Amyloids: The History of Toxicity and Functionality
Abstract
Simple Summary
Abstract
1. Introduction
2. Toxic Properties of Amyloids (the Beginning of Their History)
3. Anti-Amyloid Therapy and Its Problems
- (1)
- Block β-sheet formation
- (2)
- Prevent fibrillogenesis
- (3)
- Dissolve Aβ aggregates into non-toxic species
- (4)
- Destabilize Aβ oligomers
- (5)
- Accelerate the conversion of Aβ oligomers to Aβ aggregates (modulators of Aβ aggregation)
4. Useful Properties of Amyloids. Functional Amyloids
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| Aβ | β-amyloid protein |
| APP | amyloid precursor protein |
| AD | Alzheimer’s disease |
| CD | circular dichroism |
| FTIR | Fourier-transform infrared spectroscopy |
| NO | nitric oxide |
| PrP | prion protein |
| RC | random coil |
| RT | room temperature |
References
- Nizhnikov, A.A.; Antonets, K.S.; Inge-Vechtomov, S.G. Amyloids: From Pathogenesis to Function. Biochemistry 2015, 80, 1127–1144. [Google Scholar] [CrossRef] [PubMed]
- Kyle, R. Amyloidosis: A Convoluted Story. Br. J. Haematol. 2001, 114, 529–538. [Google Scholar] [CrossRef]
- Sipe, J.D.; Cohen, A.S. Review: History of Amyloid Fibril. J. Struct. Biol. 2000, 130, 88–89. [Google Scholar] [CrossRef]
- Westermark, P.; Benson, M.D.; Buxbaum, J.N.; Cohen, A.S.; Frangione, B.; Ikeda, S.I.; Masters, C.L.; Merlini, G.; Sipe, J.D.A.; Saraiva, M.J. Primer of Amyloid Nomenclature. Amyloid 2007, 14, 179–183. [Google Scholar] [CrossRef] [PubMed]
- Horwich, A.L.; Weissman, J.S. Deadly Conformations–Protein Misfolding in Prion Disease. Cell 1997, 89, 499–510. [Google Scholar] [CrossRef]
- Prusiner, S.B.; Scott, M.R.; DeArmond, S.J.; Cohen, F.E. Prion Protein Biology. Cell 1998, 93, 337–348. [Google Scholar] [CrossRef]
- Kushnirov, V.V.; Vishnevskaya, A.B.; Alexandrov, I.M.; Michael, D. Ter-Avanesyan Prion and Nonprion. Amyloids 2007, 1, 179–184. [Google Scholar]
- Antonets, K.S.; Belousov, M.V.; Sulatskaya, A.I.; Belousova, M.E.; Kosolapova, A.O.; Sulatsky, M.I.; Andreeva, E.A.; Zykin, P.A.; Malovichko, Y.V.; Shtark, O.Y.; et al. Accumulation of Storage Proteins in Plant Seeds is Mediated by Amyloid Formation. PLoS Biol. 2020, 18, e3000564. [Google Scholar] [CrossRef]
- Surguchov, A.; Emamzadeh, F.N.; Surguchev, A.A. Amyloidosis and Longevity: A Lesson from Plants. Biology 2019, 8, 43. [Google Scholar] [CrossRef]
- Rçmling, U.; Bian, Z.; Hammar, M.; Sierralta, W.D.; Normark, S. Curli Fibers are Highly Conserved between Salmonella typhimurium and Escherichia coli with Respect to Open Structure and Regulation. J. Bacteriol. 1998, 180, 722–731. [Google Scholar] [CrossRef]
- Claessen, D.; Rink, R.; de Jong, W.; Siebring, J.; de Vreughd, P.; Boersma, F.G.H.; Dijkhuizen, L.; Wçsten, H.A.B. A Novel Class of Secreted Hydrophobic Proteins is Involved in Aerial Hyphae Formation in Streptomyces coelicolor by Forming Amyloid-like Fibrils. Genes Dev. 2003, 17, 1714–1726. [Google Scholar] [CrossRef]
- Otzen, D.; Nielsen, P.H. We Find Them Here, We Find Them There: Functional Bacterial Amyloid. Cell Mol Life Sci. 2008, 65, 910–927. [Google Scholar] [CrossRef]
- Wöesten, H.A.B.; de Vocht, M.L. Hydrophobins, the Fungal Coat Unraveled. Biochim. Biophys. Acta 2000, 1469, 79–86. [Google Scholar] [CrossRef]
- Iconomidou, V.A.; Chryssikos, G.D.; Gionis, V.; Galanis, A.S.; Cordopatis, P.; Hoenger, A.; Hamodrakas, S.J. Amyloid Fibril Formation Propensity is Inherent into the Hexapeptidetandemly Repeating Sequence of the Central Domain of Silk Moth Chorion Proteins of the A-family. J. Struct. Biol. 2006, 156, 480–488. [Google Scholar] [CrossRef]
- Slotta, U.; Hess, S.; Spiess, K.; Stromer, T.; Serpell, L.; Scheibel, T. Spider Silk and Amyloid Fibrils: A Structural Comparison. Macromol. Biosci. 2007, 7, 183–188. [Google Scholar] [CrossRef]
- Si, K.; Lindquist, S.; Kandel, E.R. A Neuronal Isoform of the Aplysia CPEB has Prion-like Properties. Cell 2003, 115, 879–891. [Google Scholar] [CrossRef]
- Guyonnet, B.; Egge, N.; Cornwall, G.A. Functional Amyloids in the Mouse Sperm Acrosome. Mol. Cell Biol. 2014, 34, 2624–2634. [Google Scholar] [CrossRef] [PubMed]
- Fowler, D.M.; Koulov, A.V.; Alory-Jost, C.; Marks, M.S.; Balch, W.E.; Kelly, J.W. Functional Amyloid Formation within Mammalian Tissue. PLoS Biol. 2006, 4, e6. [Google Scholar] [CrossRef]
- Maji, S.K.; Perrin, M.H.; Sawaya, M.R.; Jessberger, S.; Vadodaria, K.; Rissman, R.A.; Singru, P.S.; Nilsson, K.P.; Simon, R.; Schubert, D.; et al. Functional Amyloids as Natural Storage of Peptide Hormones in Pituitary Secretory Granules. Science 2009, 325, 328–332. [Google Scholar] [CrossRef] [PubMed]
- Hardy, J.A.; Higgins, G.A. Alzheimer’s Disease: The Amyloid Cascade Hypothesis. Science 1992, 256, 184–185. [Google Scholar] [CrossRef] [PubMed]
- Mann, D.M.A. Alzheimer’s Disease and Down’s Syndrome. Histopathology 1988, 13, 125–137. [Google Scholar] [CrossRef] [PubMed]
- Shamas-Ud-Din, S.; Holmes, C. Genetics of Down’s Syndrome and Alzheimer’s Disease. Br. J. Psychiatry 2002, 181, 167–168. [Google Scholar] [CrossRef] [PubMed]
- Chartier-Harlin, M.C.; Crawford, F.; Houlden, H.; Warren, A.; Hughes, D.; Fidani, L.; Goate, A.; Rossor, M.; Roques, P.; Hardy, J.; et al. Early-onset Alzheimer’s Disease Caused by Mutations at Codon 717 of the f3-amyloid Precursor Protein Gene. Nature 1991, 353, 844–846. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.-D.; Golde, T.; Younkin, S. Release of Excess Amyloid Beta-protein from a Mutant Amyloid Beta-protein Precursor. Science 1993, 259, 514–516. [Google Scholar] [CrossRef] [PubMed]
- Citron, M.; Oltersorf, T.; Haass, C.; McConlogue, L.; Hung, A.Y.; Seubert, P.; Vigo-Pelfrey, C.; Lieberburg, I.; Selkoe, D.J. Mutation of the Beta-amyloid Precursor Protein in Familial Alzheimer’s Disease Increases Beta-protein Production. Nature 1992, 360, 672–674. [Google Scholar] [CrossRef]
- Murrell, J.; Farlow, M.; Ghetti, B.; Benson, M.D. Amutation in the Amyloid Precursor Protein Associated with Hereditary Alzheimer’s Disease. Science 1991, 254, 97–99. [Google Scholar] [CrossRef]
- Goate, A.; Chartier-Harlin, M.-C.; Mullan, M.; Brown, J.; Crawford, F.; Fidani, L.; Giuffra, L.; Haynes, A.; Irving, N.; James, L.; et al. Segregation of a Missense Mutation in the Amyloid Precursor Protein Gene with Familial Alzheimer’s Disease. Nature 1991, 349, 704–706. [Google Scholar] [CrossRef]
- Vassar, R. BACE1: The Beta-secretase Enzyme in Alzheimer’s Disease. J. Mol. Neurosci. 2004, 23, 105–114. [Google Scholar] [CrossRef]
- Benilova, I.; Karran, E.; De Strooper, B. The Toxic Aβ Oligomer and Alzheimer’s Disease: An Emperor in Need of Clothes. Nat. Neurosci. 2012, 15, 349–357. [Google Scholar] [CrossRef]
- Yankner, B.A.; Dawes, L.R.; Fisher, S.; Villa-Komaroff, L.; Oster-Granite, M.L.; Neve, R.L. Neurotoxicity of a Fragment of the Amyloid Precursor Associated with Alzheimer’s Disease. Science 1989, 245, 417–420. [Google Scholar] [CrossRef]
- Yankner, J.A.; Duffy, L.K.; Kirschner, D.A. Neurotrophic and Neurotoxic Effects of Amyloid β Protein Reversal by Tachykinin Neuropeptides. Science 1990, 250, 279–282. [Google Scholar] [CrossRef]
- Mattson, M.P.; Tomaselli, K.J.; Rydel, R.E. Calcium De-stabilizing and Neurodegenerative Effects of Aggregated β-amyloid are Attenuated by Basic FGF. Brain Res. 1993, 621, 35–49. [Google Scholar] [CrossRef]
- Barrow, C.J.; Zagorski, M.G. Solution Structures of β Peptide and Its Constituent Fragments: Relation to Amyloid Deposition. Science 1991, 253, 179–182. [Google Scholar] [CrossRef]
- Whitson, J.S.; Glabe, C.G.; Shitani, E.; Abcar, A.; Cotman, C.W. β-Amyloid Protein Promotes Neuritic Branching in Hippocampal Cultures. Neurosci. Lett. 1990, 110, 319–324. [Google Scholar] [CrossRef]
- Burdick, D.; Soreghan, B.; Kosmoski, J.; Knauer, M.; Henschen, A.; Yates, J.; Cotman, C.; Glabe, C. Assembly and Aggregation Properties of Synthetic Alzheimer’s A4/β Amyloid Peptide Analogs. J. Biol. Chem. 1992, 267, 546–554. [Google Scholar] [CrossRef]
- Hilbich, C.; Kisters-Woike, B.; Reed, J.; Masters, C.L.; Beyreuther, K. Aggregation and Secondary Structure of Synthetic Amyloid flA4 Peptides of Alzheimer’s Disease. J. Mol. Biol. 1991, 218, 149–163. [Google Scholar] [CrossRef]
- Kaltschmidt, B.; Uherek, M.; Wellman, H.; Volk, B.; Kaltschmidt, C. Inhibition of NF-KB Potentates Amyloid β-mediatated Neuronal Apoptosis. Proc. Natl. Acad. Sci. USA 1999, 96, 9409–9414. [Google Scholar] [CrossRef]
- Kuperstein, I.; Broersen, K.; Benilova, I.; Rozenski, J.; Jonckheere, W.; Debulpaep, M.; Vandersteen, A.; Segers-Nolten, I.; Van Der Werf, K.; Subramaniam, V.; et al. Neurotoxicity of Alzheimer’s Disease Aβ Peptides is Induced by Small Changes in the Aβ42 to Aβ40 Ratio. EMBO J. 2010, 29, 3408–3420. [Google Scholar] [CrossRef]
- Gibson, G.E.; Peterson, C. Calcium and the Aging Nervous System. Neurobiol. Aging 1987, 8, 329–343. [Google Scholar] [CrossRef]
- Mattson, M.P. Calcium as Sculptor and Destroyer of Neural Circuitry. Exp. Gerontol. 1992, 27, 29–49. [Google Scholar] [CrossRef]
- Mattson, M.P. Antigenic Changes Similiar to Those Seen in Neurofibrillary Tangles are Elicited by Glutamate and Calcium Influx in Cultured Hippocampal Neurons. Neuron 1990, 4, 105–117. [Google Scholar] [CrossRef]
- Mattson, M.P. Calcium and Neuronal Injury in Alzheimer’s Disease. Contributions of Beta-amyloid Precursor Protein Mismetabolism, Free Radicals, and Metabolic Compromise. Ann. N. Y. Acad. Sci. 1994, 15, 50–76. [Google Scholar]
- Weiss, J.H.; Pike, C.J.; Cotman, C.W. Ca2+ Channel Blockers Attenuate Beta-amyloid Peptide Toxicity to Cortical Neurons in Culture. J. Neurochem. 1994, 62, 372–375. [Google Scholar] [CrossRef]
- Avdulov, N.A.; Chochina, S.V.; Igbavboa, U.; Warden, C.S.; Vassiliev, A.V.; Wood, W.G. Lipid Binding to Amyloid Beta-peptide Aggregates: Preferential Binding of Cholesterol as Compared with Phosphatidylcholine and Fatty Acids. J. Neurochem. 1997, 69, 1746–1752. [Google Scholar] [CrossRef]
- Demuro, A.; Parker, I.; Stutzmann, G.E. Calcium Signaling and Amyloid Toxicity in Alzheimer Disease. J. Biol. Chem. 2010, 285, 12463–12468. [Google Scholar] [CrossRef]
- Müller, W.E.; Koch, S.; Eckert, A.; Hartmann, H.; Scheuer, K. Beta-amyloid Peptide Decreases Membrane Fluidity. Brain Res. 1995, 674, 133–136. [Google Scholar] [CrossRef]
- Mason, R.P.; Trumbore, M.W.; Pettegrew, J.W. Molecular Membrane Interactions of a Phospholipid Metabolite. Implications for Alzheimer’s Disease Pathophysiology. Ann. N. Y. Acad. Sci. 1996, 777, 368–373. [Google Scholar] [CrossRef] [PubMed]
- Green, J.D.; Kreplak, L.; Goldsbury, C.; Li Blatter, X.; Stolz, M.; Cooper, G.S.; Seelig, A.; Kistler, J.; Aebi, U. Atomic Force Microscopy Reveals Defects within Mica Supported Lipid Bilayers Induced by the Amyloidogenic Human Amylin Peptide. J. Mol. Biol. 2004, 342, 877–887. [Google Scholar] [CrossRef]
- Kawahara, M.; Kuroda, Y.; Arispe, N.; Rojas, E. Alzheimer’s Beta-amyloid, Human Islet Amylin, and Prion Protein Fragment Evoke Intracellular Free Calcium Elevations by a Common Mechanism in a Hypothalamic GnRH Neuronal Cell Line. J. Biol. Chem. 2000, 275, 14077–14083. [Google Scholar] [CrossRef]
- Kayed, R.; Sokolov, Y.; Edmonds, B.; McIntire, T.M.; Milton, S.C.; Hall, J.E.; Glabe, C.G. Permeabilization of Lipid Bilayers is a Common Conformation-dependent Activity of Soluble Amyloid Oligomers in Protein Misfolding Diseases. J. Biol. Chem. 2004, 279, 46363–46366. [Google Scholar] [CrossRef]
- Arispe, N.; Rojas, E.; Pollard, H.B. Alzheimer Disease Amyloid Beta Protein Forms Calcium Channels in Bilayer Membranes: Blockade by Tromethamine and Aluminum. Proc. Natl. Acad. Sci. USA 1993, 90, 567–571. [Google Scholar] [CrossRef]
- Arispe, N. Architecture of the Alzheimer’s A Beta P Ion Channel Pore. J. Membr. Biol. 2004, 197, 33–48. [Google Scholar] [CrossRef]
- Lin, M.C.; Kagan, B.L. Electrophysiologic Properties of Channels Induced by Abeta25-35 in Planar Lipid Bilayers. Peptides 2002, 23, 1215–1228. [Google Scholar] [CrossRef]
- Kawahara, M.; Arispe, N.; Kuroda, Y.; Rojas, E. Alzheimer’s Disease Amyloid Beta-protein Forms Zn(2+)-sensitive, Cation-selective Channels Across Excised Membrane Patches from Hypothalamic Neurons. Biophys. J. 1997, 73, 67–75. [Google Scholar] [CrossRef]
- Hirakura, Y.; Lin, M.C.; Kagan, B.L. Alzheimer Amyloid Abeta1-42 Channels: Effects of Solvent, pH, and Congo Red. J. Neurosci. Res. 1999, 57, 458–466. [Google Scholar] [CrossRef]
- Blanchard, B.J.; Chen, A.; Rozeboom, L.M.; Stafford, K.A.; Weigele, P.; Ingram, V.M. Efficient Reversal of Alzheimer’s Disease Fibril Formation and Elimination of Neurotoxicity by a Small Molecule. Proc. Natl. Acad. Sci. USA 2004, 101, 14326–14332. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Furukawa, K.; Sopher, B.L.; Pham, D.G.; Xie, J.; Robinson, N.; Martin, G.M.; Mattson, M.P. Alzheimer’s PS-1 Mutation Perturbs Calcium Homeostasis and Sensitizes PC12 Cells to Death Induced by Amyloid Beta-peptide. Neuroreport 1996, 8, 379–383. [Google Scholar] [CrossRef] [PubMed]
- Mattson, M.P.; Chan, S.L. Calcium Orchestrates Apoptosis. Nat. Cell. Biol. 2003, 5, 1041–1043. [Google Scholar] [CrossRef]
- Ferrarelli, L.K. New Connections: Amyloid-β, Calcium, and the Synapse. Sci. Signal. 2017, 10, eaao3024. [Google Scholar] [PubMed]
- Tong, B.C.; Wu, A.J.; Li, M.; Cheung, K.H. Calcium Signaling in Alzheimer’s Disease & Therapies. Biochim. Biophys. Acta Mol. Cell Res. 2018, 1865, 1745–1760. [Google Scholar]
- Angelova, P.R.; Abramov, A.Y. Alpha-synuclein and Beta-amyloid–Different Targets, Same Players: Calcium, Free Radicals and Mitochondria in the Mechanism of Neurodegeneration. Biochem. Biophys. Res. Commun. 2017, 483, 1110–1115. [Google Scholar] [CrossRef] [PubMed]
- Cristóvão, J.S.; Morris, V.K.; Cardoso, I.; Leal, S.S.; Martínez, J.; Botelho, H.M.; Göbl, C.; David, R.; Kierdorf, K.; Alemi, M.; et al. The Neuronal S100B Protein is a Calcium-tuned Suppressor of Amyloid-β Aggregation. Sci. Adv. 2018, 4, eaaq1702. [Google Scholar] [CrossRef]
- Kawahara, M.; Negishi-Kato, M.; Sadakane, Y. Calcium Dyshomeostasis and Neurotoxicity of Alzheimer’s Beta-amyloid Protein. Expert Rev. Neurother. 2009, 9, 681–693. [Google Scholar] [CrossRef]
- Sushma; Mondal, A.C. Role of GPCR Signaling and Calcium Dysregulation in Alzheimer’s Disease. Mol. Cell. Neurosci. 2019, 101, 103414. [Google Scholar] [CrossRef]
- Esteras, N.; Abramov, A.Y. Mitochondrial Calcium Deregulation in the Mechanism of Beta-Amyloid and Tau Pathology. Cells 2020, 9, 2135. [Google Scholar] [CrossRef]
- Behl, C.; Davis, J.; Cole, G.M.; Schubert, D. Vitamin E Protects Nerve Cells from Amyloid Beta Protein Toxicity. Biochem. Biophys. Res. Commun. 1992, 186, 944–950. [Google Scholar] [CrossRef]
- Hensley, K.; Carney, J.M.; Mattson, M.P.; Aksenova, M.; Harris, M.; Wu, J.F.; Floyd, R.A.; Butterfield, D.A. A Model for Beta-amyloid Aggregation and Neurotoxicity Based on Free Radical Generation by the Peptide: Relevance to Alzheimer Disease. Proc. Natl. Acad. Sci. USA 1994, 91, 3270–3274. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.A.; Richey Harris, P.L.; Sayre, L.M.; Beckman, J.S.; Perry, G. Widespread Peroxynitrite-mediated Damage in Alzheimer’s Disease. J. Neurosci. 1997, 17, 2653–2657. [Google Scholar] [CrossRef]
- Smith, M.A.; Perry, G.; Richey, P.L.; Sayre, L.M.; Anderson, V.E.; Beal, M.F.; Kowall, N. Oxidative Damage in Alzheimer’s. Nature 1996, 382, 120–121. [Google Scholar] [CrossRef]
- Goodwin, J.L.; Uemura, E.; Cunnick, J.E. Microglial Release of Nitric Oxide by the Synergistic Action of Beta-amyloid and IFN-gamma. Brain Res. 1995, 692, 207–214. [Google Scholar] [CrossRef]
- Geng, X.; Yang, B.; Li, R.; Teng, T.; Ladu, M.J.; Sun, G.Y.; Greenlief, C.M.; Lee, J.C. Effects of Docosahexaenoic Acid and Its Peroxidation Product on Amyloid-β Peptide-Stimulated Microglia. Mol Neurobiol. 2020, 57, 1085–1098. [Google Scholar] [CrossRef] [PubMed]
- Pappolla, M.A.; Omar, R.A.; Kim, K.S.; Robakis, N.K. Immunohistochemical Evidence of Oxidative (Corrected) Stress in Alzheimer’s Disease. Am. J. Pathol. 1992, 140, 621–628. [Google Scholar]
- Pereira, C.F.; Santos, A.E.; Moreira, P.I.; Pereira, A.C.; Sousa, F.J.; Cardoso, S.M.; Cruz, M.T. Is Alzheimer’s Disease an Inflammasomopathy? Ageing Res. Rev. 2019, 56, 100966. [Google Scholar] [CrossRef]
- Cheignon, C.; Tomas, M.; Bonnefont-Rousselot, D.; Faller, P.; Hureau, C.; Collin, F. Oxidative Stress and the Amyloid Beta Peptide in Alzheimer’s Disease. Redox. Biol. 2018, 14, 450–464. [Google Scholar] [CrossRef] [PubMed]
- Behl, C.; Davis, J.B.; Lesley, R.; Schubert, D. Hydrogen Peroxide Mediates Amyloid Beta Protein Toxicity. Cell 1994, 77, 817–827. [Google Scholar] [CrossRef]
- Kaltschmidt, B.; Uherek, M.; Volk, B.; Baeuerle, P.A.; Kaltschmidt, C. Transcription Factor NF-kappaB is Activated in Primary Neurons by Amyloid Beta Peptides and in Neurons Surrounding Early Plaques from Patients with Alzheimer Disease. Proc. Natl. Acad. Sci. USA 1997, 94, 2642–2647. [Google Scholar] [CrossRef]
- Pentreath, V.W. Responses of Cultured Astrocytes, C6 Glioma and 1321NI Astrocytoma Cells to Amyloid β-Peptide Fragments. Nonlinearity Biol. Toxicol. Med. 2004, 2, 45–63. [Google Scholar] [CrossRef]
- Cotman, C.W.; Anderson, A.J. A Potential Role for Apoptosis in Neurodegeneration and Alzheimer’s Disease. Mol. Neurobiol. 1995, 10, 19–45. [Google Scholar] [CrossRef] [PubMed]
- Reiss, A.B.; Arain, H.A.; Stecker, M.M.; Siegart, N.M.; Kasselman, L.J. Amyloid Toxicity in Alzheimer’s Disease. Rev. Neurosci. 2018, 29, 613–627. [Google Scholar] [CrossRef] [PubMed]
- Leong, Y.Q.; Ng, K.Y.; Chye, S.M.; Ling, A.P.K.; Koh, R.Y. Mechanisms of Action of Amyloid-beta and Its Precursor Protein in Neuronal Cell Death. Metab. Brain Dis. 2020, 35, 11–30. [Google Scholar] [CrossRef]
- Dickson, D.W. Apoptotic Mechanisms in Alzheimer Neurofibrillary Degeneration: Cause or Effect? J. Clin. Investig. 2004, 114, 23–27. [Google Scholar] [CrossRef] [PubMed]
- Stadelmann, C.; Bruck, W.; Bancher, C.; Jellinger, K.; Lassmann, H. Alzheimer Disease: DNA Fragmentation Indicates Increased Neuronal Vulnerability, but not Apoptosis. J. Neuropathol. Exp. Neurol. 1998, 57, 456–464. [Google Scholar] [CrossRef]
- Gong, Y.; Chang, L.; Viola, K.L.; Lacor, P.N.; Lambert, M.P.; Finch, C.E.; Krafft, G.A.; Klein, W.L. Alzheimer’s Disease-affected Brain: Presence of Oligomeric A Beta Ligands (ADDLs) Suggests a Molecular Basis for Reversible Memory Loss. Proc. Natl. Acad. Sci. USA 2003, 100, 10417–10422. [Google Scholar] [CrossRef]
- Su, J.H.; Anderson, A.J.; Cummings, B.J.; Cotman, C.W. Immunohistochemical Evidence for Apoptosis in Alzheimer’s Disease. Neuroreport. 1994, 5, 2529–2533. [Google Scholar] [CrossRef] [PubMed]
- Smale, G.; Nichols, N.R.; Brady, D.R.; Finch, C.E.; Horton, W.E., Jr. Evidence for Apoptotic Cell Death in Alzheimer’s Disease. Exp. Neurol. 1995, 133, 225–230. [Google Scholar] [CrossRef]
- Anderson, A.J.; Su, J.H.; Cotman, C.W. DNA Damage and Apoptosis in Alzheimer’s Disease: Colocalization with c-Jun Immunoreactivity, Relationship to Brain Area, and Effect of Postmortem Delay. J. Neurosci. 1996, 16, 1710–1719. [Google Scholar] [CrossRef]
- Obulesu, M.; Lakshmi, M.J. Apoptosis in Alzheimer’s Disease: An Understanding of the Physiology, Pathology and Therapeutic Avenues. Neurochem. Res. 2014, 39, 2301–2312. [Google Scholar] [CrossRef]
- Park, G.; Nhan, H.S.; Tyan, S.; Kawakatsu, Y.; Zhang, C.; Navarro, M.; Koo, E.H. Caspase Activation and Caspase-Mediated Cleavage of APP Is Associated with Amyloid β-Protein-Induced Synapse Loss in Alzheimer’s Disease. Cell Reports 2020, 31, 107839. [Google Scholar] [CrossRef] [PubMed]
- Bredesen, D.E.; John, V.; Galvan, V. Importance of the Caspase Cleavage Site in Amyloid-β Protein Precursor. J. Alzheimers Dis. 2010, 22, 57–63. [Google Scholar] [CrossRef]
- Rissman, R.A.; Poon, W.W.; Blurton-Jones, M.; Oddo, S.; Torp, R.; Vitek, M.P.; LaFerla, F.M.; Rohn, T.T.; Cotman, C.W. Caspase-cleavage of Tau is an Early Event in Alzheimer Disease Tangle Pathology. J. Clin. Investig. 2004, 114, 121–130. [Google Scholar] [CrossRef]
- Glabe, C. Intracellular Mechanisms of Amyloid Accumulation and Pathogenesis in Alzheimer’s Disease. J. Mol. Neurosci. 2001, 17, 137–145. [Google Scholar] [CrossRef]
- Oddo, S.; Caccamo, A.; Shepherd, J.D.; Murphy, M.P.; Golde, T.E.; Kayed, R.; Metherate, R.; Mattson, M.P.; Akbari, Y.; LaFerla, F.M. Triple-transgenic Model of Alzheimer’s Disease with Plaques and Tangles: Intracellular Abeta and Synaptic Dysfunction. Neuron 2003, 39, 409–421. [Google Scholar] [CrossRef]
- Lustbader, J.W.; Cirilli, M.; Lin, C.; Xu, H.W.; Takuma, K.; Wang, N.; Caspersen, C.; Chen, X.; Pollak, S.; Chaney, M.; et al. ABAD Directly Links Abeta to Mitochondrial Toxicity in Alzheimer’s Disease. Science 2004, 304, 448–452. [Google Scholar]
- Oda, T.; Wals, P.; Osterburg, H.H.; Johnson, S.A.; Pasinetti, G.M.; Morgan, T.E.; Rozovsky, I.; Stine, W.B.; Snyder, S.W.; Holzman, T.F.; et al. Clusterin (apoJ) Alters the Aggregation of Amyloid Beta-peptide (A beta 1-42) and Forms Slowly Sedimenting A Beta Complexes That Cause Oxidative Stress. Exp. Neurol. 1995, 136, 22–31. [Google Scholar] [CrossRef] [PubMed]
- Lambert, M.P.; Barlow, A.K.; Chromy, B.A.; Edwards, C.; Freed, R.; Liosatos, M.; Morgan, T.E.; Rozovsky, I.; Trommer, B.; Viola, K.L.; et al. Diffusible, Nonfibrillar Ligands Derived from Abeta1-42 are Potent Central Nervous System Neurotoxins. Proc. Natl. Acad. Sci. USA 1998, 95, 6448–6453. [Google Scholar] [CrossRef]
- Walsh, D.M.; Klyubin, I.; Fadeeva, J.V.; Cullen, W.K.; Anwyl, R.; Wolfe, M.S.; Rowan, M.J.; Selkoe, D.J. Naturally Secreted Oligomers of Amyloid Beta Protein Potently Inhibit Hippocampal Long-term Potentiation in vivo. Nature 2002, 416, 535–539. [Google Scholar] [CrossRef]
- McLean, C.A.; Cherny, R.A.; Fraser, F.W.; Fuller, S.J.; Smith, M.J.; Beyreuther, K.; Bush, A.I.; Masters, C.L. Soluble Pool of Abeta Amyloid as a Determinant of Severity of Neurodegeneration in Alzheimer’s Disease. Ann. Neurol. 1999, 46, 860–866. [Google Scholar] [CrossRef]
- Mc Donald, J.M.; Savva, G.M.; Brayne, C.; Welzel, A.T.; Forster, G.; Shankar, G.M.; Selkoe, D.J.; Ince, P.G.; Walsh, D.M. Medical Research Council Cognitive Function and Ageing Study. The Presence of Sodium Dodecyl Sulphate-stable Abeta Dimers is Strongly Associated with Alzheimer-type Dementia. Brain 2010, 133, 1328–1341. [Google Scholar] [CrossRef] [PubMed]
- De Strooper, B. Proteases and Proteolysis in Alzheimer Disease: A Multifactorial View on the Disease Process. Physiol. Rev. 2010, 90, 465–494. [Google Scholar] [CrossRef]
- Nilsberth, C.; Westlind-Danielsson, A.; Eckman, C.B.; Condron, M.M.; Axelman, K.; Forsell, C.; Stenh, C.; Luthman, J.; Teplow, D.B.; Younkin, S.G.; et al. The ‘Arctic’ APP Mutation (E693G) Causes Alzheimer’s Disease by Enhanced Abeta Protofibril Formation. Nat Neurosci. 2001, 4, 887–893. [Google Scholar] [CrossRef]
- Tomiyama, T.; Nagata, T.; Shimada, H.; Teraoka, R.; Fukushima, A.; Kanemitsu, H.; Takuma, H.; Kuwano, R.; Imagawa, M.; Ataka, S.; et al. A New Amyloid Beta Variant Favoring Oligomerization in Alzheimer’s-type Dementia. Ann. Neurol. 2008, 63, 377–387. [Google Scholar] [CrossRef]
- Bitan, G.; Fradinger, E.A.; Spring, S.M.; Teplow, D.B. Neurotoxic Protein Oligomers–What You See is not Always What You Get. Amyloid 2005, 12, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Hepler, R.W.; Grimm, K.M.; Nahas, D.D.; Breese, R.; Dodson, E.C.; Acton, P.; Keller, P.M.; Yeager, M.; Wang, H.; Shughrue, P.; et al. Solution State Characterization of Amyloid Beta-derived Diffusible Ligands. Biochemistry 2006, 45, 15157–15167. [Google Scholar] [CrossRef]
- Hartley, D.M.; Zhao, C.; Speier, A.C.; Woodard, G.A.; Li, S.; Li, Z.; Walz, T. Transglutaminase Induces Protofibril-like Amyloid Beta-protein Assemblies That are Protease-resistant and Inhibit Long-term Potentiation. J. Biol. Chem. 2008, 283, 16790–16800. [Google Scholar] [CrossRef]
- Smith, D.P.; Smith, D.G.; Curtain, C.C.; Boas, J.F.; Pilbrow, J.R.; Ciccotosto, G.D.; Lau, T.L.; Tew, D.J.; Perez, K.; Wade, J.D.; et al. Copper-mediated Amyloid-beta Toxicity is Associated with an Intermolecular Histidine Bridge. J. Biol. Chem. 2006, 281, 15145–15154. [Google Scholar] [CrossRef] [PubMed]
- Galeazzi, L.; Ronchi, P.; Franceschi, C.; Giunta, S. In vitro Peroxidase Oxidation Induces Stable Dimers of Beta-amyloid (1-42) through Dityrosine Bridge Formation. Amyloid 1999, 6, 7–13. [Google Scholar] [CrossRef]
- Demuro, A.; Mina, E.; Kayed, R.; Milton, S.C.; Parker, I.; Glabe, C.G. Calcium Dysregulation and Membrane Disruption as a Ubiquitous Neurotoxic Mechanism of Soluble Amyloid Oligomers. J. Biol. Chem. 2005, 280, 17294–17300. [Google Scholar] [CrossRef] [PubMed]
- Aleksis, R.; Oleskovs, F.; Jaudzems, K.; Pahnke, J.; Biverstål, H. Structural Studies of Amyloid-β Peptides: Unlocking the Mechanism of Aggregation and the Associated Toxicity. Biochimie 2017, 140, 176–192. [Google Scholar] [CrossRef]
- Karran, E.; Mercken, M.; Strooper, B. The Amyloid Cascade Hypothesis for Alzheimer’s Disease: An Appraisal for the Development of Therapeutics. Nat. Rev. Drug. Discov. 2011, 10, 698–712. [Google Scholar] [CrossRef]
- Rajasekhar, K.; Chakrabarti, M.; Govindaraju, T. Function and Toxicity of Amyloid Beta and Recent Therapeutic Interventions Targeting Amyloid Beta in Alzheimer’s Disease. Chem. Commun. 2015, 51, 13434–13450. [Google Scholar] [CrossRef]
- Kollmer, M.; Close, W.; Funk, L.; Rasmussen, J.; Bsoul, A.; Schierhorn, A.; Schmidt, M.; Sigurdson, C.J.; Jucker, M.; Fändrich, M. Cryo-EM Structure and Polymorphism of Aβ Amyloid Fibrils Purified from Alzheimer’s Brain Tissue. Nature Commun. 2019, 10, 4760. [Google Scholar] [CrossRef]
- Braak, H.; Braak, E. Neuropathological Stageing of Alzheimer-related Changes. Acta Neuropathol. 1991, 82, 239–259. [Google Scholar] [CrossRef]
- Terry, R.D.; Masliah, E.; Salmon, D.P.; Butters, N.; DeTeresa, R.; Hill, R.; Hansen, L.A.; Katzman, R. Physical Basis of Cognitive Alterations in Alzheimer’s Disease: Synapse Loss is the Major Correlate of Cognitive Impairment. Ann. Neurol. 1991, 30, 572–580. [Google Scholar] [CrossRef] [PubMed]
- ClinicalsTrails.gov. Available online: https://clinicaltrials.gov/ct2/results?cond=Alzheimer+Disease&term=&cntry=&state=&city=&dist= (accessed on 5 April 2021).
- Hardy, J.; Mayer, J. The Amyloid Cascade Hypothesis Has Misled the Pharmaceutical Industry. Biochem. Soc Trans. 2011, 39, 920–923. [Google Scholar]
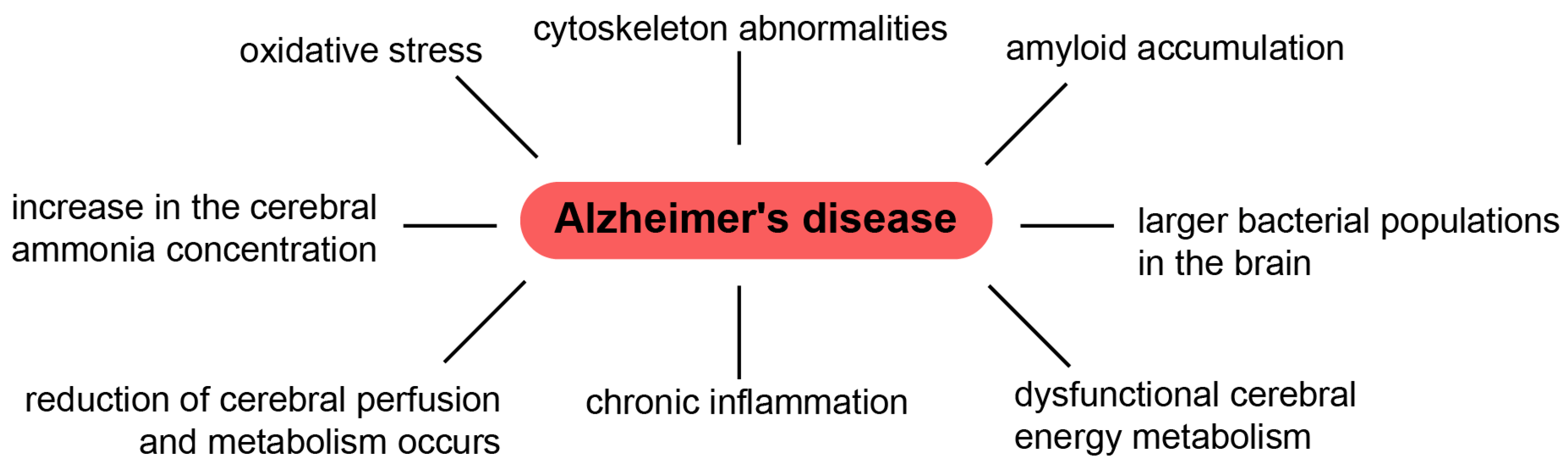
- Kosenko, E.A.; Solomadin, I.N.; Tikhonova, L.A.; Reddy, V.P.; Aliev, G.; Kaminsky, Y.G. Pathogenesis of Alzheimer Disease: Role of Oxidative Stress, Amyloid-β Peptides, Systemic Ammonia and Erythrocyte Energy Metabolism. CNS Neurol. Disord. Drug. Targets. 2014, 13, 112–119. [Google Scholar] [CrossRef]
- Schmitt, F.A.; Davis, D.G.; Wekstein, D.R.; Smith, C.D.; Ashford, J.W.; Markesbery, W.R. “Preclinical” AD Revisited: Neuropathology of Cognitively Normal Older Adults. Neurology 2000, 55, 370–376. [Google Scholar] [CrossRef] [PubMed]
- Jagust, W.J.; Mormino, E.C. Lifespan Brain Activity, β-amyloid, and Alzheimer’s Disease. Trends Cogn. Sci. 2011, 15, 520–526. [Google Scholar] [CrossRef] [PubMed]
- Hemming, M.L.; Selkoe, D.J. Amyloid Beta-protein is Degraded by Cellular Angiotensin-converting Enzyme (ACE) and Elevated by an ACE Inhibitor. J. Biol. Chem. 2005, 280, 37644–37650. [Google Scholar] [CrossRef] [PubMed]
- Holmes, C.; Boche, D.; Wilkinson, D.; Yadegarfar, G.; Hopkins, V.; Bayer, A.; Jones, R.W.; Bullock, R.; Love, S.; Neal, J.W.; et al. Long-term Effects of Abeta42 Immunisation in Alzheimer’s Disease: Follow-up of a Randomised, Placebo-controlled Phase I Trial. Lancet 2008, 372, 216–223. [Google Scholar] [CrossRef]
- Kurkinen, M. The Amyloid Hypothesis is too Good to be True. Alzheimers Dement. Cogn. Neurol. 2017, 1, 1–9. [Google Scholar] [CrossRef][Green Version]
- Orgogozo, J.M.; Gilman, S.; Dartigues, J.F.; Laurent, B.; Puel, M.; Kirby, L.C.; Jouanny, P.; Dubois, B.; Eisner, L.; Flitman, S.; et al. Subacute Meningoencephalitis in a Subset of Patients with AD after Abeta42 Immunization. Neurology 2003, 61, 46–54. [Google Scholar] [CrossRef] [PubMed]
- Nicoll, J.A.; Wilkinson, D.; Holmes, C.; Steart, P.; Markham, H.; Weller, R.O. Neuropathology of Human Alzheimer Disease after Immunization with Amyloid-beta Peptide: A Case Report. Nat. Med. 2003, 9, 448–452. [Google Scholar] [CrossRef]
- Tabira, T. Immunization Therapy for Alzheimer Disease: A Comprehensive Review of Active Immunization Strategies. Tohoku J. Exp. Med. 2010, 220, 95–106. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Blass, J.P. Immunologic Treatment of Alzheimer’s Disease. New Engl. J. Med. 1999, 341, 1694–1695. [Google Scholar] [CrossRef] [PubMed]
- Brewer, G.J.; Oelshlegel, F.J., Jr.; Moore, L.G.; Noble, N.A. In vivo Red Cell Glycolytic Control and DPG-ATP Levels. Ann. N. Y. Acad. Sci. 1974, 241, 513–523. [Google Scholar] [CrossRef]
- Bruce, D.G.; Davis, W.A.; Casey, G.P.; Clarnette, R.M.; Brown, S.G.; Jacobs, I.G.; Almeida, O.P.; Davis, T.M. Severe Hypoglycaemia and Cognitive Impairment in Older Patients with Diabetes: The Fremantle Diabetes Study. Diabetologia 2009, 52, 1808–1815. [Google Scholar] [CrossRef]
- Klepper, J.; Voit, T. Facilitated Glucose Transporter Protein Type 1 (GLUT1) Deficiency Syndrome: Impaired Glucose Transport into Brain–A Review. Eur. J. Pediatr. 2002, 161, 295–304. [Google Scholar] [CrossRef]
- Ronnback, L.; Hansson, E. On the Potential Role of Glutamate Transport in Mental Fatigue. J. Neuroinflammation 2004, 1, 22. [Google Scholar] [CrossRef]
- Llansola, M.; Rodrigo, R.; Monfort, P.; Montoliu, C.; Kosenko, E.; Cauli, O.; Piedrafita, B.; Mlili, N.; Felipo, V. NMDA Receptors in Hyperammonemia and Hepatic Encephalopathy. Metab Brain Dis. 2007, 22, 321–335. [Google Scholar] [CrossRef]
- Hoyer, S. Oxidative Metabolism Deficiencies in Brains of Patients with Alzheimer’s Disease. Acta Neurol. Scand. Suppl. 1996, 165, 18–24. [Google Scholar] [CrossRef]
- Hoyer, S.; Oesterreich, K.; Wagner, O. Glucose Metabolism as the Site of the Primary Abnormality in Early-onset Dementia of Alzheimer Type? J. Neurol. 1988, 235, 143–148. [Google Scholar] [CrossRef]
- Aliev, G.; Palacios, H.H.; Walrafen, B.; Lipsitt, A.E.; Obrenovich, M.E.; Morales, L. Brain Mitochondria as a Primary Target in the Development of Treatment Strategies for Alzheimer Disease. Int. J. Biochem. Cell Biol. 2009, 41, 1989–2004. [Google Scholar] [CrossRef] [PubMed]
- Aliev, G.; Li, Y.; Palacios, H.H.; Obrenovich, M.E. Oxidative Stress Induced Mitochondrial DNA Deletion as a Hallmark for the Drug Development in the Context of the Cerebrovascular Diseases. Rec. Pat. Cardiovasc. Drug Discov. 2011, 6, 222–241. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.G.; Casadesus, G.; Zhu, X.; Takeda, A.; Perry, G.; Smith, M.A. Challenging the Amyloid Cascade Hypothesis: Senile Plaques and Amyloid-beta as Protective Adaptations to Alzheimer Disease. Ann. N. Y. Acad. Sci. 2004, 1019, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.G.; Zhu, X.; Castellani, R.J.; Nunomura, A.; Perry, G.; Smith, M.A. Amyloid-beta in Alzheimer Disease: The Null vs. the Alternate Hypotheses. J. Pharmacol. Exp. Ther. 2007, 321, 823–829. [Google Scholar] [CrossRef] [PubMed]
- Atwood, C.S.; Robinson, S.R.; Smith, M.A. Amyloid-beta: Redox-metal Chelator and Antioxidant. J. Alzheimers Dis. 2002, 4, 203–214. [Google Scholar] [CrossRef]
- Smith, M.A.; Casadesus, G.; Joseph, J.A.; Perry, G. Amyloid-beta and Tau Serve Antioxidant Functions in the Aging and Alzheimer Brain. Free Radic. Biol. Med. 2002, 33, 1194–1199. [Google Scholar] [CrossRef]
- Soscia, S.J.; Kirby, J.E.; Washicosky, K.J.; Tucker, S.M.; Ingelsson, M.; Hyman, B.; Burton, M.A.; Goldstein, L.E.; Duong, S.; Tanzi, R.E.; et al. The Alzheimer’s Disease-associated Amyloid Beta-protein is an Antimicrobial Peptide. PLoS ONE 2010, 5, e9505. [Google Scholar] [CrossRef]
- de la Torre, J.C. Impaired Cerebromicrovascular Perfusion. Summary of Evidence in Support of Its Causality in Alzheimer’s Disease. Ann. N. Y. Acad. Sci. 2000, 924, 136–152. [Google Scholar] [CrossRef]
- de la Torre, J.C.; Aliev, G. Inhibition of Vascular Nitric Oxide after Rat Chronic Brain Hypoperfusion: Spatial Memory and Immunocytochemical Changes. J. Cereb. Blood Flow Metab. 2005, 25, 663–672. [Google Scholar] [CrossRef]
- Wen, Z.; Xie, J.; Guan, Z.; Sun, D.; Yao, W.; Chen, K.; Yan, Z.Y.; Mu, Q. A Study of Hemorheological Behaviour for Patients with Alzheimer’s Disease at the Early Stages. Clin. Hemorheol. Microcirc. 2000, 22, 261–266. [Google Scholar] [PubMed]
- Kaminsky, Y.; Poghosyan, A.; Tikhonova, L.; Palacios, H.H.; Kamal, M.A.; Kosenko, E.; Aliev, G. Glycolytic and Proteolytic Metabolism in Erythrocytes from Elderly and Demented Patients. Am. J. Neuroprotect. Neuroregener. 2012, 4, 73–77. [Google Scholar] [CrossRef]
- Kaminsky, Y.G.; Reddy, V.P.; Ashraf, G.M.; Ahmad, A.; Benberin, V.V.; Kosenko, E.A.; Aliev, G. Age-related Defects in Erythrocyte 2,3-diphosphoglycerate Metabolism in Dementia. Aging Dis. 2013, 4, 244–255. [Google Scholar] [CrossRef] [PubMed]
- Norberg, K.; Siesio, B.K. Oxidative Metabolism of the Cerebral Cortex of the Rat in Severe Insulin-induced Hypoglycaemia. J. Neurochem. 1976, 26, 345–352. [Google Scholar] [CrossRef]
- Kinney, J.W.; Bemiller, S.M.; Murtishaw, A.S.; Leisgang, A.M.; Salazar, A.M.; Lamb, B.T. Inflammation as a Central Mechanism in Alzheimer’s Disease. Alzheimers Dement. Transl. Res. Clin. Interv. 2018, 4, 575–590. [Google Scholar] [CrossRef] [PubMed]
- Rogers, J.; Cooper, N.R.; Webster, S.; Schultz, J.; McGeer, P.L.; Styren, S.D.; Civin, W.H.; Brachova, L.; Bradt, B.; Ward, P. Complement Activation by Beta-amyloid in Alzheimer Disease. Proc. Natl. Acad. Sci. USA 1992, 89, 10016–10020. [Google Scholar] [CrossRef]
- Chen, S.; Frederickson, R.C.; Brunden, K.R. Neuroglial-mediated Immunoinflammatory Responses in Alzheimer’s Disease: Complement Activation and Therapeutic Approaches. Neurobiol. Aging 1996, 17, 781–787. [Google Scholar] [CrossRef]
- Jiang, H.; Burdick, D.; Glabe, C.G.; Cotman, C.W.; Tenner, A.J. Beta-amyloid Activates Complement by Binding to a Specific Region of the Collagen-like Domain of the C1q A Chain. J. of Immunol. 1994, 152, 5050–5059. [Google Scholar]
- Afagh, A.; Cummings, B.J.; Cribbs, D.H.; Cotman, C.W.; Tenner, A.J. Localization and Cell Association of C1q in Alzheimer’s Disease Brain. Exp. Neurol. 1999, 6138, 22. [Google Scholar] [CrossRef]
- Webster, S.; Bradt, B.; Rogers, J.; Cooper, N.R. Aggregation State-dependent Activation of the Classical Complement Pathway by the Amyloid β Peptide (Aβ). J. Neurochem. 1997, 69, 388–398. [Google Scholar] [CrossRef]
- Webster, S.; Lue, L.F.; Brachova, L.; Tenner, A.; McGeer, P.L.; Walker, D.; Bradt, B.; Cooper, N.R.; Rogers, J. Molecular and Cellular Characterization of the Membrane Attack Complex, C5b-9, in Alzheimer’s Disease. Neurobiol. Aging 1997, 18, 415–421. [Google Scholar] [CrossRef]
- Veerhuis, R.; Van Breemen, M.J.; Hoozemans, J.M. Amyloid Beta Plaque-associated Proteins C1q and SAP Enhance the Abeta1-42 Peptide-induced Cytokine Secretion by Adult Human Microglia in vitro. Acta Neuropathol. 2003, 105, 135. [Google Scholar] [CrossRef] [PubMed]
- Tacnet-Delorme, P.; Chevallier, S.; Arlaud, G.J. β-Amyloid Fibrils Activate the C1 Complex of Complement Under Physiological Conditions: Evidence for a Binding Site for Aβ on the C1q Globular Regions. J. Immunol. 2001, 167, 6374. [Google Scholar] [CrossRef]
- Bradt, B.M.; Kolb, W.P.; Cooper, N.R. Complement-dependent Proinflammatory Properties of the Alzheimer’s Disease Beta-peptide. J. Exp. Med. 1998, 188, 431. [Google Scholar] [CrossRef] [PubMed]
- Strohmeyer, R.; Shen, Y.; Rogers, J. Detection of Complement Alternative Pathway mRNA and Proteins in the Alzheimer’s Disease Brain. Mol. Brain Res. 2000, 81, 7. [Google Scholar] [CrossRef]
- Watson, M.D.; Roher, A.E.; Kim, K.S.; Spiegel, K.; Emmerlin, M.R. Complement Interactions with Amyloid β1–42: A Nidus for Inflammation in AD Brains. Amyloid 1997, 4, 147–156. [Google Scholar] [CrossRef]
- Eikelenboom, P.; Stam, F.C. Immunoglobulins and Complement Factors in Senile Plaques. An Immunoperoxidase Study. Acta Neuropathol. 1982, 57, 239. [Google Scholar] [CrossRef]
- McGeer, P.L.; Akiyama, H.; Itagaki, S.; McGeer, E.G. Activation of the Classical Complement Pathway in Brain Tissue of Alzheimer Patients. Neurosci. Lett. 1989, 107, 341. [Google Scholar] [CrossRef]
- Shen, Y.; Li, R.; McGeer, E.G.; McGeer, P.L. Neuronal Expression of mRNAs for Complement Proteins of the Classical Pathway in Alzheimer Brain. Brain Res. 1997, 769, 391. [Google Scholar] [CrossRef]
- Landlinger, C.; Oberleitner, L.; Gruber, P.; Noiges, B.; Yatsyk, K.; Santic, R.; Mandler, M.; Staffler, G. Active Immunization against Complement Factor C5a: A New Therapeutic Approach for Alzheimer’s Disease. J. Neuroinflammation 2015, 12, 150. [Google Scholar] [CrossRef]
- Yakupova, E.I.; Bobyleva, L.G.; Vikhlyantsev, I.M.; Bobylev, A.G. Complement System Activation by Amyloid Aggregates of Aβ(1-40) and Aβ(1-42) Peptides: Facts and Hypotheses. Biophysics 2020, 65, 18–21. [Google Scholar] [CrossRef]
- Emery, D.C.; Shoemark, D.K.; Batstone, T.E.; Waterfall, C.M.; Coghill, J.A.; Cerajewska, T.L.; Davies, M.; West, N.X.; Allen, S.J. 16S rRNA Next Generation Sequencing Analysis Shows Bacteria in Alzheimer’s Post-Mortem Brain. Front. Aging Neurosci. 2017, 9, 195. [Google Scholar] [CrossRef]
- Kumar, D.K.; Choi, S.H.; Washicosky, K.J.; Eimer, W.A.; Tucker, S.; Ghofrani, J.; Lefkowitz, A.; McColl, G.; Goldstein, L.E.; Tanzi, R.E.; et al. Amyloid-β Peptide Protects against Microbial Infection in Mouse and Worm Models of Alzheimer’s Disease. Sci. Transl. Med. 2016, 8, 340ra72. [Google Scholar] [CrossRef]
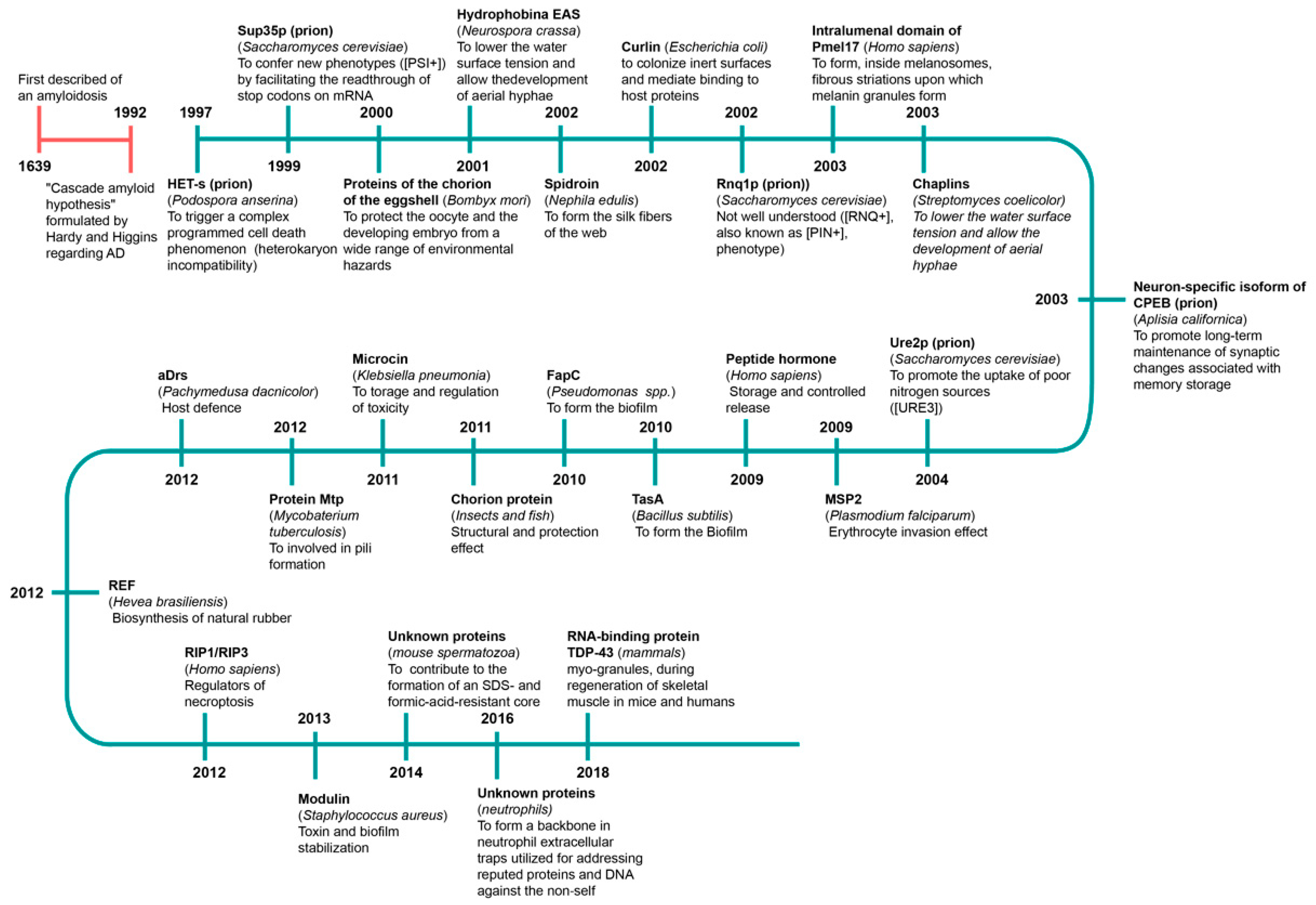
- Daskalov, A.; Saupe, S.J. The Expanding Scope of Amyloid Signalling. Prion 2021, 15, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Nicastro, L.; Tükel, Ç. Bacterial Amyloids: The Link between Bacterial Infections and Autoimmunity. Trends Microbiol. 2019, 27, 954–963. [Google Scholar] [CrossRef]
- Olsen, A.; Jonsson, A.; Normark, S. Fibronectin Binding Mediated by a Novel Class of Surface Organelles on Escherichia coli. Nature 1976, 338, 652–655. [Google Scholar] [CrossRef] [PubMed]
- Álvarez-Mena, A.; Cámara-Almirón, J.; de Vicente, A.; Romero, D. Multifunctional Amyloids in the Biology of Gram-Positive Bacteria. Microorganisms 2020, 8, 2020. [Google Scholar] [CrossRef] [PubMed]
- Pham, C.L.; Kwan, A.H.; Sunde, M. Functional Amyloid: Widespread in Nature, Diverse in Purpose. Essays Biochem. 2014, 56, 207–219. [Google Scholar] [PubMed]
- Balistreri, A.; Goetzler, E.; Chapman, M. Functional Amyloids Are the Rule Rather Than the Exception in Cellular Biology. Microorganisms 2020, 8, 1951. [Google Scholar] [CrossRef]
- Pulze, L.; Bassani, B.; Gini, E.; D’Antona, P.; Grimaldi, A.; Luini, A.; Marino, F.; Noonan, D.M.; Tettamanti, G.; Valvassori, R.; et al. NET Amyloidogenic Backbone in Human Activated Neutrophils. Clin. Exp. Immunol. 2016, 183, 469–479. [Google Scholar] [CrossRef]
- Vogler, T.O.; Wheeler, J.R.; Nguyen, E.D.; Hughes, M.P.; Britson, K.A.; Lester, E.; Rao, B.; Betta, N.D.; Whitney, O.N.; Ewachiw, T.E.; et al. TDP-43 and RNA Form Amyloid-like Myo-granules in Regenerating Muscle. Nature 2018, 563, 508–513. [Google Scholar] [CrossRef] [PubMed]
- Nizhnikov, A.A.; Antonets, K.S.; Bondarev, S.A.; Inge-Vechtomov, S.G.; Derkatch, I.L. Prions, Amyloids, and RNA: Pieces of a Puzzle. Prion 2016, 10, 182–206. [Google Scholar] [CrossRef]
- Bardin, T.; Daskalov, A.; Barrouilhet, S.; Granger-Farbos, A.; Salin, B.; Blancard, C.; Kauffmann, B.; Saupe, S.J.; Cousto, U.V. Partial Prion Cross-Seeding between Fungal and Mammalian Amyloid Signaling Motifs. mBio 2021, 12, e02782-20. [Google Scholar] [CrossRef] [PubMed]
- Kosolapova, A.O.; Antonets, K.S.; Belousov, M.V.; Nizhnikov, A.A. Biological Functions of Prokaryotic Amyloids in Interspecies Interactions: Facts and Assumptions. Int. J. Mol. Sci. 2020, 21, 7240. [Google Scholar] [CrossRef]
- Chapman, M.R.; Robinson, L.S.; Pinkner, J.S.; Roth, R.; Heuser, J.; Hammar, M.; Normark, S.; Hultgren, S.J. Role of Escherichia coli Curli Operons in Directing Amyloid Fiber Formation. Science 2002, 295, 851–855. [Google Scholar] [CrossRef] [PubMed]
- Mackay, J.P.; Matthews, J.M.; Winefield, R.D.; Mackay, L.G.; Haverkamp, R.G.; Templeton, M.D. The Hydrophobin EAS is Largely Unstructured in Solution and Functions by Forming Amyloid-like Structures. Structure 2001, 9, 83–91. [Google Scholar] [CrossRef]
- Kenney, J.M.; Knight, D.; Wise, M.J.; Vollrath, F. Amyloidogenic Nature of Spider Silk. Eur. J. Biochem. 2002, 269, 4159–4163. [Google Scholar] [CrossRef]
- Berson, J.F.; Theos, A.C.; Harper, D.C.; Tenza, D.; Raposo, G.; Marks, M.S. Proprotein Convertase Cleavage Liberates a Fibrillogenic Fragment of a Resident Glycoprotein to Initiate Melanosome Biogenesis. J. Cell Biol. 2003, 161, 521–533. [Google Scholar] [CrossRef]
- Chien, P.; Weissman, J.S.; DePace, A.H. Emerging Principles of Conformation-based Prion Inheritance. Annu. Rev. Biochem. 2004, 73, 617–656. [Google Scholar] [CrossRef] [PubMed]
- Eaglestone, S.S.; Cox, B.S.; Tuite, M.F. Translation Termination Efficiency Can be Regulated in Saccharomyces cerevisiae by Environmental Stress through a Prion-mediated Mechanism. EMBO J. 1999, 18, 1974–1981. [Google Scholar] [CrossRef]
- Giaever, G.; Chu, A.M.; Ni, L.; Connelly, C.; Riles, L.; Véronneau, S.; Dow, S.; Lucau-Danila, A.; Anderson, K.; André, B.; et al. Functional Profiling of the Saccharomyces cerevisiae Genome. Nature 2002, 418, 387–391. [Google Scholar] [CrossRef]
- Nakayashiki, T.; Kurtzman, C.P.; Edskes, H.K.; Wickner, R.B. Yeast Prions [URE3] and [PSI+] are Diseases. Proc. Natl. Acad. Sci. USA 2005, 102, 10575–10580. [Google Scholar] [CrossRef]
- Coustou, V.; Deleu, C.; Saupe, S.; Begueret, J. The Protein Product of the Het-s Heterokaryon Incompatibility Gene of the Fungus Podospora anserina Behaves as a Prion Analog. Proc. Natl. Acad. Sci. USA 1997, 94, 9773–9778. [Google Scholar] [CrossRef]
- Saupe, S.J. Molecular Genetics of Heterokaryon Incompatibility in Filamentous Ascomycetes. Microbiol. Mol. Biol. Rev. 2000, 64, 489–502. [Google Scholar] [CrossRef] [PubMed]
- Dueholm, M.S.; Petersen, S.V.; Sonderkaer, M.; Larsen, P.; Christiansen, G.; Hein, K.L.; Enghild, J.J.; Nielsen, J.L.; Nielsen, K.L.; Nielsen, P.H.; et al. Functional Amyloid in Pseudomonas. Mol. Microbiol. 2010, 77, 1009–1020. [Google Scholar] [CrossRef] [PubMed]
- Otzen, D. Functional Amyloid: Turning Swords into Plowshares. Prion 2010, 4, 256–264. [Google Scholar] [CrossRef]
- Shewmaker, F.; McGlinchey, R.P.; Wickner, R.B. Structural Insights into Functional and Pathological Amyloid. J. Biol. Chem. 2011, 286, 16533–16540. [Google Scholar] [CrossRef]
- Schwartz, K.; Boles, B.R. Microbial Amyloids: Functions and Interactions within the Host. Curr. Opin. Microbiol. 2013, 16, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Adda, C.G.; Murphy, V.J.; Sunde, M.; Waddington, L.J.; Schloegel, J.; Talbo, G.H.; Vingas, K.; Kienzle, V.; Masciantonio, R.; Howlett, G.J.; et al. Plasmodium Falciparum Merozoite Surface Protein 2 is Unstructured and Forms Amyloid-like Fibrils. Mol. Biochem. Parasitol. 2009, 166, 159–171. [Google Scholar] [CrossRef]
- Berthelot, K.; Lecomte, S.; Estevez, Y.; Coulary-Salin, B.; Bentaleb, A.; Cullin, C.; Deffieux, A.; Peruch, F. Rubber Elongation Factor (REF), a Major Allergen Component in Hevea brasiliensis latex Has Amyloid Properties. PLoS ONE 2012, 7, e48065. [Google Scholar] [CrossRef]
- Gossler-Schofberger, R.; Hesser, G.; Reif, M.M.; Friedmann, J.; Duscher, B.; Toca-Herrera, J.L.; Oostenbrink, C.; Jilek, A. A Stereochemical Switch in the aDrs Model System, a Candidate for a Functional Amyloid. Arch. Biochem. Biophys. 2012, 522, 100–106. [Google Scholar] [CrossRef]
- Li, J.; McQuade, T.; Siemer, A.B.; Napetschnig, J.; Moriwaki, K.; Hsiao, Y.S.; Damko, E.; Moquin, D.; Walz, T.; McDermott, A.; et al. The RIP1/RIP3 Necrosome Forms a Functional Amyloid Signaling Complex Required for Programmed Necrosis. Cell 2012, 150, 339–350. [Google Scholar] [CrossRef] [PubMed]
- Barnhart, M.M.; Chapman, M.R. Curli Biogenesis and Function. Annu. Rev. Microbiol. 2006, 60, 131–147. [Google Scholar] [CrossRef] [PubMed]
- Cherny, I.; Rockah, L.; Levy-Nissenbaum, O.; Gophna, U.; Ron, E.Z.; Gazit, E. The Formation of Escherichia coli Curli Amyloid Fibrils is Mediated by Prion-like Peptide Repeats. J. Mol. Biol. 2005, 352, 245–252. [Google Scholar] [CrossRef]
- Shewmaker, F.; McGlinchey, R.P.; Thurber, K.R.; McPhie, P.; Dyda, F.; Tycko, R.; Wickner, R.B. The Functional Curli Amyloid is not Based on In-register Parallel Beta-sheet Structure. J. Biol. Chem. 2009, 284, 25065–25076. [Google Scholar] [CrossRef]
- Sawyer, E.B.; Claessen, D.; Haas, M.; Hurgobin, B.; Gras, S.L. The Assembly of Individual Chaplin Peptides from Streptomyces coelicolor into Functional Amyloid Fibrils. PLoS ONE 2011, 6, e18839. [Google Scholar] [CrossRef]
- Kosolapova, A.O.; Belousov, M.V.; Sulatskaya, A.I.; Belousova, M.E.; Sulatsky, M.I.; Antonets, K.S.; Volkov, K.V.; Lykholay, A.N.; Shtark, O.Y.; Vasileva, E.N.; et al. Two Novel Amyloid Proteins, RopA and RopB, from the Root Nodule Bacterium Rhizobium leguminosarum. Biomolecules 2019, 9, 694. [Google Scholar] [CrossRef]
- Bieler, S.; Estrada, L.; Lagos, R.; Baeza, M.; Castilla, J.; Soto, C. Amyloid Formation Modulates the Biological Activity of a Bacterial Protein. J. Biol. Chem. 2005, 280, 26880–26885. [Google Scholar] [CrossRef]
- Shahnawaz, M.; Park, K.W.; Mukherjee, A.; Diaz-Espinoza, R.; Soto, C. Prion-like Characteristics of the Bacterial Protein Microcin E492. Sci. Rep. 2017, 7, 45720. [Google Scholar] [CrossRef] [PubMed]
- Arranz, R.; Mercado, G.; Martín-Benito, J.; Giraldo, R.; Monasterio, O.; Lagos, R.; Valpuesta, J.M. Structural Characterization of Microcin E492 Amyloid Formation: Identification of the Precursors. J. Struct. Biol. 2012, 178, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.; Kim, J.G.; Jeon, E.; Yoo, C.H.; Moon, J.S.; Rhee, S.; Hwang, I. Amyloidogenesis of Type III-dependent Harpins from Plant Pathogenic Bacteria. J. Biol. Chem. 2007, 282, 13601–13609. [Google Scholar] [CrossRef] [PubMed]
- Dos Reis, S.; Coulary-Salin, B.; Forge, V.; Lascu, I.; Bégueret, J.; Saupe, S.J. The HET-s Prion Protein of the Filamentous Fungus Podospora Anserina Aggregates in vitro into Amyloid-like Fibrils. J. Biol. Chem. 2002, 277, 5703–5706. [Google Scholar] [CrossRef]
- Balguerie, A.; Dos Reis, S.; Ritter, C.; Chaignepain, S.; Coulary-Salin, B.; Forge, V.; Bathany, K.; Lascu, I.; Schmitter, J.M.; Riek, R.; et al. Domain Organization and Structure-function Relationship of the HET-s Prion Protein of Podospora Anserina. EMBO J. 2003, 22, 2071–2081. [Google Scholar] [CrossRef]
- Ritter, C.; Maddelein, M.L.; Siemer, A.B.; Lührs, T.; Ernst, M.; Meier, B.H.; Saupe, S.J.; Riek, R. Correlation of Structural Elements and Infectivity of the HET-s Prion. Nature 2005, 435, 844–848. [Google Scholar] [CrossRef]
- Wan, W.; Wille, H.; Stöhr, J.; Baxa, U.; Prusiner, S.B.; Stubbs, G. Degradation of Fungal Prion HET-s(218-289) Induces Formation of a Generic Amyloid Fold. Biophys. J. 2012, 102, 2339–2344. [Google Scholar] [CrossRef]
- Baxa, U.; Cheng, N.; Winkler, D.C.; Chiu, T.K.; Davies, D.R.; Sharma, D.; Inouye, H.; Kirschner, D.A.; Wickner, R.B.; Steven, A.C. Filaments of the Ure2p Prion Protein Have a Cross-β Core Structure. J. Struct. Biol. 2005, 150, 170–179. [Google Scholar] [CrossRef]
- Sant’Anna, R.; Fernández, M.; Batlle, C.; Navarro, S.; De Groot, N.S.; Serpell, L.; Ventura, S. Characterization of Amyloid Cores in Prion Domains. Sci. Rep. 2016, 6, 34274. [Google Scholar] [CrossRef] [PubMed]
- King, C.Y.; Tittmann, P.; Gross, H.; Gebert, R.; Aebi, M.; Wüthrich, K. Prion-inducing Domain 2–114 of Yeast Sup35 Protein Transforms in vitro into Amyloid-like Filaments. Proc. Natl. Acad. Sci. USA 1997, 94, 6618–6622. [Google Scholar] [CrossRef] [PubMed]
- True, H.L.; Berlin, I.; Lindquist, S.L. Epigenetic Regulation of Translation Reveals Hidden Genetic Variation to Produce Complex Traits. Nature 2004, 431, 184–187. [Google Scholar] [CrossRef]
- Du, Z.; Park, K.W.; Yu, H.; Fan, Q.; Li, L. Newly Identified Prion Linked to the Chromatin-remodeling Factor Swi1 in Saccharomyces cerevisiae. Nat. Genet. 2008, 40, 460–465. [Google Scholar] [CrossRef]
- Alberti, S.; Halfmann, R.; King, O.; Kapila, A.; Lindquist, S. A Systematic Survey Identifies Prions and Illuminates Sequence Features of Prionogenic Proteins. Cell 2009, 137, 146–158. [Google Scholar] [CrossRef]
- Butko, P.; Buford, J.P.; Goodwin, J.S.; Stroud, P.A.; McCormick, C.L.; Cannon, G.C. Spectroscopic Evidence for Amyloid-like Interfacial Self-assembly of Hydrophobin Sc3. Biochem. Biophys. Res. Commun. 2001, 280, 212–215. [Google Scholar] [CrossRef] [PubMed]
- de Vocht, M.L.; Reviakine, I.; Wösten, H.A.; Brisson, A.; Wessels, J.G.; Robillard, G.T. Structural and Functional Role of the Disulfide Bridges in the Hydrophobin SC3. J. Biol. Chem. 2000, 275, 28428–28432. [Google Scholar] [CrossRef] [PubMed]
- Kwan, A.H.; Winefield, R.D.; Sunde, M.; Matthews, J.M.; Haverkamp, R.G.; Templeton, M.D.; Mackay, J.P. Structural Basis for Rodlet Assembly in Fungal Hydrophobins. Proc. Natl. Acad. Sci. USA 2006, 103, 3621–3626. [Google Scholar] [CrossRef] [PubMed]
- Iconomidou, V.A.; Vriend, G.; Hamodrakas, S.J. Amyloids Protect the Silkmoth Oocyte and Embryo. FEBS Lett. 2000, 479, 141–145. [Google Scholar] [CrossRef]
- Podrabsky, J.E.; Carpenter, J.F.; Hand, S.C. Survival of Water Stress in Annual Fish Embryos: Dehydration Avoidance and Egg Envelope Amyloid Fibers. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2001, 280, R123–R131. [Google Scholar] [CrossRef]
- Iconomidou, V.A.; Chryssikos, G.D.; Gionis, V.; Vriend, G.; Hoenger, A.; Hamodrakas, S.J. Amyloid-like Fibrils from an 18-residue Peptide Analogue of a Part of the Central Domain of the B-family of Silkmoth Chorion Proteins. FEBS Lett. 2001, 499, 268–273. [Google Scholar] [CrossRef]
- Hamodrakas, S.J.; Jones, C.W.; Kafatos, F.C. Secondary Structure Predictions for Silkmoth Chorion Proteins. Biochim. Biophys. Acta Protein Struct. Mol. Enzymol. 1982, 700, 42–51. [Google Scholar] [CrossRef]
- Dicko, C.; Knight, D.; Kenney, J.M.; Vollrath, F. Structural Conformation of Spidroin in Solution: A Synchrotron Radiation Circular Dichroism Study. Biomacromolecules 2004, 5, 758–767. [Google Scholar] [CrossRef]
- Watt, B.; van Niel, G.; Fowler, D.M.; Hurbain, I.; Luk, K.C.; Stayrook, S.E.; Lemmon, M.A.; Raposo, G.; Shorter, J.; Kelly, J.W.; et al. N-terminal Domains Elicit Formation of Functional Pmel17 Amyloid Fibrils. J. Biol. Chem. 2009, 284, 35543–35555. [Google Scholar] [CrossRef]
- Si, K.; Giustetto, M.; Etkin, A.; Hsu, R.; Janisiewicz, A.M.; Miniaci, M.C.; Kim, J.H.; Zhu, H.; Kandel, E.R. A Neuronal Isoform of CPEB Regulates Local Protein Synthesis and Stabilizes Synapse-specific Long-term Facilitation in Aplysia. Cell 2003, 115, 893–904. [Google Scholar] [CrossRef]
- Hervas, R.; Rau, M.J.; Park, Y.; Zhang, W.; Murzin, A.G.; Fitzpatrick, J.A.J.; Scheres, S.H.W.; Si, K. Cryo-EM Structure of a Neuronal Functional Amyloid Implicated in Memory Persistence in Drosophila. Science 2020, 367, 1230–1234. [Google Scholar] [CrossRef]
- Ulamec, S.M.; Radford, S.E. Spot the Difference: Function vs. Toxicity in Amyloid Fibrils. Trends Biochem. Sci. 2020, 45, 635–636. [Google Scholar] [CrossRef] [PubMed]
- Cervantes, S.; Bajakian, T.; Soria, M.; Falk, A.S.; Service, R.J.; Langen, R.; Siemer, A.B. Identification and Structural Characterization of the N-terminal Amyloid Core of Orb2 isoform A. Sci. Rep. 2016, 6, 38265. [Google Scholar] [CrossRef]
- Gallardo, R.; Ramakers, M.; De Smet, F.; Claes, F.; Khodaparast, L.; Khodaparast, L.; Couceiro, J.R.; Langenberg, T.; Siemons, M.; Nyström, S.; et al. De novo Design of a Biologically Active Amyloid. Science 2016, 354, aah4949. [Google Scholar] [CrossRef] [PubMed]
- Latza, V.; Guerette, P.; Ding, D.; Amini, S.; Kumar, A.; Schmidt, I.; Keating, S.; Oxman, N.; Weaver, J.C.; Masic, A. Multi-scale Thermal Stability of a Hard Thermoplastic Protein-based Material. Nat. Commun. 2015, 6, 8313. [Google Scholar] [CrossRef]
- Maji, S.K.; Schubert, D.; Rivier, C.; Lee, S.; Rivier, J.E.; Riek, R. Amyloid as a Depot for the Formulation of Long-acting Drugs. PLoS Biol. 2008, 6, e17. [Google Scholar] [CrossRef]
- Cui, M.; Qi, Q.; Gurry, T.; Zhao, T.; An, B.; Pu, J.; Gui, X.; Cheng, A.A.; Zhang, S.; Xun, D.; et al. Modular Genetic Design of Multi-domain Functional Amyloids: Insights into Self-assembly and Functional Properties. Chem. Sci. 2019, 10, 4004–4014. [Google Scholar] [CrossRef]
- Jackson, M.P.; Hewitt, E.W. Why are Functional Amyloids Non-Toxic in Humans? Biomolecules 2017, 7, 71. [Google Scholar] [CrossRef] [PubMed]
- Pavliukeviciene, B.; Zentelyte, A.; Jankunec, M.; Valiuliene, G.; Talaikis, M.; Navakauskiene, R.; Niaura, G.; Valincius, G. Amyloid β Oligomers Inhibit Growth of Human Cancer Cells. PLoS ONE 2019, 14, e0221563. [Google Scholar] [CrossRef] [PubMed]
- Kagan, B.L.; Jang, H.; Capone, R.; Teran Arce, F.; Ramachandran, S.; Lal, R.; Nussinov, R. Antimicrobial Properties of Amyloid Peptides. Mol Pharm. 2012, 9, 708–717. [Google Scholar] [CrossRef] [PubMed]
| Species or Organisms | Protein or Peptide | Function | Mol. Weight | Structure | Evidence of Cross β-Structure Presence | Secondary Structure Changes | Congo Red and ThT Binding | Condition of in vitro Amyloid Fibril Forming | References |
|---|---|---|---|---|---|---|---|---|---|
| Bacteria | |||||||||
| Escherichia coli, Salmonella spp. | Curli | Biofilm formation, host invasion. | CsgA (main damain of curlin) ~17.5 kDa. | Previously β-structure. | X-ray diffraction for CsgA. | CD method: CsgA fibrils are as follows: 16 ± 2% α-helix, 40 ± 2% β-sheet, 13 ± 2% β-turn and 31 ± 2% remainder. | CR, ThT | CsgA fibrils were prepared by dialyzing purified protein into 25 mM Tris, pH 7.5, 100 mM NaCl and 0.5 mM EDTA and incubating at room temperature (RT) for several days. | [176,193,194,195] |
| Streptomyces coelicolor | Chaplins | Modulation of water surface tension (i.e., development of aerial structures). | ChpD-H up to 6 kDa ChpA-C ~17–20 kDa. | ChpD and ChpF comprise β-sheet; ChpE is random coil (RC); ChpG and ChpH have mixed secondary structure comprising elements of both β-sheet and RC. | X-ray diffraction. | CD method: the protein mixture adopted a conformation rich in β-sheet. | ThT | Synthetic chaplin peptides were dissolved at a final concentration of 0.5 mg/mL in water and the pH adjusted by titration of NaOH/HCl. | [11,196] |
| Rhizobium leguminosarum | RopA and RopB | Possibility role in the control of plant-microbial symbiosis. | RopA 38.97 kDa RopB 22 kDa. | Previously β- structure. | none | CD method: Before aggregation: RopA more than 40% β-structure, RopB more than 30% β-structure After aggregation: 42% and 38% β-structure for RopA and RopB aggregates respectively. | CR, ThT | Proteins were dissolved in 1,1,1,3,3,3-hexafluoro-2-propanol (HFIP) and incubated for seven days. Afterward, HFIP was evaporated under a stream of nitrogen, and the samples were stirred for an additional seven days. | [197] |
| Klebsiella pneumoniae | Microcin E492 (Mcc) | Bacteriocin, membrane pore-forming peptide, amyloid form is inactive. | ~7.8 kDa. | RC conformation in aqueous buffer and α-helix in methanol. | X-Ray diffraction. | CD method: Aggregated Mcc is rich in β-sheet structures. | CR, ThT | Purified Mcc a (400 μg/mL) were incubated in aggregation buffer (50 mM PIPES-NaOH, pH 6.5, 0.5 M NaCl) for 48 h at 37 °C with vigorous shaking. | [198,199,200] |
| Xanthomonas species | Harpins (HpaG) | Secreted by plant pathogenic bacteria, destabilize plant membranes, induce cell death. | 15.6 kDa. | Previously α-helix. | non | CD method: After 3 days, the CD spectrum of HpaG changed to a minimum at 220 nm, which is indicative of transition to a β-sheet. | CR | Harpin samples were incubated without agitation in 20 mm Tris-HCl (pH 8.0) containing 10 mm NaCl to mimic the salt concentration in the intercellular space of plant tissues at 27 °C for 14 days. | [201] |
| Fungi | |||||||||
| Podospora anserine | HET-s | Regulation of heterokaryon formation. | ~32 kDa. | Estimated content of 34% α-helical, 16% β-sheet and 50% RC structure. | X-ray diffraction of HET-s (218–289). | CD method: 17% α-helix, 32% β-sheet and 50% random coil. FTIR: In the infrared spectrum of the soluble form the amide I′ band reached a maximum at 1650 cm−1. In the spectrum of the aggregated form this maximum was shifted at 1643 cm−1 and a shoulder around 1625 cm−1 was observed. | CR, ThT of HET-s (218–289). | The HET-s (218–289) peptide was soluble at pH 2.5 in 150 mM acetic acid, but, under non-denaturing conditions at pH 8.0, in a time course of a few hours, the peptide spontaneously formed aggregates. | [202,203,204,205] |
| Saccharomyces cerevisiae | URE2p | Regulation of nitrogen catabolism. | ~38 kDa. | β-strands, α-helix and RC. | Electron diffraction, X-ray diffraction and X-ray diffraction (PFD domain). | CD method of PFD domain: Switching from an initially disordered, RC structure, to a β-sheet enriched conformation. FTIR of PFD domain: A band at ∼1625 cm−1 dominates the spectrum (the presence of intermolecular β-sheet structure). | CR, ThT | Filaments were made by incubation of protein solutions (usually at about 1 mg/mL) on a shaker for 16 h at 4 °C. | [206,207] |
| Sup35p (Prion-inducing domain 2–114 and PFD domain) | Regulation of stop-codon read-through. | ~75 kDa. | A freshly prepared solution exhibits a far UV CD spectrum that indicates little α-helix or β-sheet content. | X-ray diffraction (PFD domain). | CD method (PFD domain): Switching from an initially disordered, RC structure, to a β-sheet enriched conformation. FTIR: A band at ∼1625 cm−1 dominates the spectrum. | CR | Filaments of Sup35pN (Prion-inducing domain 2–114) were prepared in 0.1% (vol/vol) TFA/40% (vol/vol) acetonitrile using reverse-phase HPLC fractions containing isocratically eluted Sup35pN. Preparation of a 100 μM solution of Sup35pN yielded filaments after 1 week of incubation at 4 °C. Spontaneous filament formation exists in 50 mM sodium phosphate buffer (pH 2.0) with 40% acetonitrile. | [207,208,209] | |
| Swi1p | Chromatin remodeling factor, prion form inactive. | ~140 kDa. | none | X-ray diffraction (PFD domain). | CD method (PFD domain): Switching from an initially disordered, RC structure, to a β-sheet enriched conformation. FTIR: A band at ∼1625 cm−1 dominates the spectrum. | none | none | [209,210] | |
| Mot3 | Transcriptional regulator of cell wall remodeling genes, prion form is inactive. | ~55 kDa. | none | X-ray diffraction (PFD domain). | CD method (PFD domain) Switching from an initially disordered, random coil structure, to a β-sheet enriched conformation. FTIR (PFD domain) A band at ∼1625 cm−1 dominates the spectrum. | CR, ThT. | none | [207,211] | |
| Most fungi | Hydrophobins | Fungal coat formation, modulation of adhesion and surface tension. | 7–9 kDa. | Previously RC and small core of antiparallel β-sheet. | X-ray. | CD method: β sheet is the predominant element of secondary structure in polymerized hydrophobin rodlets. | CR, ThT | For Hydrophobin SC3 Schizophyllum commune: Upon binding to a hydrophobic solid surface, the protein is arrested in an intermediate α-helical state, whereas, upon self-assembly at the air–water interface, rodlets are formed in a β-sheet conformation. | [176,212,213,214] |
| Animal | |||||||||
| Insects and fish | Chorion proteins (central domain of silkmoth chorion proteins of the A and B -family) | Structural and protective functions in the eggshell. | 34 and 24 kDa. | In both families of proteins β-sheet structure predominates. | X-ray diffraction | FTIR method ATR FT-IR supports the presence of uniform β-sheets in the structure of cA_m1 peptide fibrils; β-sheet structure also suggested by X-ray diffraction ATR FT-IR data: 64% antiparallel β-sheet and 30% β-turns in the central domain of silkmoth chorion proteins. | CR | cA peptide (central domain of the A class of silkmoth chorion proteins) was dissolved in a 50 mM sodium acetate buffer (pH 5) at a concentration of 9 mg/mL to produce amyloid-like fibrils after 3–4 weeks incubation. | [14,215,216,217,218] |
| Nephila clavipes Nephila edulis Araneus diadematus | Spidroins and Araneus diadematus fibroin | Structural (i.e., spider silk). | ~320 kDa (spidroin). | β-sheet or β-turn and RC. | X-Ray diffraction. | CD method: increasing of β-sheet structures. | CR, ThT | Lyophilized protein was dissolved in 6 M guanidinium thiocyanate at a concentration of 10 mg/mL−1 and dialyzed against 10 × 10−3 M potassium phosphate for several days at RT. For acceleration of fibril formation, 10 vol.-% methanol was added. | [15,177,219] |
| All mammalians including Homo sapiens | Non-glycosylated, 442-residue lumenal fragment of Pmel17 (rMα) | Pmel17 amyloid templates and accelerates the covalent polymerization of reactive small molecules into melanin. | 110 kDa (28-kDa transmembrane fragment (Mβ) and an 80-kDa lumenal fragment (Mα)). | β-strands, α-helix and RC | X-ray diffraction | CD and FTIR: Mα aggregates are approximately 11% α-helix, 32% β-sheet, 23% β-turn and 33% disordered, based on curve fitting with a basis set of 43 soluble proteins. | CR, ThT | rMα fibers were generated by diluting (from concentrated 8 M GdmCl, 50 mM KH2PO4/K2HPO 4 [pH 7.4], 100 mM KCl stock) rMα into 125 mM CH3COOH/ CH3COOK buffer (pH 5.0) at a final concentration of 10 μM and allowing it to stand at RT for 24 h. | [18,220] |
| Drosophila melanogaster | CPEEB (Orb2) | Memory consolidation Cytoplasmic polyadenylation element-binding protein regulates mRNA translation. | ~62 kDa | The protofilament core adopts a simple hairpin-like fold, composed of two β-strands, b1 (residues 176 to 186) and b2 (residues 197 to 206). | CryoEM | Only 31 residues (176–206) of the 704-residue protein form the amyloid core. N650 residues are dynamically disordered. | ThT | Recombinant Orb2A and Orb2A88 samples were exchanged into 10 mM HEPES, pH 7.6, 100 mM KCl, 1 M Urea and 1 mM DTT using dialysis and a PD-10 desalting column, respectively. Samples were then incubated on a shaker at RT for up to 2 weeks. | [221,222,223,224] |
| Plants | |||||||||
| Pisum sativum L. | Vicilin (Cupin-1.1 ((19–166 aa) and Cupin-1.2 (229–394 aa)) | Amyloid formation in charge of the accumulation of storage proteins in plant seeds. | ~50 kDa. | β-barrel domains. | X-Ray diffraction. | CD method Before aggregation Cupin-1.1 and Cupin-1.2 (4–12% β-content), Vicilin (39% β-content) After aggregation Cupin-1.1,Cupin-1.2 and Vicilin (40–42% β-content). | CR, ThT. | 1,1,1,3,3,3-Hexafluoro-2-propanol (HFIP) solvent for the proteins dissolution with its subsequent removal from the sample and incubation of dissolved proteins in the distilled water at 37 °C for 7 days for Vicilin, Cupin-1.1, Cupin-1.2 and 5 mM phosphate buffered saline (PBS) [pH 7.4]) for one day at 25 °C) for Cupin-1.2 | [8] |
| Synthetic amyloid aggregates | |||||||||
| Synthesized peptides (Homo sapiens and Mouse) | Vascin (Peptide based on an amyloidogenic sequence in the vascular endothelial growth factor receptor (VEGFR2) | Inhibited VEGFR2-dependent tumor growth. | 2272.15 Da. | Secondary structures are absent. | X-Ray diffraction. | FTIR method b-sheet structure change in b-structured conformation. | ThT. | 300 mM vascin in 1% (w/v) NH4CO3 after 24 h incubation at room temperature. | [225] |
| Dosidicus gigas (D. gigas) | Sucker ring teeth (SRT) from squid. SRT are assembled entirely from a protein family | Molecular design of biomimetic protein- and peptide-based thermoplastic structural biopolymers with potential biomedical and 3D printing applications. | none | Previously β-structure. | X-Ray diffraction. | FTIR method: the β-sheet-specific infrared band was centered at 1.235 cm−1 from RT up to 150 °C, at which point it shifted gradually to 1.220 cm−1, which was still within the β-sheet- region. | none | none | [226] |
| Synthesized peptides | Gonadotropin-releasing hormone analog (GnRH) | Use of amyloids in the formulation of long-acting drugs. Sorting, storage, and release of diverse hormones. | 1183.27 Da. | Secondary structures are absent. | none | none | CR, ThT. | GnRH analogs were dissolved in a glass tube in 1 mL of 5% D-mannitol and 0.01% sodium azide at a concentration of 1 mg/mL. The GnRH analogs were then incubated at RT without stirring. | [227] |
| Escherichia coli and Bacillus circus and Mytilus galloprovincialis | CsgA (as amyloidogenic cores) + chitin-binding domains (CBDs) + mussel foot proteins (Mfp3/Mfp5) two-domain and three-domain constructions with constant presence of CsgA | Development of multifunctional molecular materials with individual structure and characteristics based on amyloid. | CsgA ~17.5 kDa Mfp3 5–7.5 kDa Mfp5 9.5 CBD 6 kDa. | β-strands and RC. | X-Ray diffraction. | The two-domain proteins contained 60% of β-sheet/β-turn structures and 40% of RC, owing to the introduction of RC Mfps. Compared with their two-domain counterparts, the three-domain fibrils possess more β-sheet structures. | CR, ThT | Proteins were either dialyzed against PBS solutions (pH = 5.0 or 2.5) for 2 days or were incubated at 4 °C under acidic conditions for 3 days to promote the formation of amyloid fibers, followed by redissolving in hexafluoro-2-propanol (HFIP) solvent. | [228] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yakupova, E.I.; Bobyleva, L.G.; Shumeyko, S.A.; Vikhlyantsev, I.M.; Bobylev, A.G. Amyloids: The History of Toxicity and Functionality. Biology 2021, 10, 394. https://doi.org/10.3390/biology10050394
Yakupova EI, Bobyleva LG, Shumeyko SA, Vikhlyantsev IM, Bobylev AG. Amyloids: The History of Toxicity and Functionality. Biology. 2021; 10(5):394. https://doi.org/10.3390/biology10050394
Chicago/Turabian StyleYakupova, Elmira I., Liya G. Bobyleva, Sergey A. Shumeyko, Ivan M. Vikhlyantsev, and Alexander G. Bobylev. 2021. "Amyloids: The History of Toxicity and Functionality" Biology 10, no. 5: 394. https://doi.org/10.3390/biology10050394
APA StyleYakupova, E. I., Bobyleva, L. G., Shumeyko, S. A., Vikhlyantsev, I. M., & Bobylev, A. G. (2021). Amyloids: The History of Toxicity and Functionality. Biology, 10(5), 394. https://doi.org/10.3390/biology10050394