66- and NgR-mediated inhibition of regenerating axons in the zebrafish optic nerve. J. Neurosci. 2009, 29, 15489–15498. [Google Scholar] [CrossRef]
- Zou, S.; Tian, C.; Ge, S.; Hu, B. Neurogenesis of Retinal Ganglion Cells Is Not Essential to Visual Functional Recovery after Optic Nerve Injury in Adult Zebrafish. PLoS ONE 2013, 8, e57280. [Google Scholar] [CrossRef]
- Diekmann, H.; Kalbhen, P.; Fischer, D. Characterization of optic nerve regeneration using transgenic zebrafish. Front. Cell. Neurosci. 2015, 9. [Google Scholar] [CrossRef]
- Gonzalez, D.; Allende, M. Current Advances in Comprehending Dynamics of Regenerating Axons and Axon–Glia Interactions after Peripheral Nerve Injury in Zebrafish. Int. J. Mol. Sci. 2021, 22, 2484. [Google Scholar] [CrossRef]
- Hu, B.-B.; Chen, M.; Huang, R.-C.; Huang, Y.-B.; Xu, Y.; Yin, W.; Li, L.; Hu, B. In vivo imaging of Mauthner axon regeneration, remyelination and synapses re-establishment after laser axotomy in zebrafish larvae. Exp. Neurol. 2018, 300, 67–73. [Google Scholar] [CrossRef]
- Magill-Solc, C.; Mcmahan, U.J. Motor neurons contain agrin-like molecules. J. Cell Biol. 1988, 107, 1825–1833. [Google Scholar] [CrossRef]
- Neuhuber, B.; Daniels, M.P. Targeting of recombinant agrin to axonal growth cones. Mol. Cell. Neurosci. 2003, 24, 1180–1196. [Google Scholar] [CrossRef]
- Suzuki, K.; Lovera, M.; Schmachtenberg, O.; Couve, E. Axonal Degeneration in Dental Pulp Precedes Human Primary Teeth Exfoliation. J. Dent. Res. 2015, 94, 1446–1453. [Google Scholar] [CrossRef] [PubMed]
- De Felipe, C.; Hunt, S.P. The differential control of c-jun expression in regenerating sensory neurons and their associated glial cells. J. Neurosci. 1994, 14, 2911–2923. [Google Scholar] [CrossRef] [PubMed]
- Parkinson, D.B.; Bhaskaran, A.; Droggiti, A.; Dickinson, S.; D’Antonio, M.; Mirsky, R.; Jessen, K.R. Krox-20 inhibits Jun-NH2-terminal kinase/c-Jun to control Schwann cell proliferation and death. J. Cell Biol. 2004, 164, 385–394. [Google Scholar] [CrossRef]
- Parkinson, D.B.; Bhaskaran, A.; Arthur-Farraj, P. c-Jun is a negative regulator of myelination. J. Cell Biol. 2008, 181, 625–637. [Google Scholar] [CrossRef] [PubMed]
- Matsuno, K. Notch signaling. Dev. Growth Differ. 2019, 62, 3. [Google Scholar] [CrossRef]
- Jessen, K.R.; Mirsky, R. The repair Schwann cell and its function in regenerating nerves. J. Physiol. 2016, 594, 3521–3531. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hui, T.-K.; Lai, X.-S.; Dong, X.; Jing, H.; Liu, Z.; Fei, E.; Chen, W.-B.; Wang, S.; Ren, D.; Zou, S.; et al. Ablation of Lrp4 in Schwann Cells Promotes Peripheral Nerve Regeneration in Mice. Biology 2021, 10, 452. https://doi.org/10.3390/biology10060452
Hui T-K, Lai X-S, Dong X, Jing H, Liu Z, Fei E, Chen W-B, Wang S, Ren D, Zou S, et al. Ablation of Lrp4 in Schwann Cells Promotes Peripheral Nerve Regeneration in Mice. Biology. 2021; 10(6):452. https://doi.org/10.3390/biology10060452
Chicago/Turabian StyleHui, Tian-Kun, Xin-Sheng Lai, Xia Dong, Hongyang Jing, Ziyang Liu, Erkang Fei, Wen-Bing Chen, Shunqi Wang, Dongyan Ren, Suqi Zou, and et al. 2021. "Ablation of Lrp4 in Schwann Cells Promotes Peripheral Nerve Regeneration in Mice" Biology 10, no. 6: 452. https://doi.org/10.3390/biology10060452
APA StyleHui, T.-K., Lai, X.-S., Dong, X., Jing, H., Liu, Z., Fei, E., Chen, W.-B., Wang, S., Ren, D., Zou, S., Wu, H.-T., & Pan, B.-X. (2021). Ablation of Lrp4 in Schwann Cells Promotes Peripheral Nerve Regeneration in Mice. Biology, 10(6), 452. https://doi.org/10.3390/biology10060452








