Proteomics-Based Identification of Salivary Changes in Patients with Burning Mouth Syndrome
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Subjects
2.2. Sample Description
2.3. Saliva Collection
2.4. Two-Dimensional Electrophoresis, Computational Image, and Mass Tandem Spectrometry
2.5. Statistical and Bioinformatics Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Headache Classification Committee of the International Headache Society (HIS). III edición de la Clasificación internacional de las cefaleas. Cephalalgia 2018, 38, 1–211. [Google Scholar]
- Klein, B.; Thoppay, J.R.; Rossi, S.S.D.; Ciarrocca, K. Burning mouth syndrome. Dermatol. Clin. 2020, 38, 477–483. [Google Scholar] [CrossRef] [PubMed]
- Ariyawardana, A.; Chmieliauskaite, M.; Farag, A.M.; Albuquerque, R.; Forssell, H.; Nasri-Heir, C.; Klasser, G.D.; Sardella, A.; Mignogna, M.D.; Ingram, M.; et al. World Workshop on Oral Medicine VII: Burning mouth syndrome: A Systematic Review of Disease Definitions and Diagnostic Criteria Utilized in Randomized Clinical Trials. Oral Dis. 2019, 25, 141–156. [Google Scholar] [CrossRef]
- Ritchie, A.; Kramer, J.M. Recent advances in the etiology and treatment of burning mouth syndrome. J. Dent. Res. 2018, 97, 1193–1199. [Google Scholar] [CrossRef] [PubMed]
- Imura, H.; Shimada, M.; Yamazaki, Y.; Sugimoto, K. Characteristic changes of saliva and taste in burning mouth syndrome patients. J. Oral Pathol. Med. 2016, 45, 231–236. [Google Scholar] [CrossRef]
- Hershkovich, O.; Nagler, R.M. Biochemical analysis of saliva and taste acuity evaluation in patients with burning mouth syndrome, xerostomia and/or gustatory disturbances. Arch. Oral Biol. 2004, 49, 515–522. [Google Scholar] [CrossRef] [PubMed]
- Nasri-Heir, C.; Zagury, J.G.; Thomas, D.; Ananthan, S. Burning mouth syndrome: Current concepts. J. Indian Prosthodont. Soc. 2015, 15, 300–307. [Google Scholar] [CrossRef]
- Dym, H.; Lin, S.; Thakkar, J. Neuropathic pain and burning mouth syndrome: An overview and current update. Dent. Clin. 2020, 64, 379–399. [Google Scholar]
- Sardella, A.; Lodi, G.; Demarosi, F.; Bez, C.; Cassano, S.; Carrassi, A. Burning mouth syndrome: A retrospective study investigating spontaneous remission and response to treatments. Oral Dis. 2006, 12, 152–155. [Google Scholar] [CrossRef]
- Roi, A.; Rusu, L.C.; Roi, C.I.; Luca, R.E.; Boia, S.; Munteanu, R.I. A new approach for the diagnosis of systemic and oral diseases based on salivary biomolecules. Dis. Markers 2019, 2019, 8761860. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Jornet, P.; Castillo Felipe, C.; Pardo-Marin, L.; Ceron, J.J.; Pons-Fuster, E.; Tvarijonaviciute, A. Salivary biomarkers and their correlation with pain and stress in patients with burning mouth syndrome. J. Clin. Med. 2020, 9, 929. [Google Scholar] [CrossRef]
- Yoshizawa, J.M.; Schafer, C.A.; Schafer, J.J.; Farrell, J.J.; Paster, B.J.; Wong, D.T.W. Salivary biomarkers: Toward future clinical and diagnostic utilities. Clin. Microbiol. Rev. 2013, 26, 781–791. [Google Scholar] [CrossRef]
- Melguizo-Rodríguez, L.; Costela-Ruiz, V.J.; Manzano-Moreno, F.J.; Ruiz, C.; Illescas-Montes, R. Salivary biomarkers and their application in the diagnosis and monitoring of the most common oral pathologies. Int. J. Mol. Sci. 2020, 21, 5173. [Google Scholar] [CrossRef] [PubMed]
- Ji, E.H.; Diep, C.; Liu, T.; Li, H.; Merrill, R.; Messadi, D.; Hu, S. Potential protein biomarkers for burning mouth syndrome discovered by quantitative proteomics. Mol. Pain 2017, 13, 1744806916686796. [Google Scholar] [CrossRef] [PubMed]
- Cabras, T.; Manconi, B.; Castagnola, M.; Sanna, M.T.; Arba, M.; Acharya, S.; Ekström, J.; Carlén, A.; Messana, I. Proteomics of the acid-soluble fraction of whole and major gland saliva in burning mouth syndrome patients. Arch. Oral Biol. 2019, 98, 148–155. [Google Scholar] [CrossRef] [PubMed]
- Krief, G.; Haviv, Y.; Deutsch, O.; Keshet, N.; Almoznino, G.; Zacks, B.; Palmon, A.; Aframian, D.J. Proteomic profiling of whole-saliva reveals correlation between burning mouth syndrome and the neurotrophin signaling pathway. Sci. Rep. 2019, 9, 1–9. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Carreira, L.; Castelo, P.M.; Simões, C.; Capela E Silva, F.; Viegas, C.; Lamy, E. Changes in salivary proteome in response to bread odour. Nutrients 2020, 12, 1002. [Google Scholar] [CrossRef]
- Franco-Martínez, L.; Martínez-Subiela, S.; Escribano, D.; Schlosser, S.; Nöbauer, K.; Razzazi-Fazeli, E.; Romero, D.; Cerón, J.J.; Tvarijonaviciute, A. Alterations in haemolymph proteome of Mytilus galloprovincialis mussel after an induced injury. Fish Shellfish Immunol. 2018, 75, 41–47. [Google Scholar] [CrossRef]
- Jiménez, C.R.; Huang, L.; Qiu, Y.; Burlingame, A.L. In-gel digestion of proteins for MALDI-MS fingerprint mapping. Curr. Protoc. Protein Sci. 2001, 14, 16.4.1–16.4.5. [Google Scholar] [CrossRef]
- Gharahdaghi, F.; Weinberg, C.R.; Meagher, D.A.; Imai, B.S.; Mische, S.M. Mass spectrometric identification of proteins from silver-stained polyacrylamide gel: A method for the removal of silver ions to enhance sensitivity. Electrophoresis 1999, 20, 601–605. [Google Scholar] [CrossRef]
- Cox, S.W.; Rodriguez-Gonzalez, E.M.; Booth, V.; Eley, B.M. Secretory leukocyte protease inhibitor and its potential interactions with elastase and cathepsin B in gingival crevicular fluid and saliva from patients with chronic periodontitis. J. Periodontal Res. 2006, 41, 477–485. [Google Scholar] [CrossRef]
- Chan, H.H.; Rahim, Z.H.A.; Jessie, K.; Hashim, O.H.; Taiyeb-Ali, T.B. Salivary proteins associated with periodontitis in patients with type 2 diabetes mellitus. Int. J. Mol. Sci. 2012, 13, 4642–4654. [Google Scholar] [CrossRef] [PubMed]
- Ngamchuea, K.; Chaisiwamongkhol, K.; Batchelor-McAuley, C.; Compton, R.G. Chemical analysis in saliva and the search for salivary biomarkers—A tutorial review. Analyst 2017, 143, 81–99. [Google Scholar] [CrossRef]
- Nagler, R.M.; Hershkovicht, O. Sialochemical and gustatory analysis in patients with oral sensory complaints. J. Pain 2004, 5, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Hirtz, C.; Chevalier, F.; Centeno, D.; Rofidal, V.; Egea, J.C.; Rossignol, M.; Sommerer, N.; Périère, D.D.D. MS characterization of multiple forms of alpha-amylase in human saliva. Proteomics 2005, 5, 4597–4607. [Google Scholar] [CrossRef] [PubMed]
- Henskens, Y.M.C.; Van der Velden, U.; Veerman, E.C.I.; Amerongen, A.V.N. Protein, albumin and cystatin concentrations in saliva of healthy subjects and of patients with gingivitis periodonitis. J. Periodontal Res. 1993, 28, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Koh, D.; Ng, V.; Lee, C.Y.; Chan, G.; Dong, F.; Goh, S.H.; Anantharaman, V.; Chia, S.E. Self perceived work related stress and the relation with salivary IgA and lysozyme among emergency department nurses. Occup. Environ. Med. 2002, 59, 836–841. [Google Scholar] [CrossRef]
- Granot, M.; Nagler, R.M. Association between regional idiopathic neuropathy and salivary involvement as the possible mechanism for oral sensory complaints. J. Pain 2005, 6, 581–587. [Google Scholar] [CrossRef]
- Lauritano, D.; Spadari, F.; Formaglio, F.; Artini, M.Z.; Salvato, A. Etiopathogenic, clinical-diagnostic and therapeutic aspects of the burning mouth syndrome. Research and treatment protocols in a patient group. Minerva Stomatol. 1998, 47, 239–251. [Google Scholar]
- Chanakankun, R.; Proungvitaya, T.; Chua-On, D.; Limpaiboon, T.; Roytrakul, S.; Jusakul, A.; Titapun, A.; Jarearnrat, A.; Proungvitaya, S. Serum coiled-coil domain containing 25 protein as a potential screening/diagnostic biomarker for cholangiocarcinoma. Oncol. Lett. 2020, 19, 930–942. [Google Scholar] [CrossRef] [PubMed]
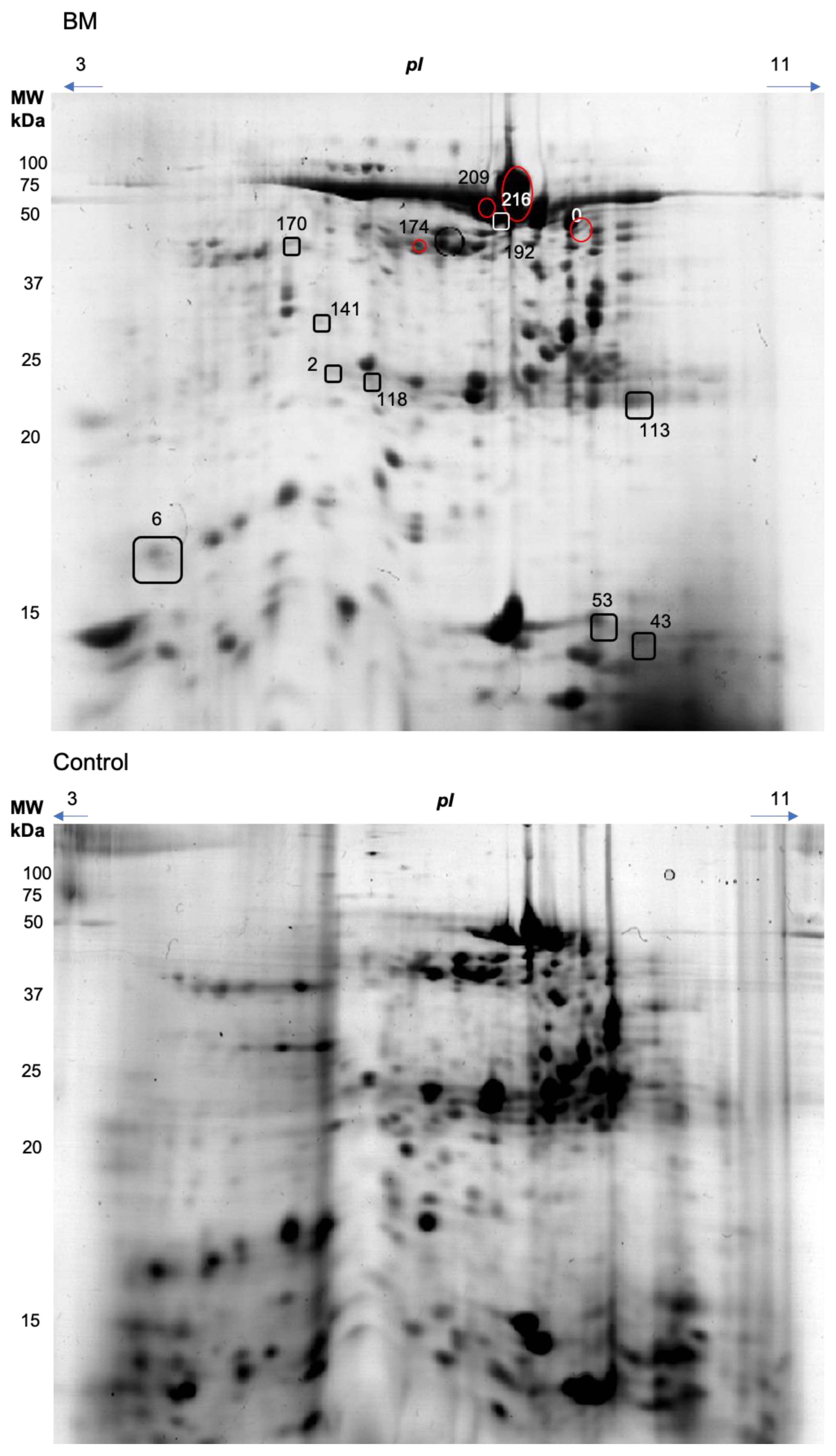
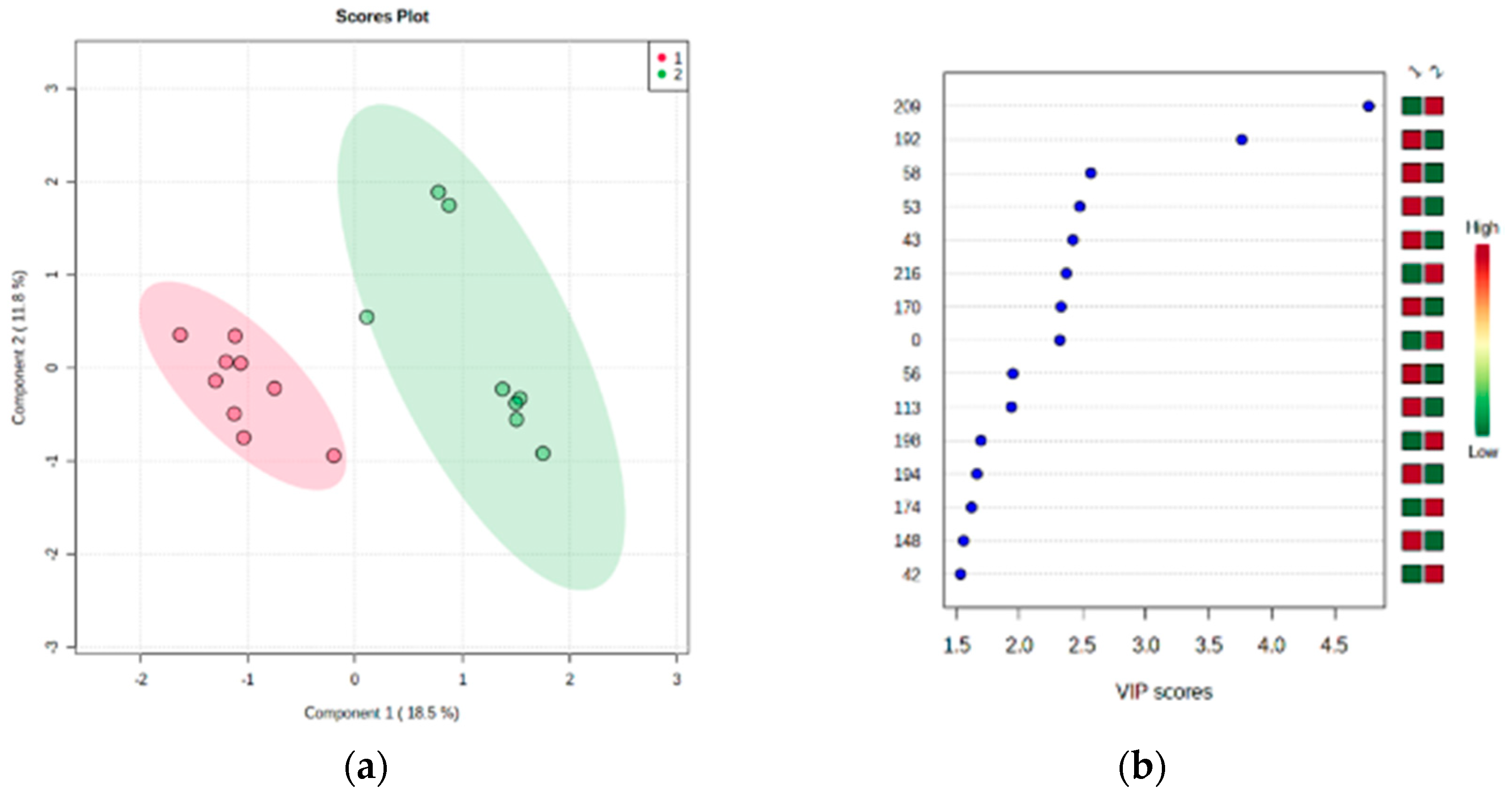
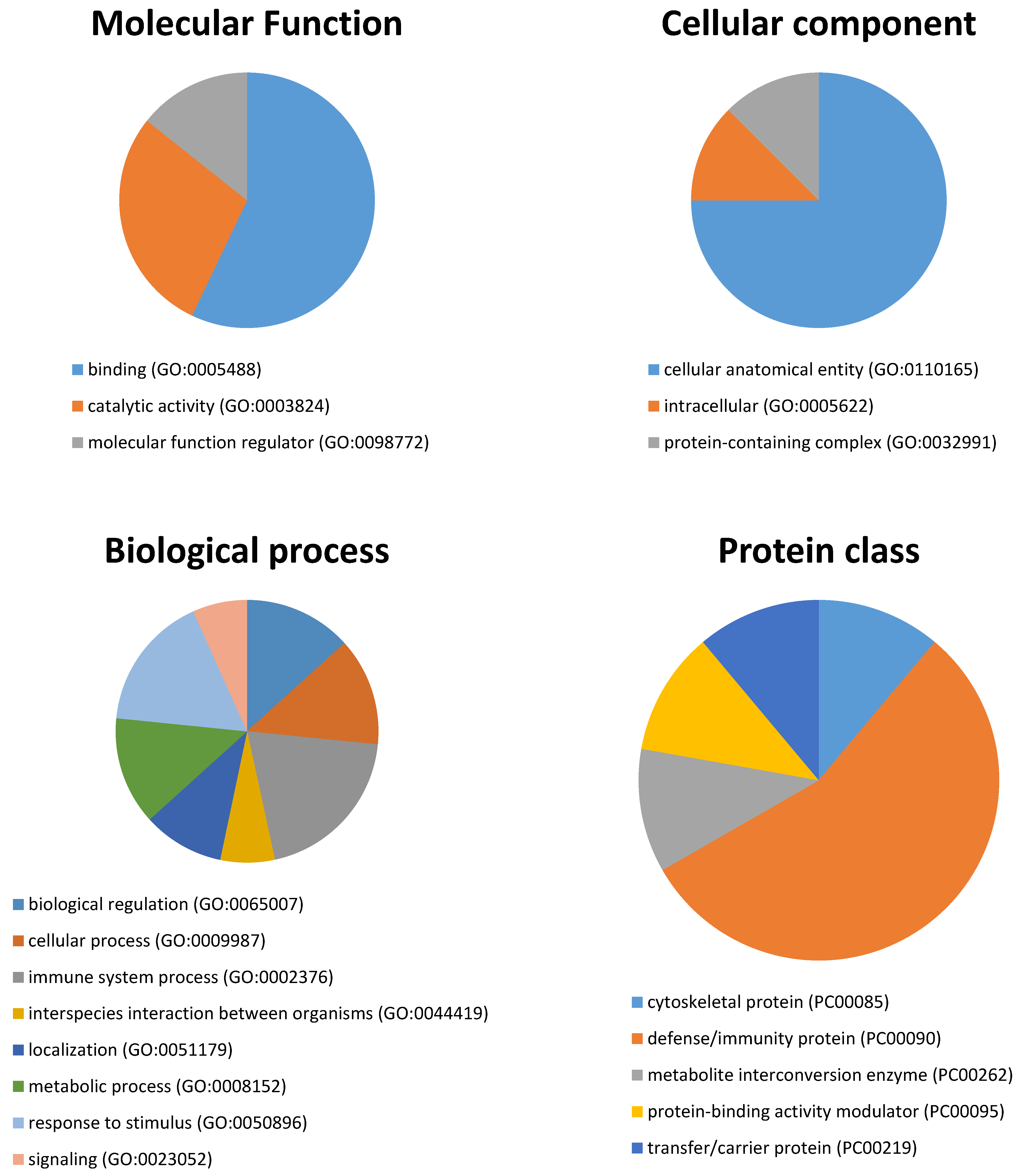
| Spot | Spectra | Peptides | Score | Coverage | Intensity | Protein mw | Accession Number | Entry Name | Control | Burning Mouth Syndrome | p |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 58 | 5 | 4 | 34.68 | 4.9 | 2.22 × 104 | 69,365.5 | P02768 | Albumin | 0.684 (0426–0.684) | 0.086 (0.029–0.421) | 0.001 |
| 5 | 3 | 31.59 | 17.8 | 6.65 × 104 | 16,572.2 | P12273 | Prolactin-inducible protein | ||||
| 5 | 3 | 30.68 | 25.4 | 2.44 × 104 | 11,283.7 | P81605 | Dermcidin | ||||
| 2 | 1 | 14.54 | 4.3 | 2.43 × 104 | 24,478.4 | Q86WR0 | Coiled-coil domain-containing protein 25 | ||||
| 118 | 12 | 7 | 84.61 | 12.9 | 1.35 × 105 | 57,766.8 | P04745 | Alpha-amylase 1 | 0.207 (0.074–0.207) | 0.005 (0.005–0.056) | 0.023 |
| 6 | 3 | 32.42 | 47.6 | 6.03 × 104 | 11,764.8 | P01834 | Immunoglobulin kappa constant | ||||
| 192 | 81 | 24 | 362.45 | 51.6 | 3.20 × 106 | 57,766.8 | P04745 | Alpha-amylase 1 | 1.038 (0.595–1.285) | 0.121 (0.326–1.090) | 0.002 |
| 5 | 3 | 36.75 | 8.7 | 4.16 × 104 | 37,654 | P01876 | Immunoglobulin heavy constant alpha 1 | ||||
| 2 | 1 | 11.96 | 4.3 | 2.33 × 104 | 24,478.4 | Q86WR0 | Coiled-coil domain-containing protein 25 | ||||
| 2 | 4 | 3 | 28.1 | 3.4 | 2.45 × 104 | 69,365.5 | P02768 | Albumin | 0.118 (0.052–0.198) | 0.007 (0.007–0.051) | 0.004 |
| 1 | 1 | 15.97 | 4.3 | 9.20 × 103 | 24,478.4 | Q86WR0 | Coiled-coil domain-containing protein 25 | ||||
| 141 | 7 | 4 | 49.52 | 24.5 | 5.03 × 104 | 11,283.7 | P81605 | Dermcidin | 0.116 (0.070–0.132) | 0.007 (0.007–0.028) | 0.0001 |
| 1 | 1 | 11.92 | 4.3 | 2.11 × 104 | 24,478.4 | Q86WR0 | Coiled-coil domain-containing protein 25 | ||||
| 2 | 1 | 10.93 | 0.4 | 3.13 × 104 | 21,6853.7 | Q8TF72 | Protein Shroom3 | ||||
| 170 | 9 | 9 | 97.49 | 24.2 | 5.68 × 104 | 42,741 | P30740 | Leukocyte elastase inhibitor | 0.476 (0.157–0.600) | 0.012 (0.012–0.047) | 0.0004 |
| 43 | 7 | 5 | 49.82 | 34.5 | 6.48 × 104 | 11,283.7 | P81605 | Dermcidin | 0.330 (0.266–0.658) | 0.013 (0.013–0.053) | 0.0004 |
| 6 | 3 | 34.96 | 33.3 | 9.31 × 104 | 16,387.4 | P01037 | Cystatin-SN | ||||
| 209 | 19 | 13 | 147.03 | 25.8 | 2.05 × 105 | 57,766.8 | P04745 | Alpha-amylase 1 | 0.049 (0.049–0.147) | 1.586 (0.841–3.629) | 0.001 |
| 4 | 3 | 36.45 | 7.9 | 2.69 × 104 | 37,654 | P01876 | Immunoglobulin heavy constant alpha 1 | ||||
| 2 | 2 | 16.14 | 15.4 | 1.08 × 104 | 11,283.7 | P81605 | Dermcidin | ||||
| 174 | 8 | 6 | 79.02 | 11.3 | 9.81 × 104 | 57,766.8 | P04745 | Alpha-amylase 1 | 0.011 (0.011–0.147) | 0.206 (0.159–0.427) | 0.00005 |
| 7 | 4 | 56.48 | 9.9 | 7.86 × 104 | 37,654 | P01876 | Immunoglobulin heavy constant alpha 1 | ||||
| 2 | 1 | 12.29 | 4.3 | 1.16 × 104 | 24,478.4 | Q86WR0 | Coiled-coil domain-containing protein 25 | ||||
| 0 | 56 | 22 | 298.52 | 51.2 | 2.01 × 106 | 57,766.8 | P04745 | Alpha-amylase 1 | 0.027 (0.027–0.027) | 0.382 (0.142–0.539) | 0.049 |
| 113 | 12 | 4 | 55.29 | 60.7 | 2.95 × 105 | 11,764.8 | P01834 | Immunoglobulin kappa constant | 0.512 (0.024–0.890) | 0.024 (0.024–0.094) | 0.030 |
| 2 | 2 | 22.19 | 13.4 | 1.74 × 104 | 13,147.6 | A0A0C4DH55 | Immunoglobulin kappa variable 3D-7 | ||||
| 2 | 2 | 18.76 | 13.9 | 1.32 × 104 | 12,625 | A0A0A0MRZ8 | Immunoglobulin kappa variable 3D-11 | ||||
| 3 | 2 | 15.57 | 13.7 | 2.04 × 104 | 12,556.9 | P01619 | Immunoglobulin kappa variable 3-20 | ||||
| 216 | 143 | 27 | 468.85 | 66.9 | 2.37 × 107 | 57,766.8 | P04745 | Alpha-amylase 1 | 0.013 (0.013–0.013) | 0.105 (0.025–0.146) | 0.016 |
| 53 | 27 | 7 | 91.17 | 63.1 | 1.16 × 106 | 16,387.4 | P01037 | Cystatin-SN | 1.914 (1.351–3.299) | 0.709 (0.211–1.823) | 0.035 |
| 18 | 4 | 50.6 | 34 | 1.05 × 106 | 16,214.1 | P01036 | Cystatin-S |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Castillo-Felipe, C.; Franco-Martínez, L.; Tvarijonaviciute, A.; Lopez-Jornet, P.; Lamy, E. Proteomics-Based Identification of Salivary Changes in Patients with Burning Mouth Syndrome. Biology 2021, 10, 392. https://doi.org/10.3390/biology10050392
Castillo-Felipe C, Franco-Martínez L, Tvarijonaviciute A, Lopez-Jornet P, Lamy E. Proteomics-Based Identification of Salivary Changes in Patients with Burning Mouth Syndrome. Biology. 2021; 10(5):392. https://doi.org/10.3390/biology10050392
Chicago/Turabian StyleCastillo-Felipe, Candela, Lorena Franco-Martínez, Asta Tvarijonaviciute, Pia Lopez-Jornet, and Elsa Lamy. 2021. "Proteomics-Based Identification of Salivary Changes in Patients with Burning Mouth Syndrome" Biology 10, no. 5: 392. https://doi.org/10.3390/biology10050392
APA StyleCastillo-Felipe, C., Franco-Martínez, L., Tvarijonaviciute, A., Lopez-Jornet, P., & Lamy, E. (2021). Proteomics-Based Identification of Salivary Changes in Patients with Burning Mouth Syndrome. Biology, 10(5), 392. https://doi.org/10.3390/biology10050392







