Simple Summary
Craniofacial development is an intricate and delicate process in normal embryogenesis requiring spatiotemporal release of various mediators/proteins that provide communication between different cell/tissue types, like epithelial cells, connective tissue, and endothelial cells. If this sequence is impaired or blocked due to genetic or environmental factors, it can lead to clefting. A cleft is an abnormal space or gap in the upper lip, alveolus, or palate that occurs due to failure of completion of fusion processes in the midline during facial development. Previous studies have identified various genetic factors (genes) that can lead to clefting. The most promising candidates amongst them are FGF/FGFR (fibroblast growth factor/FGF receptor) signaling genes and FOX (forkhead box protein) genes. We investigated the expression of these genes in tissue material obtained from cleft-affected patients. Our results indicate that these genes profoundly affect the pathogenesis and manifestation of clefts, especially by enhancing local site inflammation and fibrosis. Further, they play a vital role in angiogenesis, apoptosis, and cell proliferation.
Abstract
Orofacial clefts affect hundreds of thousands of children worldwide annually and are usually corrected by a series of surgeries extending to childhood. The underlying mechanisms that lead to clefts are still unknown, mainly because of the multifactorial etiology and the myriad of interactions between genes and environmental factors. In the present study, we investigated the role and expression of candidate genes belonging to the FGF/FGFR signaling pathway and FOX family in tissue material obtained from 12 pediatric patients undergoing cleft correction surgery. The expression was investigated using immunohistochemistry (IHC) and chromogenic in-situ hybridization (CISH) in three cell/tissue types—epithelial cells, connective tissue, and endothelial cells. We found elevated expression of FGFR1 in epithelial cells while no expression was observed in endothelial cells. Further, our results elucidate the potential pathogenetic role of FGFR1 in cellular proliferation, local site inflammation, and fibrosis in cleft patients. Along with bFGF (also called FGF2), FGFR1 could play a pro-inflammatory role in clefts. Over-amplification of FGFR2 in some patients, along with bFGF, could potentially suggest roles for these genes in angiogenesis. Additionally, increased expression of FOXE1 (also called TTF2) contributes to local site inflammation. Finally, zero to low amplification of FOXO1 could suggest its potential role in inducing oxidative stress in the endothelium along with reduced epithelial apoptosis.
Keywords:
cleft palate; cleft lip; immunohistochemistry; in-situ hybridization; inflammation; FGF/FGFR; FOXE1; FOXO1 1. Introduction
Congenital cleft lip and palate is one of the most commonly reported birth defects, affecting hundreds of thousands of children worldwide [1]. A cleft is defined as an abnormal space or gap in the upper lip, alveolus, or palate that occurs due to failure of completion of fusion processes in the midline during facial development [2]. Orofacial clefts have been reported to occur as isolated cases or as associated manifestations in over 500 recognized Mendelian syndromes (Online Mendelian Inheritance in Man (OMIM), www.ncbi.nlm.nih.gov/omim/, accessed on 25 March 2021).
Due to its complex etiology, clinical manifestations of clefting are not limited to only dental complications but also encompass speech difficulties, ear infections, feeding problems, as well as behavioral complications [3]. The treatment of such patients, naturally, is also complex, multidisciplinary, long term, exhaustive (both mentally and physically), and generally involves multiple phases of surgical intervention [2,4]. To achieve the optimum results, both in terms of aesthetic restoration and functional normality, it is extremely important that the surgical care is provided at the right age and right time of development [5].
The process of palatogenesis (embryonic development of lip and palate) is a multistep process that begins at the fourth gestational week [6]. It is a very tightly regulated sequalae of events that involves the development, maturation, and fusion of facial processes in the midline [6]. For this fusion to occur, the bilateral palatal shelves must grow beside the tongue and then elevate towards the midline to form the medial edge epithelium (MEE) [7,8]. This is followed by degeneration of the MEE and disappearance of the midline epithelial seam [9]. This programmed cell death of MEE is mediated by the mesenchymal cells (embryonic connective tissue), which ensure that a continuity can be established across the horizontal plate [7]. The mesenchyme in the palate traces its origin to the cranial neural crest (CNC) cells, a subset of neural crest cells (NCCs). During craniofacial development, CNC cells migrate from the lateral ridges of the neural plate to branchial arches [8]. Similarly, part of the epithelium in the palate traces its origin to pharyngeal ectoderm-derived epithelial cells [8,10].
The crosstalk (mediated by cytokines) between the epithelium and the mesenchyme is crucial for normal palatogenesis (especially for degeneration of the MEE) and any disruption in the mesenchyme–epithelial crosstalk eventually leads to clefts of varying severity [6,7,8,9,10]. The crosstalk can be disrupted by various genetic and environmental factors, like maternal smoking, nutrition, alcohol consumption, and so on. Further, defects, either in facial mesenchyme patterning, growth or in epithelium fusion, also result in cleft palate [8]. Occasionally, a fibrous band, known as Simonart’s band, is observed attached to the cleft, suggesting that occlusive epithelial adhesions can also result in clefting [11].
Therefore, it is imperative to study the genes/pathways that regulate or control this crosstalk during palatogenesis in order to completely understand the underlying pathogenesis. Many candidate genes have been suggested to play a role in crosstalk (directly or indirectly) and carry mutations in patients with non-syndromic clefts. While the roles of genes like IRF6 (interferon regulatory factor 6) [12,13] and VAX1 (ventral anterior homeobox 1) [14,15] have been confirmed after extensive research, the roles of other genes are yet to be fully substantiated and confirmed. Other promising candidates include the FGF (fibroblast growth factor) signaling family genes [16,17,18], FOX (forkhead box protein) family genes [19,20], MSX1 (msh homeobox 1) [21,22], and BMP4 (bone morphogenetic protein 4) [23].
The FGF family includes 18 members that mediate their actions via four distinct receptors (FGFRs). These receptors show differential binding properties and, upon activation, regulate various cellular processes such as cell proliferation, differentiation, apoptosis, and mobility [24,25]. The FGF/FGFR family plays a vital role in maintenance of bone homeostasis and mutations in these genes are associated with various congenital bone diseases, including chondrodysplasia syndromes, craniosynostosis syndromes, and syndromes with dysregulated phosphate metabolism [25,26]. FOX family genes, on the other hand, consist of transcriptional regulators, divided into 19 classes from FOXA to FOXS [11], which are involved in the development of various organs, regulation of senescence or proliferation, and metabolic homeostasis [27].
Due to their crucial roles in mesenchymal–epithelial crosstalk and in overall craniofacial development, in the present study we investigated the expression and role of FGF/FGFR signaling pathway genes and the FOX family genes in orofacial clefts. Further, we investigated the expression of these genes in different tissues/cells—epithelial cells, connective tissue, and endothelial cells. Finally, we investigated their possible roles in subsequent local inflammation, along with possible gene–gene interactions and the role of environmental factors, to allow for better understanding, prediction, and diagnosis of clefts, as well as better treatment modalities.
2. Materials and Methods
2.1. Profile of Study Participants
In the present study, tissue samples were obtained from 12 pediatric patients (10 male children and 2 female children) who presented for consultation and treatment at the Department of Oral and Maxillofacial Surgery, Institute of Stomatology, Riga Stradiņš University (RSU), Latvia. The tissue for all patients was collected from the site of clefting by the same surgeon. The study was approved by the Research Ethics Committee (REC) of RSU with the approvals dated 22 May 2003, 17 January 2013, and 28 June 2018, in accordance with the 1975 Helsinki Declaration (as revised in 2008). Written informed consent was obtained from all patients (given by the parents) for participation in the study and publication of the study data.
Children in the study were aged between 3 and 18 months at the time of tissue collection and were scheduled for plastic surgery of either bilateral or unilateral clefts. None of the children were previously diagnosed with coexisting genetic syndromes, chromosomal abnormalities, or immune deficiencies. Briefly, mothers of two infants were reported to have threats of miscarriage in pregnancy; two other infants were reported to have parents with histories of smoking; three infants had mothers who used paracetamol in pregnancy and two infants presented with family histories of genetic disorders (however, the children were not affected). Table 1 summarizes the clinical information of the patients.

Table 1.
Profile of study participants and clinical diagnosis.
2.2. Data and Sample Collection
Tissue samples were collected immediately after the plastic surgery and fixed for a day in a mixture of 2% formaldehyde and 0.2% picric acid in 0.1 M phosphate buffer (pH 7.2). Next, the samples were rinsed in Tyrode buffer (content: NaCl, KCl, CaCl2·2H2O, MgCl2·6H2O, NaHCO3, NaH2PO4·H2O, glucose) containing 10% saccharose for 12 h followed by paraffin embedment. Samples were registered and assigned randomized sequence tags. Patient identity was not disclosed at any time to the researchers and/or lab assistants, in accordance with protocol. Only patient history (as shown in Table 1) was kept with the sequence tags.
2.3. Routine Histological Investigation
In accordance with standard laboratory procedures, 3–4 µm of serial tissue sections were prepared from the solidified paraffin block for histological staining and immunohistochemistry (IHC). Tissue sections placed on the slides were kept at 56 °C for 20–60 min in a thermostat. De-paraffinization of the sections was undertaken using xylene solution and 96% ethanol alcohol. Hematoxylin and eosin staining (Mayer’s; Bio Optica Milano, Milan, Italy) was undertaken using standard procedure.
Slides were dehydrated with ethanol and clarified with carboxylol and xylene. Finally, a drop of histological Pertex glue (Histolab Products AB, Askim, Sweden) was applied and slides were covered with a cover glass. Slides were visualized using brightfield light microscopy with a Leica DC 300F camera microscope (Leica DM500RB; Leica Biosystems Richmond, Richmond, IL, USA).
2.4. Immunohistochemistry (IHC)
De-paraffinized, washed, and cleared tissue sections were rinsed with TRIS buffer (Diapath, Martinengo, Italy) for 10 min followed by boiling in EDTA buffer in a microwave for 10 min. The tissue samples were cooled to 65 °C and then placed again in TRIS wash buffer. Endogenous peroxidase was blocked with 3% peroxidase (Dako, Naestved, Denmark). FGF basic (ab16828, working dilution 1:200, rabbit, Abcam, Cambridge, UK), FGFR1 (orb38277, working dilution 1:50, rabbit, Biorbyt Limited, Cambridge, UK), and FOXE1 (ab5080, working dilution 1:500, goat, Abcam, Cambridge, UK) antibodies were used for biotin–streptavidin immunohistochemistry.
All antibodies were diluted with antibody diluent (Cell MarqueTM, Rocklin, CA, USA). Incubation with primary antibody was performed for 2 h followed by washing in TRIS wash buffer. The HiDef DetectionTM HRP polymer system (Cell MarqueTM, Rocklin, CA, USA) was used for rabbit antibodies as per the manufacturer’s guidelines. The ImmunoCruzTM ABC staining system was used for goat antibody (Santa Cruz Biotechnology, Dallas, TX, USA) as per the manufacturer’s guidelines. Tissue sections were incubated with biotin-containing secondary antibody for 30 min and rinsed again for 10 min in TRIS wash buffer, followed by another round of incubation and washing with biotin-containing tertiary antibody and TRIS wash buffer.
The tissue sections were then coated with the DAB+ chromogenic liquid using a DAB Substrate Kit (Cell MarqueTM, Rocklin, CA, USA) and incubated at room temperature for up to 10 min to obtain brown staining of immunoreactive structures. The sections were then washed in distilled water and contrast-stained with hematoxylin for 2 min. The antibody-treated tissue material was dehydrated with ethanol solutions and clarified with carboxylol and xylene. The slides were prepared and viewed under a light microscope (as described previously). Negative and positive IHC controls were prepared for each sample in the study.
2.5. Chromogenic In-Situ Hybridization (CISH)
CISH is a relatively new technique that utilizes a chromogen-labeled DNA probe which is often visualized using peroxidase reaction. The technique is based on the principle of subtractive hybridization. The technique allows for simultaneous assessment of tissue morphology and CISH signals and is standardized with complete kits, thereby eliminating the need to perform the more expensive fluorescent microscopy [28,29]. Due to easy interpretation of results and the technique’s superiority when compared with IHC, we decided to detect the signals of the candidate genes using CISH.
CISH was performed using ZytoDot 2C CISH Implementation Kit (ZytoVision GmbH, Bremerhaven, Germany). Probes of FGFR1, FGFR2, and FOXO1 were used in this study. Pretreatment was performed using standard laboratory methods. Denaturation and hybridization were undertaken using 10 μL of each probe placed on each pretreated specimen with a pipette. Slides were covered with an 18 mm × 18 mm coverslip and placed on a hot plate for 5 min at 79 °C, then transferred to a humidity chamber and hybridized overnight at 37 °C.
To proceed with the detection process, the coverslips were removed by submerging the slides in SSC wash buffer followed by TBS wash buffer. Then, the slides underwent the next steps in the CISH procedure as per the manufacturer’s guidelines. Slides were transferred into a staining jar and washed for 2 min under cold running tap water. Dehydration was undertaken with 100% ethanol and the slides were then incubated in xylene. The coverslips were re-attached while avoiding air bubbles and the slides visualized under a light microscope.
Under the microscope, red-colored dots indicated control, whereas green-colored dots indicated target. After counterstaining the nucleus with a nuclear dye, hybridized probe fragments were visualized. Two signals per probe were expected to appear in the cell nuclei of normal cells in interphase or metaphase or in the nuclei of cells without aberrations in the examined chromosomes.
2.6. Visualization and Statistical Analysis
The images obtained using microscopy were analyzed with Image Pro Plus 6.0 (Media Cybernetics, Rockville, MD, USA). Cells with nucleus/cytoplasm marked brown in the IHC reaction were considered as immunoreactive, i.e., as showing immunopositivity. For CISH, the signals from the green probes in the cells/nuclei were evaluated. Epithelium, Connective Tissue, and wall of mucosal microcirculation blood vessels (i.e., Endothelium) were assessed. Semi-quantitative counting was undertaken by two independent morphologists in at least five randomly selected vision fields, each at 400× magnification, for each tissue section in order to quantify the immunoreactive and probe-containing cells (Table 2) [30,31].

Table 2.
Semi-quantitative grading scale used in the present study.
Statistical analysis was performed using non-parametric Kruskal–Wallis ANOVA with appropriate post hoc tests and Bonferroni correction for inter-group comparison. Spearman’s Rho was used for correlation analysis. The data were stored and analyzed using MS Excel (MS Office 365) and SPSS v26.0 (IBM Corp., Armonk, NY, USA). For statistical analysis, the numbers of “+” values were considered as equivalent to absolute whole numbers (e.g., “+” corresponded to 1; “++” corresponded to 2, and so on). Statistical significance was set at p < 0.05.
3. Results
3.1. Immunohistochemistry Analysis
As shown in Figure 1 and Table 3, low to moderate numbers of epithelial cells in the tissue demonstrated bFGF, FGFR1, and FOXE1 protein expression (mean IHC semi-quantitative grade: 1.2+, 1.4+, and 1.5+, respectively). All three proteins, however, showed high variation in expression amongst the patients, as evidenced by the high coefficient of variation (CV%). In the connective tissue, expression of bFGF and FGFR1 was undetectable, with only a few cells showing a positive reaction (except in patients 3 and 8). In contrast, FOXE1 expression was seen in numerous cells, with small variation amongst the patients (CV% = 29%).
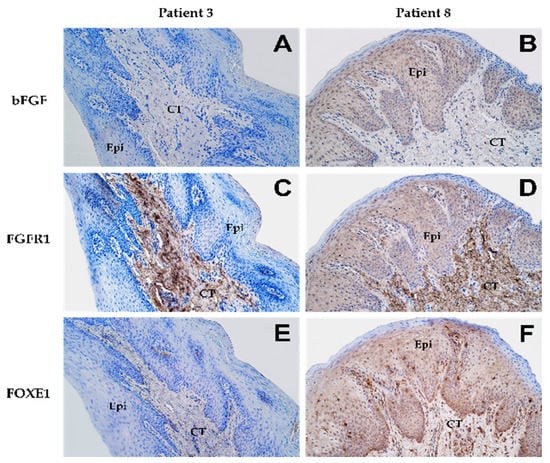
Figure 1.
Expression patterns of various proteins as shown by immunohistochemical staining in lip mucosal tissue obtained from patient 3 (A,C,E) and patient 8 (B,D,F) at 200× magnification. “Epi” denotes epithelium while “CT” denotes connective tissue. Expression of bFGF in the epithelium shown by (A) none of the cells (0) and (B) a moderate number of cells (++). Connective tissue in both patients (A,B) showed no expression (0) of bFGF. Expression of FGFR1 in the epithelium shown by (C) none of the cells (0) and (D) a moderate number of cells (++). In connective tissue, both patients (C,D) presented numerous cells (+++) showing expression of FGFR1. Expression of FOXE1 in the epithelium shown by (E) none of the cells (0) and (F) numerous cells (+++). However, in connective tissue, patient 3 (E) showed few positive cells (+) while patient 8 (F) showed numerous positive cells (+++) for FOXE1.

Table 3.
Results by patient (semi-quantitative grading scale) for the IHC staining.
None of the endothelial cells showed a positive reaction for FGFR1 protein in any of the patients while bFGF protein was evident only in a few cells. A high abundance of cells showed positive reaction for FOXE1 in the endothelium (mean IHC semi-quantitative grade: 3.75+). Further, expression of FOXE1 protein was also found to be the least variable (low CV%) amongst patients, indicating its near-universally high positive reaction in endothelial cells.
Kruskal–Wallis ANOVA post hoc analysis revealed that there were significant differences in the number of cells expressing FGFR1 and FOXE1 proteins amongst the three cell types (Figure 2B,C). A significantly greater number of both connective tissue and epithelial cells showed positive reactions for FGFR1 compared to the endothelial cells (p = 0.018 and < 0.001, respectively). For FOXE1, a significantly higher number of epithelial cells showed positive reactions compared to both connective tissue and endothelial cells (p = 0.028 and < 0.001, respectively). No significant differences were obtained for the number of cells positive for bFGF protein expression amongst the tissue types (Figure 2A).
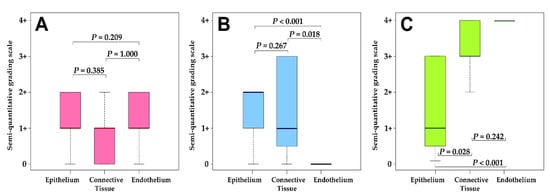
Figure 2.
Distribution of semi-quantitative grading for IHC staining for (A) bFGF, (B) FGFR1, and (C) FOXE1. The adjusted P values indicated were obtained using the post hoc tests from the Kruskal–Wallis ANOVA test. An interpretation of the grading scale is shown in Table 2.
3.2. Chromogenic In-situ Hybridization Analysis
As shown in Figure 3 and Table 4, zero to low levels of amplification were observed for FGFR1 and FGFR2 in the epithelial tissue (mean CISH semi-quantitative grade: 0.67+ and 0.75+, respectively). However, the level of amplification was not uniform across the patients and varied greatly, indicating the role of other potential influencing factors (CV% > 100%). In the connective tissue, FGFR1 demonstrated a low level of amplification while FGFR2 demonstrated no amplification.
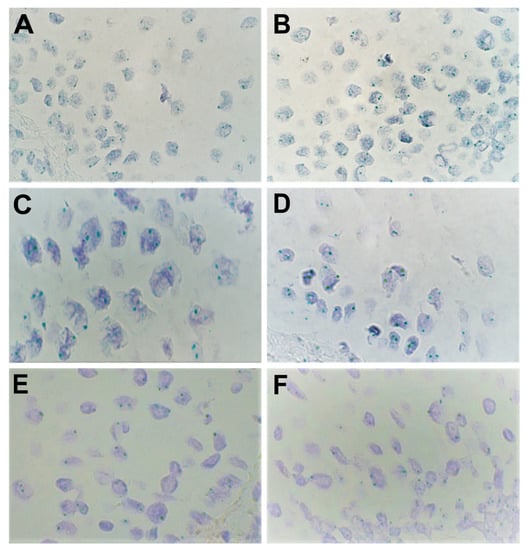
Figure 3.
Expression of various genes as shown by chromogenic in-situ hybridization (CISH) in lip mucosal tissue at 1000× magnification (visualized using immersion oil). Amplification was visualized through the number of green signals (dots) per nucleus per cell. (A,B) Moderate amplification (++) was found for levels of FGFR1 in the epithelium in patients 2 and 5, respectively. (C) Weak amplification (+) and (D) no amplification (0) were found for levels of FGFR2 in the epithelium in patients 9 and 1, respectively. (E,F) No amplification (0) was found for levels of FOXO1 in the epithelium in patients 8 and 12, respectively.

Table 4.
Results by patient (semi-quantitative grading scale) for the CISH analysis.
In the endothelium, almost no amplification was noted for both FGFR1 and FGFR2. No amplification was detected for FOXO1 in any of the three tissues. Further, the Kruskal–Wallis test showed no significant difference in amplification levels of FGFR1 and FGFR2 between the three cell types (p = 0.0583 and 0.0581, respectively). Significant differences in amplification levels amongst the cells were noted for FOXO1 (p = 0.0415).
3.3. Correlation Analysis.
With regard to the immunohistochemistry results, FGFR1 in connective tissue showed a significant (p < 0.05) and very strong positive correlation with bFGF in connective tissue (ρ = 0.762) and FOXE1 in endothelial cells (ρ = 0.771), while a strong positive relationship was noted with FOXE1 in connective tissue (ρ = 0.668). As shown in Figure 4, in the epithelium all three proteins showed very strong significant positive correlations with each other (bFGF–FGFR1 with ρ = 0.775; bFGF–FOXE1 with ρ = 0.725; and FOXE1–FGFR1 with ρ = 0.720). A strong positive correlation was observed for bFGF in connective tissue and the endothelium (ρ = 0.645). A very strongly association was found for FOXE1 in connective tissue and the endothelium (ρ = 0.708).
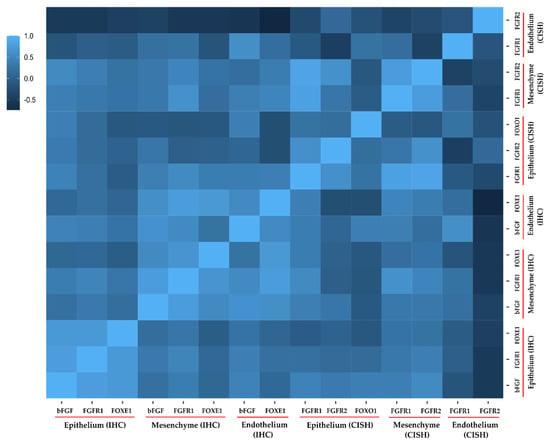
Figure 4.
Correlation heatmap for different genes/proteins studied in the present study. The legend shows the correlation strength as determined using Spearman’s rho (ρ). Mesenchyme refers to connective tissue. A standard interpretation scale was used, i.e., 0.01 to 0.19 indicated no correlation; 0.20 to 0.29 indicated a weak correlation; 0.30 to 0.39 indicated a moderate correlation; 0.40 to 0.69 indicated a strong correlation; and ≥0.70 indicated a very strong correlation. IHC denotes immunohistochemistry; CISH denotes chromogenic in-situ hybridization.
In regard to the chromogenic in-situ hybridization results, a strong to very strong positive correlation was observed between FGFR1 in epithelial cells and FGFR2 in both epithelium and connective tissue, respectively (ρ = 0.596 and 0.855, respectively). Further, FGFR1 in the epithelium was also very strongly correlated with FGFR1 in connective tissue (ρ = 0.821). A similar observation was noted for FGFR2 in the epithelium and connective tissue (ρ = 0.637). In connective tissue, both FGFR1 and FGFR2 were also very strongly associated (ρ = 0.767). Utilizing both the CISH and IHC results, a very strong and significant negative correlation was observed between FOXE1 and FGFR2 in endothelial cells (ρ = −0.739). Similarly, a strong positive association was noted for bFGF and FGFR1 (CISH) in endothelial cells (ρ = 0.590).
4. Discussion
A multifactorial pathoetiology combined with high incidence and translated downstream high socio-economic burden makes orofacial clefts of particular interest to various research groups working to elucidate the factors/cellular pathways that play a role in pathogenesis of clefts. Understanding these interactions, which can range from gene–gene (GxG) to gene–environment (GxE), is crucial for creating models and systems that can aid in predication, diagnosis, prognosis, and treatment of cleft pathology. Over 70% of cases of cleft lip and palate are non-syndromic while the remainder comprise of syndromic cases and of those syndromes that usually arise secondary to chromosomal or teratogenic effects [11]. The etiology of syndromic forms is traceable by genetic analysis, which reveals the underlying genetic mutation responsible. However, in cases of isolated, non-syndromic clefts, the gene–environment interactions (GxE), which are thought to be the main reasons for clefting, are difficult to confirm. This warrants a search for possible gene–gene interactions (GxG) and their interactions with environmental teratogens in order to isolate the underlying cause of clefting [32].
4.1. Fibroblast Growth Factor Receptor 1 (FGFR1)
The FGF/FGFR signaling genes are expressed in a spatiotemporally specific manner in the palatal tissue and constitute a directional regulatory axis between the stromal and epithelial compartments [8,17]. FGFR1, which is highly expressed in the CNC-derived connective tissue (mesenchyme) in the palatal shelf, is usually responsible for mediating epithelium-to-mesenchyme signaling while FGFR2 mediates the reciprocal communication (hence FGFR2 is more abundant in the epithelium). In a study on mice conducted by Wang et al. [8], the authors demonstrated that ablation of FGFR1 in NCCs delayed (not inhibited) cellular proliferation of both the mesenchyme and epithelium and impeded development of medial nasal processes. Further, they reported impeded elevation of palatal shelves prior to midline fusion (a key event in normal palatogenesis). Additionally, FGFR1 was also shown to play an important role in controlling MEE degeneration during palate fusion [8]. In contrast, it has also been shown that increased expression of FGFR1 leads to increased cellular proliferation in the palate shelves and cleft palate [33].
Therefore, both excess and deficiency of FGFR1 signaling leads to clefting and a precise balance in protein and gene expression are needed for normal palatogenesis. In the endothelium, FGFR1 plays a key role in inhibiting the endothelium–mesenchyme transition (EndMT) via the inhibition of TGFβ/Smad signaling pathway, thereby preventing tissue fibrosis and maintaining a normal state of vascularity [34]. Further, loss of endothelial FGFR1 and FGFR2 has been shown to result in impaired neovascularization after injury in adult mice [35]. FGFR1 has also been shown to be widely expressed in the myofibroblasts of injured palates, suggesting that FGFR1 signaling is also important for palate repair during injury [8,36]. Finally, it has been demonstrated that disruption of FGFR1 signaling reduces local inflammation by restraining activation of the NF-κB signaling cascades [37,38]. Clearly, FGFR1 plays a key role in maintaining normal development processes and in preventing fibrosis and inflammation.
Overall, our IHC and CISH results indicate that more FGFR1 receptors are present in epithelial cells, followed by connective tissue cells, and none in the endothelial cells. The presence of elevated expression of FGFR1 in cleft epithelium thus, leads to increased cellular proliferation that may lead to clefting and local site inflammation. This is in line with our previous studies in which we demonstrated moderate expression of proliferation marker Ki67 in epithelial cells from cleft tissue (compared with no Ki67 expression in control samples) [39].
Further, the fact that FGFR1 was not detected in the endothelium indicates its potential role in promoting fibrosis [34]. This finding potentially also correlates clinically with the post-operative complications, like slow healing and hypertrophic scars, that are frequently reported in cleft patients [40]. Further, elevated expression of FGFR1 leads to a significant decrease in TGFβ1 expression, a potent anti-inflammatory and immunosuppressive factor, which brings our findings in line with those of our previous study [41].
4.2. Fibroblast Growth Factor Receptor 2 (FGFR2)
FGFR2, another associated FGF receptor, is primarily expressed in the developing palatal epithelium that binds FGF10, a factor known to be localized in the adjacent underlying mesenchyme [42]. Deficiency of either FGFR2 or FGF10, or both, leads to clefting and a thin epithelium due to the severe reduction in cell proliferation [42]. Since FGF10/FGFR2 signaling affects the epithelium, there is a lack of FGFR1-mediated reciprocal signaling from the epithelium, which consequently leads to proliferation defects in the mesenchyme [42]. Further, loss of FGFR2 has been shown to compromise the organization of the rugae (the thickened lines on the secondary palate) [43]. FGFR2 has two main epithelial isoforms, namely FGFR2b and FGFR2c. While FGFR2b has been associated with tumor suppression, amplification of FGFR2c has been linked with various epithelial tumors [44,45]. Further, it has been shown that an abnormal switch from FGFR2b to the FGFR2c isoform could be the main triggering event leading to epithelium-to-mesenchyme transition (EMT) in normal human keratinocytes [46].
In a study on human keratinocyte cell line HaCaT, Ranieri et al. concluded that increased expression of FGFR2c leads to morphological and cytoskeletal changes, gene reprogramming, and invasive behavior, reminiscent of type III EMT (seen in carcinogenesis in which epithelial cells may completely lose their vestiges and become fully mesenchymal) [45]. Further, higher expression of FGFR2b has been associated with physiological type II EMT (seen in adult tissue regeneration). Since our FGFR2 CISH probe could not distinguish between the two isoforms, it was difficult to conclude, for patients 2, 5, and 11, who showed moderate amplification, and patients 7 and 3, who showed low amplification in the epithelium, which splicing isoform was more abundant and, hence, the type of EMT. Therefore, we can conclude that FGFR2 in general demonstrates a specific expression pattern and that more intensive studies investigating the role of FGFR2 isoforms are needed to further our understanding in predicting the clinical course of clefts.
4.3. Basic Fibroblast Growth Factor/Fibroblast Growth Factor 2 (bFGF/FGF2)
bFGF has been described as a biological mediator that regulates connective tissue cell migration, proliferation, and synthesis of intracellular proteins and extracellular matrix [47,48]. It has been shown to induce angiogenesis via VEGF (vascular endothelial growth factor) in vitro [49], while in vivo it has been demonstrated to accelerate the healing process [50]. Additionally, Choi et al. found that bFGF suppressed collagen type I generation and subsequent scar tissue formation [48]. Like FGFR2b, bFGF has also been associated with the potential to accelerate type II EMT, thereby promoting wound closure [51]. Elevation of bFGF levels in our patients was in line with previous findings in which bFGF was reported to be increased in concentration in the serum and affected tissue of patients with chronic inflammation and rheumatoid arthritis [52,53]. bFGF has been shown in vivo to enhance the recruitment of monocytes, T cells (due to elevated IFN-γ (interferon gamma)), and PMN (polymorphonuclear) cells in response to elevated TNF-α (tumor necrosis factor alpha) and IFN-γ [52]. These findings are in line with our previous study in which we reported elevated TNF-α and IFN-γ levels [41], indicating that pro-inflammatory cytokines mediate their inflammatory effects via elevation of bFGF in cleft tissue. Thus, our findings also indicate that bFGF, along with other factors, induces inflammatory and immune cell recruitment, which chronically leads to an angiogenic environment (supported by results showing elevated bFGF protein expression in endothelium from IHC) [41,52].
4.4. Potential Roles Mediated By FGF/FGFR Family Genes in Cleft Lip/Palate Pathogenesis
It has been reported that the bFGF/FGFR1 axis can exert an anti-inflammatory response in astrocyte-mediated neuroinflammation, especially after infrasound exposure [54]. However, other authors have reported that the axis exerts a pro-inflammatory effect [38,40,55]. Hence, in the case of orofacial clefts, we postulate that this axis indeed exerts a pro-inflammatory effect. The elevated expression of FGFR1 and bFGF and the high correlation between them in the epithelium, connective tissue, and endothelium cells supports our hypothesis. The correlation indicates a positive feedback loop between them, as previously it has been demonstrated that bFGF can upregulate the expression of FGFR1 on astrocytes [54]. It has also been shown that endothelial cells can be stimulated to produce and release bFGF by pro-inflammatory mediators like IFN-γ in combination with IL-2, IL-1β, and nitric oxide (NO) [56,57]. This creates a vicious cycle whereby damaged mucosal tissue (due to improper palatal closure, shear stress, or shock) secretes IFN-γ, TNF-α, IL-2, and so on, which upregulate bFGF, which then upregulates FGFR1 protein expression that then recruits PMNs, T cells, and so on. These cells, in turn, secrete more bFGF [58,59] which sustains inflammation (via FGFR1) and angiogenesis (via FGFR2) at the cleft site, leading to chronic inflammation and fibrotic changes and tissue scarring.
We expected that in almost half of our patients—those with no detectable FGFR2 amplification (patients 1, 4, 6, 8, 9, 10, and 12)—there would be low or zero amplification/protein levels for FGFR1 since the crosstalk was impaired. However, our findings regarding FGFR1 amplification and the moderate number of cells expressing FGFR1 protein confirm the possible amplifying role of bFGF in FGFR1 expression and the importance of these receptors in the pathogenesis of clefts. Additionally, hypoxia has been associated with the pathogenesis of cleft lip and palate; however, the exact mechanism is not yet known. Recently, polymorphisms of the HIF-1A (hypoxia inducible factor 1A) gene were ruled out as a cause of clefting [60]. We propose that hypoxia-induced clefting is probably rather related more directly with the FGF/FGFR signaling pathway (this needs further validation). Conte et al. found that, in the early hypoxia phase, bFGF upregulation does not occur at the mRNA level but at the protein level. This upregulation (in vascular smooth muscle cells) is due to increased ribosomal translational activity via IRES (internal ribosome entry site, a transcriptional regulator) rather than HREs (hypoxia response elements), which are used by HIF-1A to regulate bFGF expression [61]. However, in later stages of hypoxia, bFGF induces HIF-1A protein expression which then regulates bFGF expression (via HREs), thereby creating an autocrine amplification loop [62].
Interestingly, hypoxia along with cell damage and fluid/plasma protein exudation can also manifest as an effect of clefting in tissue due to chronic inflammation. Hypoxia can increase the sensitivity of endothelial cells via increased HSPG (heparan sulfate proteoglycan) synthesis in endothelial cells [63] along with increased bFGF production by vascular pericytes [64]. Finally, regarding angiogenesis induced by the bFGF/FGFR2 pathway, although autocrine amplification has been reported [65], its implications and effects are relatively unknown. Additionally, negative regulators that bind excessive bFGF have also been reported. PF4 (platelet factor 4) and PTX3 (pattern recognition receptor pentraxin 3, synthesized in response to TNF-α and IL-1β) both bind bFGF, leading to its decreased interaction with FGFR1/2 [66,67]. In fact, the roles of PTX3 and PF4 in cleft lip and palate have never been reported before and we suggest this as a potential new avenue for investigation since they modulate the binding of bFGF with its receptors.
4.5. Forkhead Box Protein E1/Thyroid Transcription Factor 2 (FOXE1/TTF2)
Also known as thyroid transcription factor 2 (TTF2), FOXE1 is usually expressed in later stages of development in tissues derived from pharyngeal arches and the pharyngeal wall, including the thyroid, tongue, epiglottis, palate, and esophagus [68]. Further, it has been shown to be co-expressed with Shh (sonic hedgehog) in these tissues, usually around the same time, and to mediate the epithelial–mesenchymal crosstalk [68]. Recently, analysis revealed that amplification of FOXE1 affects MSX1 and TGF-β3 downstream in the exertion of its action in the cleft tissue [69]. Interestingly, both these targets are found to be abundant at the sites where epithelial–mesenchymal interactions take place, including the cellular primordia involved in craniofacial morphogenesis [69,70]. To add to this, MSX1 is another commonly associated gene with isolated non-syndromic clefts [21,22]. TGF-β3, on the other hand, is expressed in the MEE of pre-fusion shelves, with its expression decreasing shortly after the midline epithelial seam is formed [69,71].
It has been shown in thyroid tissue that FOXE1 regulates the activity of NR4A2 (nuclear receptor subfamily 4 group A member 2). The authors showed that there is an indirect regulation between FOXE1 and NR4A2 whereby silencing of FOXE1 leads to downregulation of NR4A2 [72]. The gene NR4A2 belongs to the family of ligand-independent early response genes, which are involved in proliferation, apoptosis, and inflammation and have been demonstrated to enhance migration of mesenchymal stromal cells [73]. Additionally, it has been shown that TGF-β1/2 significantly downregulates the expression of NR4A2 in palatal mesenchymal cells [74]. NR4A2 has also been shown to be induced by TNF-α, IL-1β, and VEGF in synoviocytes, chondrocytes, endothelial cells, and immune cells [75,76]. Elevated expression of FOXE1 in our patients and the fact that VEGF can be induced by elevated bFGF (as seen in our patients), coupled with elevated TNF-α and downregulated TGF-β1 in cleft patients [41], suggests that local site inflammation is alternatively maintained in part by upregulation of the FOXE1/NR4A2 pathway in cleft patients.
4.6. Forkhead Box Protein O1 (FOXO1)
FOXO1, another transcriptional regulator, has been shown to play a crucial role in palatogenesis. It is needed for transcriptional promotion of the pro-apoptotic Fas ligand (FasL)/caspase-3 pathway in MEE cells, which paves the way for correct palate fusion [77]. However, for FOXO1 to function it needs to undergo acetylation by p300 in the presence of BAG6 (Bcl-2-associated anthanogene 6) [78]. BAG6 has also been recently described as another candidate gene for non-syndromic clefts. Non-detection of FOXO1 amplification in our patients indicates upstream signaling impairment which leads to clefting. BAG6 has also been shown to induce TGF-β3 transcriptional target expression [79], which shows the complex web of interactions taking place in cleft tissue. FOXO1 also plays a crucial role as a gatekeeper in endothelial cells. It restricts endothelial growth, thereby lowering endothelial metabolism and supporting the function of endothelial cells [80]. This helps the cells to consume less energy, nutrients, and O2, thereby reducing oxidative stress in endothelial cells. Deletion of FOXO1 has been shown to cause a profound increase in endothelial proliferation that interferes with coordinated sprouting, thereby causing hyperplasia and vessel enlargement [80]. Hence, no to low amplification of FOXO1 (as seen in our patients) leads to increased oxidative stress and damage in the cells, leading to promotion of inflammatory changes in the tissue.
4.7. Clinical Diagnostic Techniques and Advances for Cleft-Affected Patients
Cleft lip and/or palate have been reported to be associated with aplasia of the salivary glands, which leads to xerostomia, multiple dental caries, and early tooth loss [81,82]. The characteristics of the tissue replacing the defective sites can also be evaluated without surgical intervention using ultrasound, CT, MRI, or FNA (fine needle aspiration) [83]. Since salivary glands and the oral epithelium share the same ectodermal origin, it has been demonstrated that genes involved in clefting are also expressed in ductal epithelium during embryogenesis [84,85,86]. Further, it has been reported that the absence of FGFR2b is associated with an absence of salivary glands in mouse models [87]. Since more than half of our patients did not show detectable amplification of FGFR2b, it would be interesting to further study and investigate the salivary gland tissue in cleft-affected patients.
Further, with the advent of new modern technologies, like microtomography and peripheral quantitative computed tomography (pQCT), it is easier than ever to precisely map and reconstruct in 3D the morphology of the oral structures, both quantitatively and qualitatively [88]. Such techniques could, in the future, have the potential to aid and predict the outcome and severity of clefting, as well as predict the outcomes of treatments, i.e., post-operative complications like inflammation, scarring, and so on. Other digital techniques, like CAD/CAM technology and intra-oral digital scanners, can aid in the appropriate diagnostics and management of cleft-affected patients [89,90].
These techniques are more efficient compared to conventional impression modeling techniques since they get rid of the need to send impression models physically. A single electronic file can be sent to laboratories for investigation, thereby saving time, resources, and space. They also eliminate distortions as well as volumetric variations related to impression material [91,92].
4.8. Surgical Management of Cleft-Affected Patients
The main aim of surgical management is the early reconstruction of normal anatomy to allow for physiologic growth of the midface structures and enable children to develop undisturbed mastication, speech, hearing, and esthetics [93,94]. The generally accepted time for intervention has historically been reported as 10 weeks (2.5 months) after birth, since earlier intervention risks increased chances of post-operative complications during the neonatal period [95]. Advances in neonatal care and technology have made it possible to provide surgical care even at earlier stages (within the first 28 days post-birth) [96]; however, no significant differences have been reported in terms of cosmetic attraction or the success of the surgical outcome, even when the surgery is not done in the neonatal period [97]. For patients with clefting of the hard palate, the optimal time of surgery is within 18 months post-birth, since repair at later stages, i.e., beyond three years of age, has been associated with severely restricted language development and stereotyped cleft speech characteristics [95,98].
Cleft-affected patients usually undergo reconstruction of the alveolar ridge (gingiva-peri-osteoplasty) in combination with closure of the lip. However, despite this, 60–80% of patients require a second bone graft or sinus lift to allow for implant placement [93]. In these patients, apart from plastic surgery, it is important to preserve the bone volume along with augmentation of the vertical aspect of the bone. This could be followed by localized sinus elevation with minimal surgical trauma, thereby increasing the available bone for implant placement [99,100,101]. The crestal bone is displaced toward the sinus floor and the apical portion of the implant is placed in the augmented space [99,100,101]. Additionally, human maxillary sinus membrane tissue represents a potential source of multipotent mesenchymal stem cells that can promote a natural healing process [99,102,103]. Further, since the incidence of missing teeth in cleft patients is close to sixfold higher than in non-cleft patients, insertion of endosseous dental implants to replace missing teeth is greatly preferred [93,104,105,106]. Finally, maintenance of oral hygiene is of high importance in such patients to prevent the inserted implant from failing and to prevent systemic spread of infection from the oral cavity [107,108,109].
4.9. Relevance and Limitations of the Present Study
The main aim of the present study was to explore and understand the expression levels of FGF/FGFR and FOX family proteins in tissue from cleft-affected children. Firstly, the expression analysis of these genes allows an understanding of their role in promoting post-operative complications, like scarring, inflammation, and fibrosis. Secondly, since cleft lip and palate is a hereditary condition, gathering knowledge about the chances of the gene being passed on in a family and the nature and severity of the condition caused by the causative genes is inevitably a genuine concern for affected families. Hence, evaluation of such genes enables us to be better positioned to predict the heritability and the course of the manifestation of the clefting. Thirdly, since humans are diphyodonts (i.e., have two sets of dentitions—primary and permanent), it is of utmost importance to understand the gene expression of these genes in cleft-affected tissues, since 60–80% of cleft patients regularly require more than two rounds of corrective surgery and the incidence of missing teeth in cleft children is nearly six times higher than in non-cleft children [93,104,105,106].
Nonetheless, the present study has its limitations, including the limited number of patients studied and the lack of control samples. However, we must point out that the number of studies on human tissue is limited due to the non-availability of tissue material. This is understandable, especially since tissue material needs to be taken during surgery, which happens at a very tender age and is accompanied by genuine parental concerns. Further, decisions regarding procurement of tissue from patients are made with the best interests of patients in mind (if the obtainment of tissue is possible at all). Finally, we cannot at this stage predict or state whether neonatal gene expression resembles embryonic gene expression, especially at the time of palatogenesis. This is firstly because of technological limitations and secondly because of ethical guidelines prohibiting experimentation on human embryos. However, studies in rat models suggest that, during normal palatogenesis, expression of the studied genes, like FGFR1 and FGFR2, is elevated before fusion of the palate but ceases to exist after palatal plate fusion and is exhibited only by MEE cells [110]. Since it is known that MEE cells eventually degenerate due to epithelial–mesenchymal crosstalk, it can be naturally expected that expression of these genes also ceases to exist in palatal tissue. However, detection of such genes (at higher levels of expression) in palatal tissue in our samples makes us to postulate that the palate failed to fuse in these cases (and hence that the patients had clefting); thus, is the closest representation of gene expression during the events of palatal fusion. We suggest that future studies should investigate phenotypic and molecular data from animal models to further bolster our findings.
5. Conclusions
The complex myriad of interactions between various factors and environmental teratogens discussed above needs more investigation and in-depth studies. The most significant findings of the present study can be summarized as follow:
- Elevated expression of FGFR1 in cleft epithelium indicates its role in mediating cellular proliferation and local site inflammation. No to low expression in the endothelium indicates its role in fibrosis. Coupled together, this indicates that FGFR1 expression can help in predicting the sequalae and intensity of post-operative complications like scarring.
- bFGF (or FGF2) elevation may induce local site inflammation (via FGFR1) which chronically leads to creation and promotion of an environment suitable for angiogenic activity (via FGFR2). Additionally, over-amplification of FGFR2 in some patients points to its possible disordered role in epithelial–mesenchymal transition in cleft patients.
- High expression of FOXE1 possibly exerts a pro-inflammatory effect via involvement of the NR4A2/VEGF pathway (also induced by bFGF), while the lack or low level of amplification of FOXO1 can lead to retention of the midline epithelium coupled with increased endothelial oxidative stress and tissue inflammation.
Author Contributions
The present study was conceptualized, designed, and implemented by M.P., N.J. and Z.V.-V. Statistical analysis, data curation, the literature search, and visualization of the data were undertaken by N.J. Funding and project supervision were the responsibility of M.P. The initial draft was written by N.J. while reviewing and editing was undertaken by M.P., N.J. and Z.V.-V. All authors contributed equally. All authors have read and agreed to the published version of the manuscript.
Funding
The present study was funded by Riga Stradinš University (RSU), Project Nb. 5-1/384/2020 (dated 10 September 2020).
Institutional Review Board Statement
The study was approved by the Research Ethics Committee (REC) of Riga Stradinš University (RSU), with approvals dated 22 May 2003, 17 January 2013, and 28 June 2018. The protocol was designed in accordance with the Declaration of Helsinki guidelines.
Informed Consent Statement
All participants included in the study gave written as well as oral informed consent (provided by the parents) for participation in the study. The participants further consented to publication of the results.
Data Availability Statement
All datasets used/analyzed in the present study are presented in the results sections of the article.
Acknowledgments
We would like to extend our gratitude to Ilze Akota, Department of Oral and Maxillofacial Surgery, Faculty of Dentistry, RSU for providing the patient material and to Laboratory Assistant Natalija Moroza for tissue processing and tissue material preparation. We would also like to acknowledge the parents of the patients for consenting to participate in the present study.
Conflicts of Interest
The authors declare no competing interests in the present study. Furthermore, neither the funders nor the funding institution had a role in the design of the study; in the collection, analysis, or interpretation of the data; in the writing of the manuscript; or in the decision to publish the results.
References
- Mathes, D. Grabb and Smith’s plastic surgery. JAMA 2008, 299, 2450. [Google Scholar] [CrossRef]
- Vyas, T.; Gupta, P.; Kumar, S.; Gupta, R.; Gupta, T.; Singh, H.P. Cleft of lip and palate: A review. J. Fam. Med. Prim. Care 2020, 9, 2621–2625. [Google Scholar] [CrossRef]
- Mitchell, J.C.; Wood, R.J. Management of cleft lip and palate in primary care. J. Pediatr. Health Care 2000, 14, 13–19. [Google Scholar] [CrossRef]
- American Cleft Palate-Craniofacial Association (ACPA). Parameters for evaluation and treatment of patients with cleft lip/palate or other craniofacial differences. Cleft Palate Craniofacial J. 2018, 55, 137–156. [Google Scholar] [CrossRef]
- Banerjee, M.; Dhakar, A. Epidemiology-clinical profile of cleft lip and palate among children in india and its surgical consideration. CIBTech J. Surg. 2013, 2, 45–51. [Google Scholar]
- Muhamad, A.H.; Azzaldeen, A.; Watted, N. Cleft lip and palate: A comprehensive review. Int. J. Basic Appl. Med. Sci. 2014, 4, 338–355. [Google Scholar]
- Ferguson, M.W. Palate development. Development 1988, 103, 41–60. [Google Scholar] [CrossRef]
- Wang, C.; Chang, J.Y.; Yang, C.; Huang, Y.; Liu, J.; You, P.; McKeehan, W.L.; Wang, F.; Li, X. Type 1 fibroblast growth factor receptor in cranial neural crest cell-derived mesenchyme is required for palatogenesis. J. Biol. Chem. 2013, 288, 22174–22183. [Google Scholar] [CrossRef] [PubMed]
- Snyder-Warwick, A.K.; Perlyn, C.A. Coordinated events: FGF signaling and other related pathways in palatogenesis. J. Craniofacial Surg. 2012, 397–400. [Google Scholar] [CrossRef]
- Wilkie, A.O.; Morriss-Kay, G.M. Genetics of craniofacial development and malformation. Nat. Rev. Genet. 2001, 2, 458–468. [Google Scholar] [CrossRef] [PubMed]
- Dixon, M.J.; Marazita, M.L.; Beaty, T.H.; Murray, J.C. Cleft lip and palate: Understanding genetic and environmental influences. Nat. Rev. Genet. 2011, 12, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Weinberg, S.M.; Brandon, C.A.; McHenry, T.H.; Neiswanger, K.; Deleyiannis, F.W.; De Salamanca, J.E.; Castilla, E.E.; Czeizel, A.E.; Vieira, A.R.; Marazita, M.L. Rethinking isolated cleft palate: Evidence of occult lip defects in a subset of cases. Am. J. Med. Genet. A 2008, 146A, 1670–1675. [Google Scholar] [CrossRef] [PubMed]
- Scapoli, L.; Palmieri, A.; Martinelli, M.; Pezzetti, F.; Carinci, P.; Tognon, M.; Carinci, F. Strong evidence of linkage disequilibrium between polymorphisms at the IRF6 locus and nonsyndromic cleft lip with or without cleft palate, in an Italian population. Am. J. Human Genet. 2005, 76, 180–183. [Google Scholar] [CrossRef] [PubMed]
- Mangold, E.; Ludwig, K.U.; Birnbaum, S.; Baluardo, C.; Ferrian, M.; Herms, S.; Reutter, H.; de Assis, N.A.; Al Chawa, T.; Mattheisen, M.; et al. Genome-wide association study identifies two susceptibility loci for nonsyndromic cleft lip with or without cleft palate. Nat. Genet. 2010, 42, 24–26. [Google Scholar] [CrossRef]
- Beaty, T.H.; Murray, J.C.; Marazita, M.L.; Munger, R.G.; Ruczinski, I.; Hetmanski, J.B.; Liang, K.Y.; Wu, T.; Murray, T.; Fallin, M.D.; et al. A genome-wide association study of cleft lip with and without cleft palate identifies risk variants near MAFB and ABCA4. Nat. Genet. 2010, 42, 525–529. [Google Scholar] [CrossRef]
- Osoegawa, K.; Vessere, G.M.; Utami, K.H.; Mansilla, M.A.; Johnson, M.K.; Riley, B.M.; L’Heureux, J.; Pfundt, R.; Staaf, J.; Van Der Vliet, W.A.; et al. Identification of novel candidate genes associated with cleft lip and palate using array comparative genomic hybridisation. J. Med. Genet. 2008, 45, 81–86. [Google Scholar] [CrossRef]
- Nie, X.; Luukko, K.; Kettunen, P. FGF signalling in craniofacial development and developmental disorders. Oral. Dis. 2006, 12, 102–111. [Google Scholar] [CrossRef] [PubMed]
- Menezes, R.; Letra, A.; Ruff, J.; Granjeiro, J.M.; Vieira, A.R. Studies of genes in the FGF signaling pathway and oral clefts with or without dental anomalies. Am. J. Med. Genet. A 2008, 146A, 1614–1617. [Google Scholar] [CrossRef][Green Version]
- Marazita, M.L.; Lidral, A.C.; Murray, J.C.; Field, L.L.; Maher, B.S.; McHenry, T.G.; Cooper, M.E.; Govil, M.; Daack-Hirsch, S.; Riley, B.; et al. Genome scan, fine-mapping, and candidate gene analysis of non-syndromic cleft lip with or without cleft palate reveals phenotype-specific differences in linkage and association results. Human Hered. 2009, 68, 151–170. [Google Scholar] [CrossRef] [PubMed]
- Moreno, L.M.; Mansilla, M.A.; Bullard, S.A.; Cooper, M.E.; Busch, T.D.; Machida, J.; Johnson, M.K.; Brauer, D.; Krahn, K.; Daack-Hirsch, S.; et al. FOXE1 association with both isolated cleft lip with or without cleft palate, and isolated cleft palate. Human Mol. Genet. 2009, 18, 4879–4896. [Google Scholar] [CrossRef]
- van den Boogaard, M.J.; Dorland, M.; Beemer, F.A.; van Amstel, H.K. MSX1 mutation is associated with orofacial clefting and tooth agenesis in humans. Nat. Genet. 2000, 24, 342–343. [Google Scholar] [CrossRef] [PubMed]
- Jezewski, P.A.; Vieira, A.R.; Nishimura, C.; Ludwig, B.; Johnson, M.; O’Brien, S.E.; Daack-Hirsch, S.; Schultz, R.E.; Weber, A.; Nepomucena, B.; et al. Complete sequencing shows a role for MSX1 in non-syndromic cleft lip and palate. J. Med. Genet. 2003, 40, 399–407. [Google Scholar] [CrossRef]
- Suzuki, S.; Marazita, M.L.; Cooper, M.E.; Miwa, N.; Hing, A.; Jugessur, A.; Natsume, N.; Shimozato, K.; Ohbayashi, N.; Suzuki, Y.; et al. Mutations in BMP4 are associated with subepithelial, microform, and overt cleft lip. Am. J. Human Genet. 2009, 84, 406–411. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.E.; Williams, L.T. Structural and functional diversity in the FGF receptor multigene family. Adv. Cancer Res. 1993, 60, 1–41. [Google Scholar] [PubMed]
- Su, N.; Jin, M.; Chen, L. Role of FGF/FGFR signaling in skeletal development and homeostasis: Learning from mouse models. Bone Res. 2014, 2, 14003. [Google Scholar] [CrossRef] [PubMed]
- Roscioli, T.; Flanagan, S.; Kumar, P.; Masel, J.; Gattas, M.; Hyland, V.; Glass, I. Clinical findings in a patient with FGFR1 P252R mutation and comparison with the literature. Am. J. Med. Genet. 2000, 93, 22–28. [Google Scholar] [CrossRef]
- Jackson, B.C.; Carpenter, C.; Nebert, D.W.; Vasiliou, V. Update of human and mouse forkhead box (FOX) gene families. Human Genom. 2010, 4, 345. [Google Scholar] [CrossRef]
- Ali, A.H.M.; Yahya, A.Q.; Mohammed, H.L. Chromogenic in-situ hybridization technique versus immunohistochemistry in assessment of HER2/neu Status in 448 Iraqi patients with invasive breast carcinoma. Open Access Maced. J. Med. Sci. 2019, 7, 1917–1925. [Google Scholar] [CrossRef]
- Reliable and Simple Detection of Genomic Alterations Using Light Microscopy. ZytoDotR 2CTM-2-Color CISH for the Detection of Genomic Alterations. A User Manual Provided by ZytoVision GmbH-Fischkai 1, 27572 Bremerhaven- Germany. ZytoVision Molecular Diagnostics Simplified. 2019, pp. 182–183. Available online: www.Zytovision.com (accessed on 25 March 2021).
- Pilmane, M.; Rumba, I.; Sundler, F.; Luts, A. Patterns of distribution and occurrence of neuroendocrine elements in lungs of humans with chronic lung disease. Proc. Latv. Acad. Sci. 1998, 52, 144–152. [Google Scholar]
- van de Vijver, M.; Bilous, M.; Hanna, W.; Hofmann, M.; Kristel, P.; Penault-Llorca, F.; Rüschoff, J. Chromogenic in-situ hybridisation for the assessment of HER2 status in breast cancer: An international validation ring study. Breast Cancer Res. 2007, 9, R68. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, T.; Wu, T.; Hetmanski, J.B.; Ruczinski, I.; Schwender, H.; Liang, K.Y.; Murray, T.; Fallin, M.D.; Redett, R.J.; et al. The FGF and FGFR gene family and risk of cleft lip with or without cleft palate. Cleft Palate Craniofacial J. 2013, 50, 96–103. [Google Scholar] [CrossRef] [PubMed]
- Welsh, I.C.; Hagge-Greenberg, A.; O’Brien, T.P. A dosage-dependent role for Spry2 in growth and patterning during palate development. Mech. Dev. 2007, 124, 746–761. [Google Scholar] [CrossRef]
- Li, J.; Shi, S.; Srivastava, S.; Kitada, M.; Nagai, T.; Nitta, K.; Kohno, M.; Kanasaki, K.; Koya, D. FGFR1 is critical for the anti-endothelial mesenchymal transition effect of N-acetyl-seryl-aspartyl-lysyl-proline via induction of the MAP4K4 pathway. Cell Death Dis. 2017, 8, e2965. [Google Scholar] [CrossRef] [PubMed]
- Oladipupo, S.S.; Smith, C.; Santeford, A.; Park, C.; Sene, A.; Wiley, L.A.; Osei-Owusu, P.; Hsu, J.; Zapata, N.; Liu, F.; et al. Endothelial cell FGF signaling is required for injury response but not for vascular homeostasis. Proc. Natl. Acad. Sci. USA 2014, 111, 13379–13384. [Google Scholar] [CrossRef] [PubMed]
- Kanda, T.; Funato, N.; Baba, Y.; Kuroda, T. Evidence for fibroblast growth factor receptors in myofibroblasts during palatal mucoperiosteal repair. Arch. Oral. Biol. 2003, 48, 213–221. [Google Scholar] [CrossRef]
- Huang, Y.; Wang, F.; Li, H.; Xu, S.; Xu, W.; Pan, X.; Hu, Y.; Mao, L.; Qian, S.; Pan, J. Inhibition of fibroblast growth factor receptor by AZD4547 protects against inflammation in septic mice. Inflammation 2019, 42, 1957–1967. [Google Scholar] [CrossRef]
- Wang, C.; Li, Y.; Li, H.; Zhang, Y.; Ying, Z.; Wang, X.; Zhang, T.; Zhang, W.; Fan, Z.; Li, X.; et al. Disruption of FGF Signaling ameliorates inflammatory response in hepatic stellate cells. Front. Cell Dev. Biol. 2020, 8, 601. [Google Scholar] [CrossRef]
- Pilmane, M.; Sidhoma, E.; Akota, I.; Kazoka, D. Characterization of cytokines and proliferation marker Ki67 in cleft affected lip tissue. Medicina (Kaunas) 2019, 55, 518. [Google Scholar] [CrossRef]
- Soltani, A.M.; Francis, C.S.; Motamed, A.; Karatsonyi, A.L.; Hammoudeh, J.A.; Sanchez-Lara, P.A.; Reinisch, J.F.; Urata, M.M. Hypertrophic scarring in cleft lip repair: A comparison of incidence among ethnic groups. Clin. Epidemiol. 2012, 4, 187–191. [Google Scholar] [CrossRef]
- Pilmane, M.; Jain, N.; Jain, S.; Akota, I.; Kroiča, J. Quantification of cytokines in lip tissue from infants affected by congenital cleft lip and palate. Children (Basel) 2021, 8, 140. [Google Scholar] [CrossRef]
- Rice, R.; Spencer-Dene, B.; Connor, E.C.; Gritli-Linde, A.; McMahon, A.P.; Dickson, C.; Thesleff, I.; Rice, D.P. Disruption of Fgf10/Fgfr2b-coordinated epithelial-mesenchymal interactions causes cleft palate. J. Clin. Investig. 2004, 113, 1692–1700. [Google Scholar] [CrossRef] [PubMed]
- Hosokawa, R.; Deng, X.; Takamori, K.; Xu, X.; Urata, M.; Bringas, P., Jr.; Chai, Y. Epithelial-specific requirement of FGFR2 signaling during tooth and palate development. J. Exp. Zool. B Mol. Dev. Evol. 2009, 312B, 343–350. [Google Scholar] [CrossRef] [PubMed]
- Turner, N.; Grose, R. Fibroblast growth factor signalling: From development to cancer. Nat. Rev. Cancer 2010, 10, 116–129. [Google Scholar] [CrossRef]
- Ranieri, D.; Rosato, B.; Nanni, M.; Magenta, A.; Belleudi, F.; Torrisi, M.R. Expression of the FGFR2 mesenchymal splicing variant in epithelial cells drives epithelial-mesenchymal transition. Oncotarget 2016, 7, 5440–5460. [Google Scholar] [CrossRef]
- Ranieri, D.; Belleudi, F.; Magenta, A.; Torrisi, M.R. HPV16 E5 expression induces switching from FGFR2b to FGFR2c and epithelial-mesenchymal transition. Int. J. Cancer 2015, 137, 61–72. [Google Scholar] [CrossRef]
- Wahl, S.M.; Wong, H.; McCartney-Francis, N. Role of growthfactors in inflammation and repair. J. Cell Biochem. 1989, 40, 193–199. [Google Scholar] [CrossRef]
- Choi, W.; Kawanabe, H.; Sawa, Y.; Taniguchi, K.; Ishikawa, H. Effects of bFGF on suppression of collagen type I accumulation and scar tissue formation during wound healing after mucoperiosteal denudation of rat palate. Acta Odontol. Scand. 2008, 66, 31–37. [Google Scholar] [CrossRef]
- Ribatti, D.; Nico, B.; Vacca, A.; Roncali, L.; Presta, M. Endogenous and exogenous fibroblast growth factor-2 modulate wound healing in the chick embryo chorio-allantoic membrane. Angiogenesis 1999, 3, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Kawaguchi, H.; Kurokawa, T.; Hanada, K.; Hiyama, Y.; Tamura, M.; Ogata, E.; Matsumoto, T. Stimulation of fracture repair by recombinant human basic fibroblast growth factor in normal and streptozotocin-diabetic rats. Endocrinology 1994, 135, 774–781. [Google Scholar] [CrossRef]
- Koike, Y.; Yozaki, M.; Utani, A.; Murota, H. Fibroblast growth factor 2 accelerates the epithelial–mesenchymal transition in keratinocytes during wound healing process. Sci. Rep. 2020, 10, 18545. [Google Scholar] [CrossRef]
- Zittermann, S.I.; Issekutz, A.C. Basic fibroblast growth factor (bFGF, FGF-2) potentiates leukocyte recruitment to inflammation by enhancing endothelial adhesion molecule expression. Am. J. Pathol. 2006, 168, 835–846. [Google Scholar] [CrossRef] [PubMed]
- Kanazawa, S.; Tsunoda, T.; Onuma, E.; Majima, T.; Kagiyama, M.; Kikuchi, K. VEGF, basic-FGF, and TGF-beta in Crohn’s disease and ulcerative colitis: A novel mechanism of chronic intestinal inflammation. Am. J. Gastroenterol. 2001, 96, 822–828. [Google Scholar] [CrossRef]
- Shi, Y.J.; Shi, M.; Xiao, L.J.; Li, L.; Zou, L.H.; Li, C.Y.; Zhang, Q.J.; Zhou, L.F.; Ji, X.C.; Huang, H.; et al. Inhibitive Effects of FGF2/FGFR1 Pathway on Astrocyte-Mediated Inflammation in vivo and in vitro After Infrasound Exposure. Front. Neurosci. 2018, 12, 582. [Google Scholar] [CrossRef]
- Wang, C.; Ke, Y.; Liu, S.; Pan, S.; Liu, Z.; Zhang, H.; Fan, Z.; Zhou, C.; Liu, J.; Wang, F. Ectopic fibroblast growth factor receptor 1 promotes inflammation by promoting nuclear factor-kappaB signaling in prostate cancer cells. J. Biol. Chem. 2018, 293, 14839–14849. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.T.; Lee, J.G.; Na, M.; Kay, E.P. FGF-2 induced by interleukin-1 beta through the action of phosphatidylinositol 3-kinase mediates endothelial mesenchymal transformation in corneal endothelial cells. J. Biol. Chem. 2004, 279, 32325–32332. [Google Scholar] [CrossRef] [PubMed]
- Cozzolino, F.; Torcia, M.; Lucibello, M.; Morbidelli, L.; Ziche, M.; Platt, J.; Fabiani, S.; Brett, J.; Stern, D. Interferon-alpha and interleukin 2 synergistically enhance basic fibroblast growth factor synthesis and induce release, promoting endothelial cell growth. J. Clin. Investig. 1993, 91, 2504–2512. [Google Scholar] [CrossRef]
- Kuwabara, K.; Ogawa, S.; Matsumoto, M.; Koga, S.; Clauss, M.; Pinsky, D.J.; Lyn, P.; Leavy, J.; Witte, L.; Joseph-Silverstein, J. Hypoxia-mediated induction of acidic/basic fibroblast growth factor and platelet-derived growth factor in mononuclear phagocytes stimulates growth of hypoxic endothelial cells. Proc. Natl. Acad. Sci. USA 1995, 92, 4606–4610. [Google Scholar] [CrossRef]
- Presta, M.; Andrés, G.; Leali, D.; Dell’Era, P.; Ronca, R. Inflammatory cells and chemokines sustain FGF2-induced angiogenesis. Eur. Cytokine Netw. 2009, 20, 39–50. [Google Scholar] [CrossRef]
- Küchler, E.C.; Silva, L.A.D.; Nelson-Filho, P.; Sabóia, T.M.; Rentschler, A.M.; Granjeiro, J.M.; Oliveira, D.; Tannure, P.N.; Silva, R.A.D.; Antunes, L.S.; et al. Assessing the association between hypoxia during craniofacial development and oral clefts. J. Appl. Oral. Sci. 2018, 26, e20170234. [Google Scholar] [CrossRef]
- Conte, C.; Riant, E.; Toutain, C.; Pujol, F.; Arnal, J.F.; Lenfant, F.; Prats, A.C. FGF2 translationally induced by hypoxia is involved in negative and positive feedback loops with HIF-1alpha. PLoS ONE 2008, 3, e3078. [Google Scholar] [CrossRef]
- Calvani, M.; Rapisarda, A.; Uranchimeg, B.; Shoemaker, R.H.; Melillo, G. Hypoxic induction of an HIF-1alpha-dependent bFGF autocrine loop drives angiogenesis in human endothelial cells. Blood 2006, 107, 2705–2712. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Shworak, N.W.; Simons, M. Increased responsiveness of hypoxic endothelial cells to FGF2 is mediated by HIF-1 alpha-dependent regulation of enzymes involved in synthesis of heparan sulfate FGF2-binding sites. J. Cell Sci. 2002, 115, 1951. [Google Scholar] [CrossRef]
- Wang, L.; Xiong, M.; Che, D.; Shaochun, L.; Chunrong, H.; Xiaojing, Z. The effect of hypoxia on expression of basic fibroblast growth factor in pulmonary vascular pericytes. J. Tongji Med. Univ. 2000, 20, 265. [Google Scholar] [PubMed]
- Yang, J.; Zhang, D.; Yu, Y.; Zhang, R.J.; Hu, X.L.; Huang, H.F.; Lu, Y.C. Binding of FGF2 to FGFR2 in an autocrine mode in trophectoderm cells is indispensable for mouse blastocyst formation through PKC-p38 pathway. Cell Cycle 2015, 14, 3318–3330. [Google Scholar] [CrossRef]
- Breviario, F.; d’Aniello, E.M.; Golay, J.; Peri, G.; Bottazzi, B.; Bairoch, A.; Saccone, S.; Marzella, R.; Predazzi, V.; Rocchi, M. Interleukin-1-induciblegenes in endothelial cells. Cloning of a new gene related to C-reactive protein and serum amyloid P component. J. Biol. Chem. 1992, 267, 22190. [Google Scholar] [CrossRef]
- Perollet, C.; Han, Z.C.; Savona, C.; Caen, J.P.; Bikfalvi, A. Platelet factor 4 modulates fibroblast growth factor 2 (FGF-2) activity and inhibits FGF-2dimerization. Blood 1998, 91, 3289. [Google Scholar] [CrossRef]
- Dathan, N.; Parlato, R.; Rosica, A.; De Felice, M.; Di Lauro, R. Distribution of the titf2/foxe1 gene product is consistent with an important role in the development of foregut endoderm, palate, and hair. Dev. Dyn. 2002, 224, 450–456. [Google Scholar] [CrossRef]
- Venza, I.; Visalli, M.; Parrillo, L.; De Felice, M.; Teti, D.; Venza, M. MSX1 and TGF-beta3 are novel target genes functionally regulated by FOXE1. Human Mol Genet. 2011, 20, 1016–1025. [Google Scholar] [CrossRef] [PubMed]
- Adab, K.; Sayne, J.R.; Carlson, D.S.; Opperman, L.A. Tgf-beta1, Tgf-beta2, Tgf-beta3 and Msx2 expression is elevated during frontonasal suture morphogenesis and during active postnatal facial growth. Orthod. Craniofacal Res. 2002, 5, 227–237. [Google Scholar] [CrossRef]
- Fitzpatrick, D.R.; Denhez, F.; Kondaiah, P.; Akhurst, R.J. Differential expression of TGF beta isoforms in murine palatogenesis. Development 1990, 109, 585–595. [Google Scholar] [CrossRef]
- Fernández, L.P.; López-Márquez, A.; Martínez, A.M.; Gómez-López, G.; Santisteban, P. New insights into FoxE1 functions: Identification of direct FoxE1 targets in thyroid cells. PLoS ONE 2013, 8, e62849. [Google Scholar] [CrossRef] [PubMed]
- Maijenburg, M.W.; Gilissen, C.; Melief, S.M.; Kleijer, M.; Weijer, K.; Ten Brinke, A.; Roelofs, H.; Van Tiel, C.M.; Veltman, J.A.; de Vries, C.J.; et al. Nuclear receptors Nur77 and Nurr1 modulate mesenchymal stromal cell migration. Stem Cells Dev. 2012, 21, 228–238. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Ozturk, F.; Pandey, S.; Guda, C.B.; Nawshad, A. Implications of TGFβ on transcriptome and cellular biofunctions of palatal mesenchyme. Front Physiol. 2012, 3, 85. [Google Scholar] [CrossRef]
- McEvoy, A.N.; Murphy, E.A.; Ponnio, T.; Conneely, O.M.; Bresnihan, B.; FitzGerald, O.; Murphy, E.P. Activation of nuclear orphan receptor {NURR1} transcription by {NF-kappa} B and cyclic adenosine 5′-monophosphate response element-binding protein in rheumatoid arthritis synovial tissue. J. Immunol. 2002, 168, 2979–2987. [Google Scholar] [CrossRef] [PubMed]
- McCoy, J.M.; Walkenhorst, D.E.; McCauley, K.S.; Elaasar, H.; Everett, J.R.; Mix, K.S. Orphan nuclear receptor NR4A2 induces transcription of the immunomodulatory peptide hormone prolactin. J. Inflamm. 2015, 12, 13. [Google Scholar] [CrossRef]
- Ke, C.Y.; Xiao, W.L.; Chen, C.M.; Lo, L.J.; Wong, F.H. IRF6 is the mediator of TGFβ3 during regulation of the epithelial mesenchymal transition and palatal fusion. Sci. Rep. 2015, 5, 12791. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Liu, F.; Xiong, Z.; Huo, J.; Li, W.; Jiang, B.; Mao, W.; He, B.; Wang, X.; Li, G. The cleft palate candidate gene BAG6 supports FoxO1 acetylation to promote FasL-mediated apoptosis during palate fusion. Exp. Cell Res. 2020, 396, 112310. [Google Scholar] [CrossRef]
- Kwak, J.H.; Kim, S.I.; Kim, J.K.; Choi, M.E. BAT3 interacts with transforming growth factor-beta (TGF-beta) receptors and enhances TGF-beta1-induced type I collagen expression in mesangial cells. J. Biol. Chem. 2008, 283, 19816–19825. [Google Scholar] [CrossRef] [PubMed]
- Wilhelm, K.; Happel, K.; Eelen, G.; Schoors, S.; Oellerich, M.F.; Lim, R.; Zimmermann, B.; Aspalter, I.M.; Franco, C.A.; Boettger, T.; et al. FOXO1 couples metabolic activity and growth state in the vascular endothelium. Nature 2016, 529, 216–220. [Google Scholar] [CrossRef]
- Matsuda, C.; Matsui, Y.; Ohno, K.; Michi, K. Salivary gland aplasia with cleft lip and palate: A case report and review of the literature. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 1999, 87, 594–599. [Google Scholar] [CrossRef]
- Ashley, L.M.; Richardson, G.E. Multiple congenital anomalies in astillborn infant. Anat. Rec. 1943, 86, 457–471. [Google Scholar] [CrossRef]
- Salgarelli, A.C.; Capparè, P.; Bellini, P.; Collini, M. Usefulness of fine-needle aspiration in parotid diagnostics. Oral Maxillofacial Surg. 2009, 13, 185–190. [Google Scholar] [CrossRef]
- Fakhouri, W.D.; Rhea, L.; Du, T.; Sweezer, E.; Morrison, H.; Fitzpatrick, D.; Yang, B.; Dunnwald, M.; Schutte, B.C. MCS9.7 enhancer activity is highly, but not completely, associated with expression of Irf6 and p63. Dev. Dyn. 2012, 241, 340–349. [Google Scholar] [CrossRef]
- Laugel-Haushalter, V.; Langer, A.; Marrie, J.; Fraulob, V.; Schuhbaur, B.; Koch-Phillips, M.; Dollé, P.; Bloch-Zupan, A. From the transcription of genes involved in ectodermal dysplasias to the understanding of associated dental anomalies. Mol. Syndromol. 2012, 3, 158–168. [Google Scholar] [CrossRef] [PubMed]
- Tamasas, B.; Cox, T.C. Massively increased caries susceptibility in an Irf6 cleft lip/palate model. J. Dent. Res. 2017, 96, 315–322. [Google Scholar] [CrossRef] [PubMed]
- De Moerlooze, L.; Spencer-Dene, B.; Revest, J.; Hajihosseini, M.; Rosewell, I.; Dickson, C. An important role for the IIIb isoform of fibroblast growth factor receptor 2 (FGFR2) in mesenchymal-epithelial signalling during mouse organogenesis. Development 2000, 127, 483–492. [Google Scholar] [CrossRef] [PubMed]
- Sberna, M.T.; Rizzo, G.; Zacchi, E.; Capparè, P.; Rubinacci, A. A preliminary study of the use of peripheral quantitative computed tomography for investigating root canal anatomy. Int. Endod. J. 2009, 42, 66–75. [Google Scholar] [CrossRef]
- Ferrini, F.; Sannino, G.; Chiola, C.; Capparé, P.; Gastaldi, G.; Gherlone, E.F. Influence of Intra-Oral Scanner (I.O.S.) on The Marginal Accuracy of CAD/CAM Single Crowns. Int. J. Environ. Res. Public Health 2019, 16, 544. [Google Scholar] [CrossRef]
- Cattoni, F.; Teté, G.; Calloni, A.M.; Manazza, F.; Gastaldi, G.; Capparè, P. Milled versus moulded mock-ups based on the superimposition of 3D meshes from digital oral impressions: A comparative in vitro study in the aesthetic area. BMC Oral Health 2019, 19, 230. [Google Scholar] [CrossRef]
- Joda, T.; Zarone, F.; Ferrari, M. The complete digital workflow in fixed prosthodontics: A systematic review. BMC Oral Health 2017, 17, 124. [Google Scholar] [CrossRef]
- Patzelt, S.B.; Emmanouilidi, A.; Stampf, S.; Strub, J.R.; Att, W. Accuracy of full-arch scans using intraoral scanners. Clin. Oral Investig. 2014, 18, 1687–1694. [Google Scholar] [CrossRef]
- Wermker, K.; Jung, S.; Joos, U.; Kleinheinz, J. Dental implants in cleft lip, alveolus, and palate patients: A systematic review. Int. J. Oral Maxillofac. Implant. 2014, 29, 384–390. [Google Scholar] [CrossRef]
- Anastassov, G.E.; Joos, U. Comprehensive management of cleft lip and palate deformities. J. Oral Maxillofacial Surg. 2001, 59, 1062–1075. [Google Scholar] [CrossRef] [PubMed]
- Pang, J.; Broyles, J.; Redett, R. Cleft lip and palate. Eplasty 2013, 13, ic25. [Google Scholar] [PubMed]
- Harris, P.A.; Oliver, N.K.; Slater, P.; Murdoch, L.; Moss, A.L. Safety of neonatal cleft lip repair. J. Plast. Surg. Hand Surg. 2010, 44, 231–236. [Google Scholar] [CrossRef] [PubMed]
- Goodacre, T.E.; Hentges, F.; Moss, T.L.; Short, V.; Murray, L. Does repairing a cleft lip neonatally have any effect on the longer-term attractiveness of the repair? Cleft Palate Craniofacial J. 2004, 41, 603–608. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.F.; Cole, T.J.; Mars, M. Hard palate repair timing and facial growth in unilateral cleft lip and palate: A longitudinal study. Cleft Palate Craniofacial J. 2006, 43, 547–556. [Google Scholar] [CrossRef]
- Crespi, R.; Capparè, P.; Gherlone, E. Sinus floor elevation by osteotome: Hand mallet versus electric mallet. A prospective clinical study. Int. J. Oral Maxillofac. Implant. 2012, 27, 1144–1150. [Google Scholar]
- Vinci, R.; Teté, G.; Lucchetti, F.R.; Capparé, P.; Gherlone, E.F. Implant survival rate in calvarial bone grafts: A retrospective clinical study with 10 year follow-up. Clin. Implant. Dent. Relat. Res. 2019, 21, 662–668. [Google Scholar] [CrossRef]
- Bruschi, G.B.; Crespi, R.; Capparè, P.; Bravi, F.; Bruschi, E.; Gherlone, E. Localized management of sinus floor technique for implant placement in fresh molar sockets. Clin. Implant. Dent. Relat. Res. 2013, 15, 243–250. [Google Scholar] [CrossRef]
- Kim, S.W.; Lee, I.K.; Yun, K.I.; Kim, C.H.; Park, J.U. Adult stem cells derived from human maxillary sinus membrane and their osteogenic differentiation. Int. J. Oral Maxillofac. Implant. 2009, 24, 991–998. [Google Scholar]
- Capparè, P.; Tetè, G.; Sberna, M.T.; Panina-Bordignon, P. The emerging role of stem cells in regenerative dentistry. Curr. Gene Ther. 2020, 20, 259–268. [Google Scholar] [CrossRef]
- Capparé, P.; Teté, G.; Romanos, G.E.; Nagni, M.; Sannino, G.; Gherlone, E.F. The ‘All-on-four’ protocol in HIV-positive patients: A prospective, longitudinal 7-year clinical study. Int. J. Oral Implantol. (Berl.) 2019, 12, 501–510. [Google Scholar]
- Gherlone, E.F.; Capparé, P.; Tecco, S.; Polizzi, E.; Pantaleo, G.; Gastaldi, G.; Grusovin, M.G. A prospective longitudinal study on implant prosthetic rehabilitation in controlled HIV-positive patients with 1-year follow-up: The role of CD4+ level, smoking habits, and oral hygiene. Clin. Implant. Dent. Relat. Res. 2016, 18, 955–964. [Google Scholar] [CrossRef]
- Gherlone, E.F.; Capparé, P.; Tecco, S.; Polizzi, E.; Pantaleo, G.; Gastaldi, G.; Grusovin, M.G. Implant prosthetic rehabilitation in controlled HIV-positive patients: A prospective longitudinal study with 1-year follow-up. Clin. Implant. Dent. Relat. Res. 2016, 18, 725–734. [Google Scholar] [CrossRef] [PubMed]
- Tecco, S.; Grusovin, M.G.; Sciara, S.; Bova, F.; Pantaleo, G.; Capparé, P. The association between three attitude-related indexes of oral hygiene and secondary implant failures: A retrospective longitudinal study. Int. J. Dent. Hyg. 2018, 16, 372–379. [Google Scholar] [CrossRef]
- Gherlone, E.F.; Capparé, P.; Pasciuta, R.; Grusovin, M.G.; Mancini, N.; Burioni, R. Evaluation of resistance against bacterial microleakage of a new conical implant-abutment connection versus conventional connections: An in vitro study. New Microbiol. 2016, 39, 49–56. [Google Scholar]
- Bruschi, G.B.; Crespi, R.; Capparè, P.; Grande, N.; Bruschi, E.; Gherlone, E. Radiographic evaluation of crestal bone levels of delayed implants at medium-term follow-up. Int. J. Oral Maxillofac. Implant. 2014, 29, 441–447. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fujiwara, K.; Yamada, T.; Mishima, K.; Imura, H.; Sugahara, T. Morphological and immunohistochemical studies on cleft palates induced by 2,3,7,8-tetrachlorodibenzo-p-dioxin in mice. Congenit Anom. (Kyoto) 2008, 48, 68–73. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).