Comprehensive Transcriptome Analysis of mRNA Expression Patterns of Early Embryo Development in Goat under Hypoxic and Normoxic Conditions
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Collection and Culture of Embryos
2.2. RNA Library Construction and Sequencing
2.3. Read Alignment and Differential Expression Analysis
2.4. Functional Annotation of Differentially Expressed Genes
2.5. Whole Transcriptome Amplification of Single-Cell Embryo and Quantitative Real-Time PCR
2.6. Cell Staining
2.7. Statistical and Data Analysis
3. Results
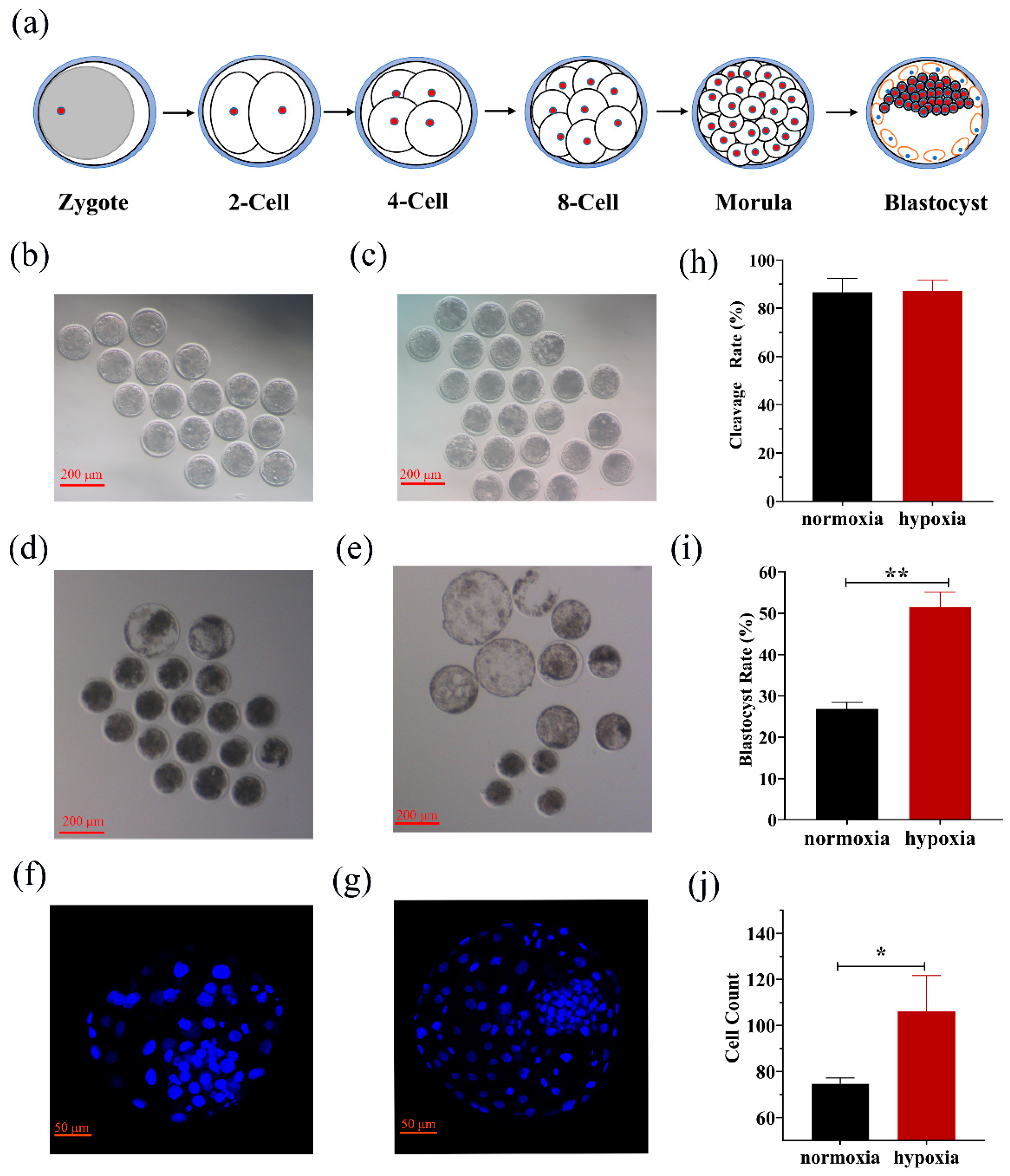
3.1. Effects of Oxygen Concentration on Embryonic Growth and Development
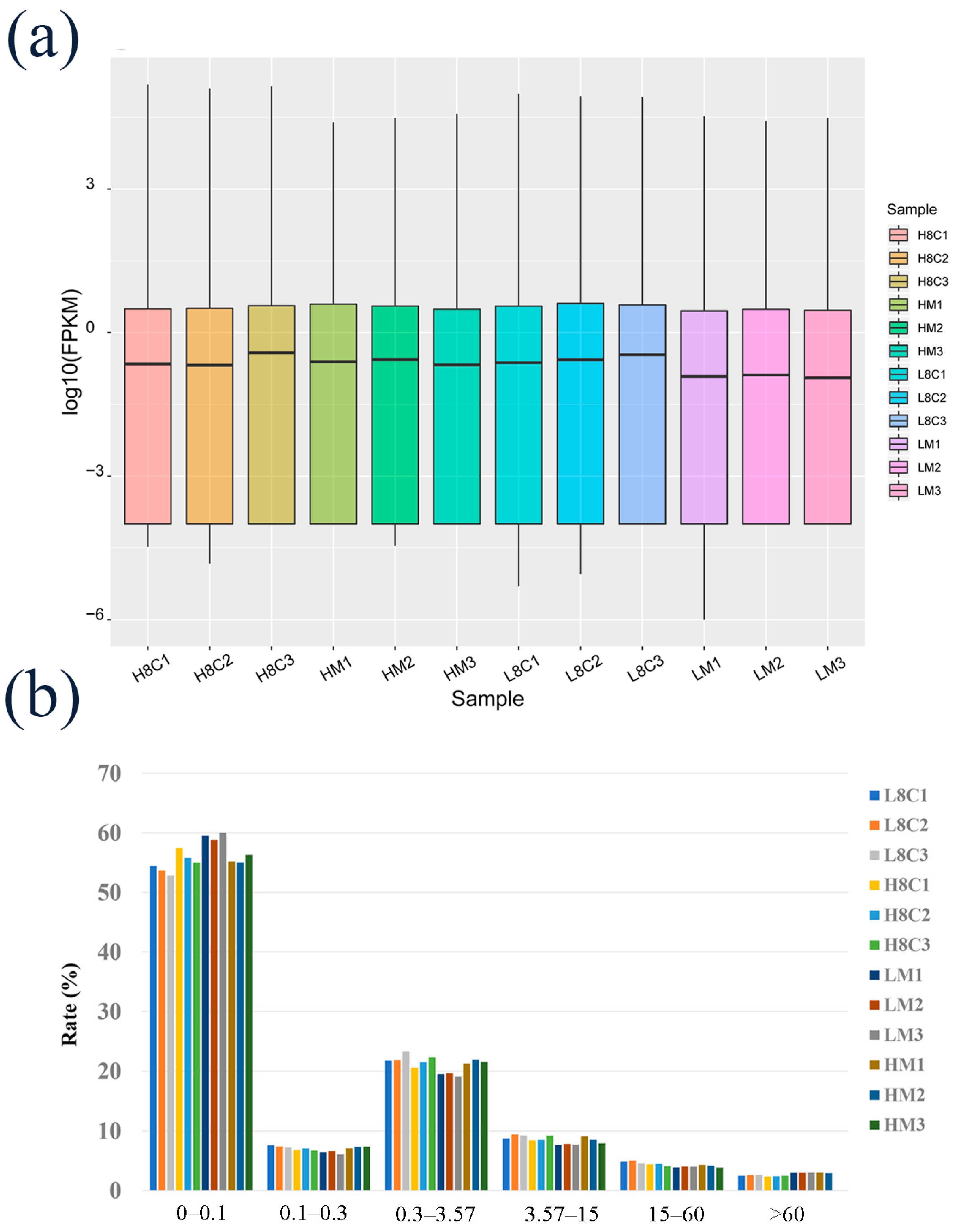
3.2. Summary of Transcriptomic Profiles of Goat Embryos
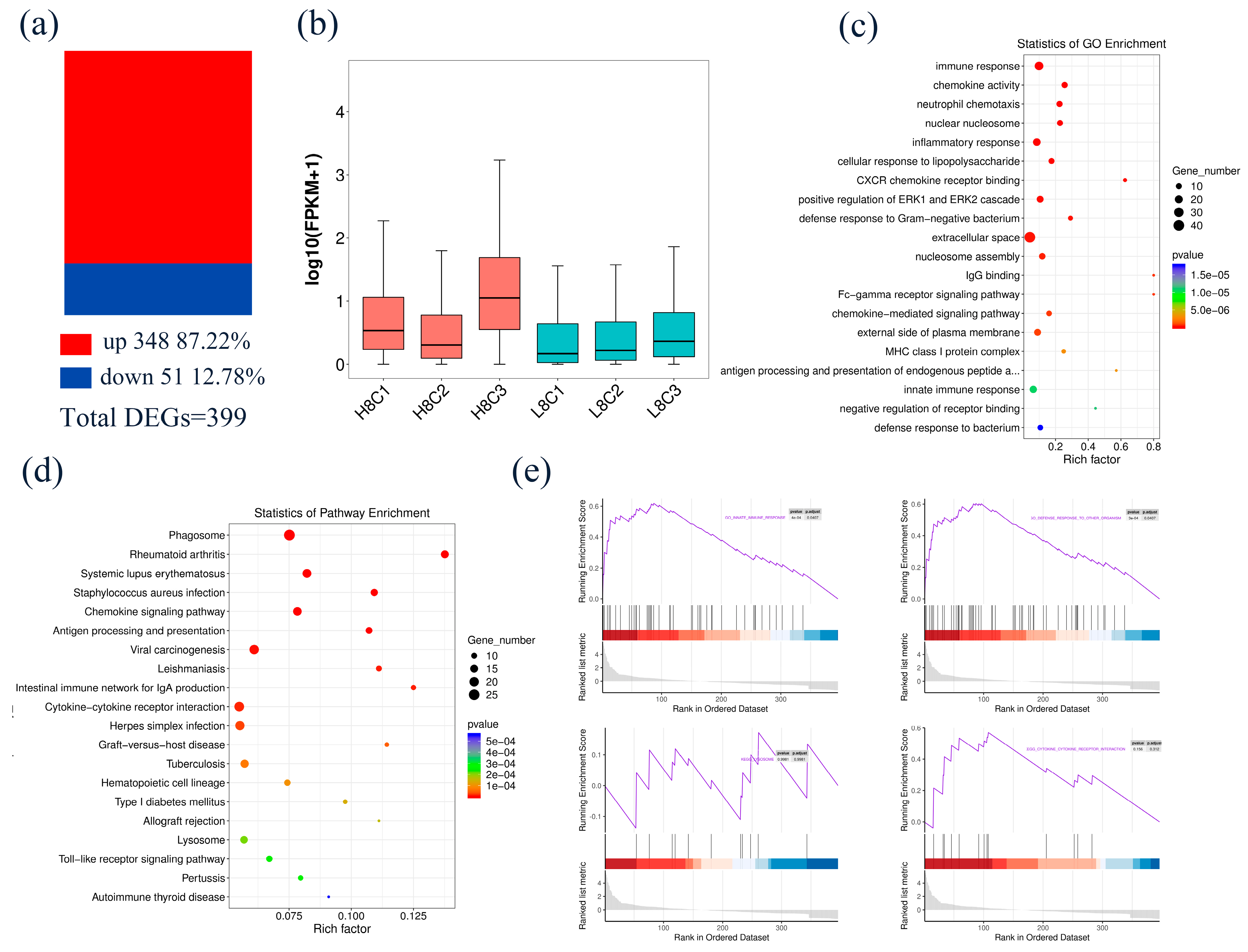
3.3. Identification and Functional Analysis of DEGs in 8-Cell-Stage Goat Embryo
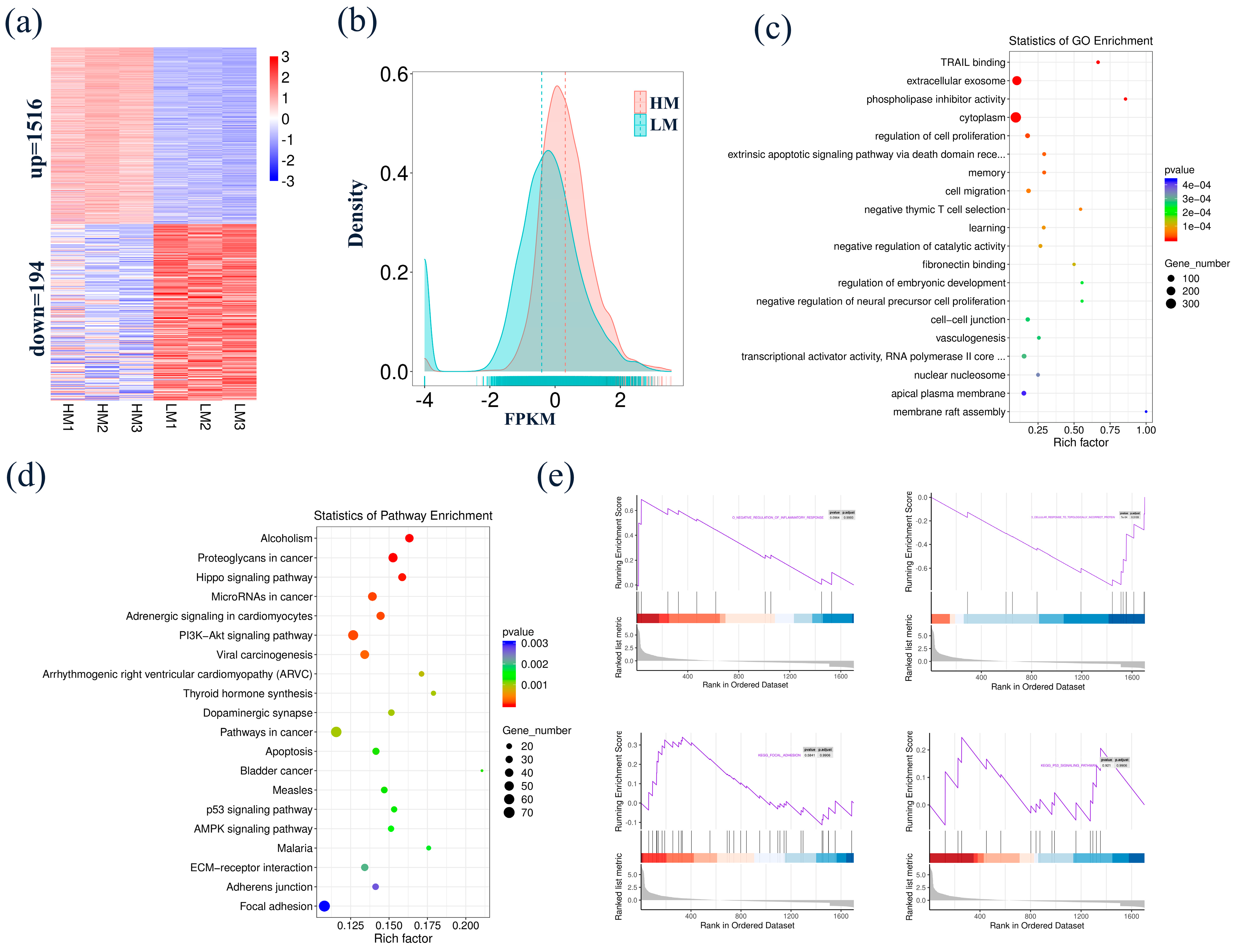
3.4. Identification and Functional Analysis of DEGs in Blastocyst-Stage Goat Embryo
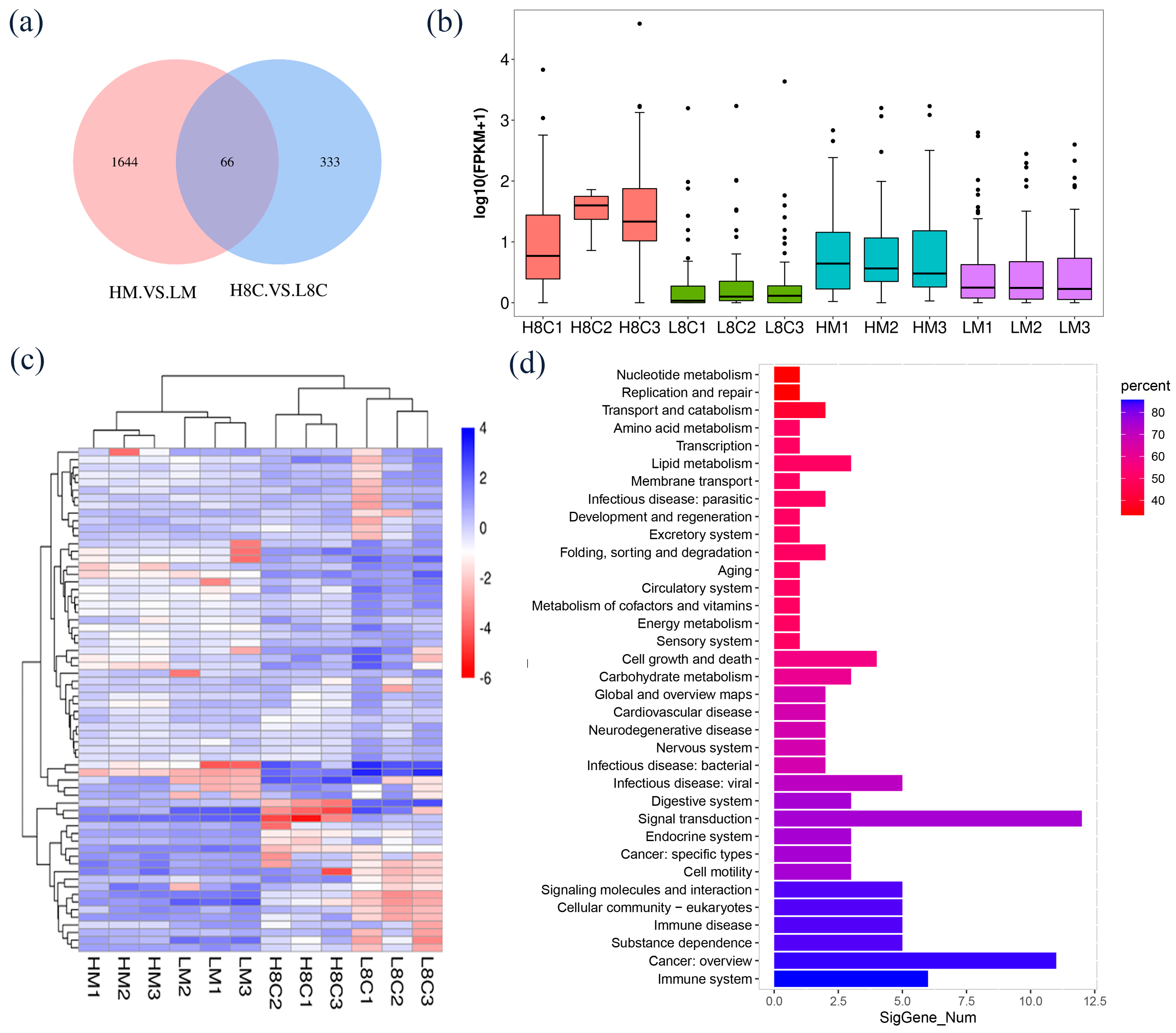
3.5. Identification and Functional Analysis of Common DEGs between H8C and L8C Groups and HM and LM Groups
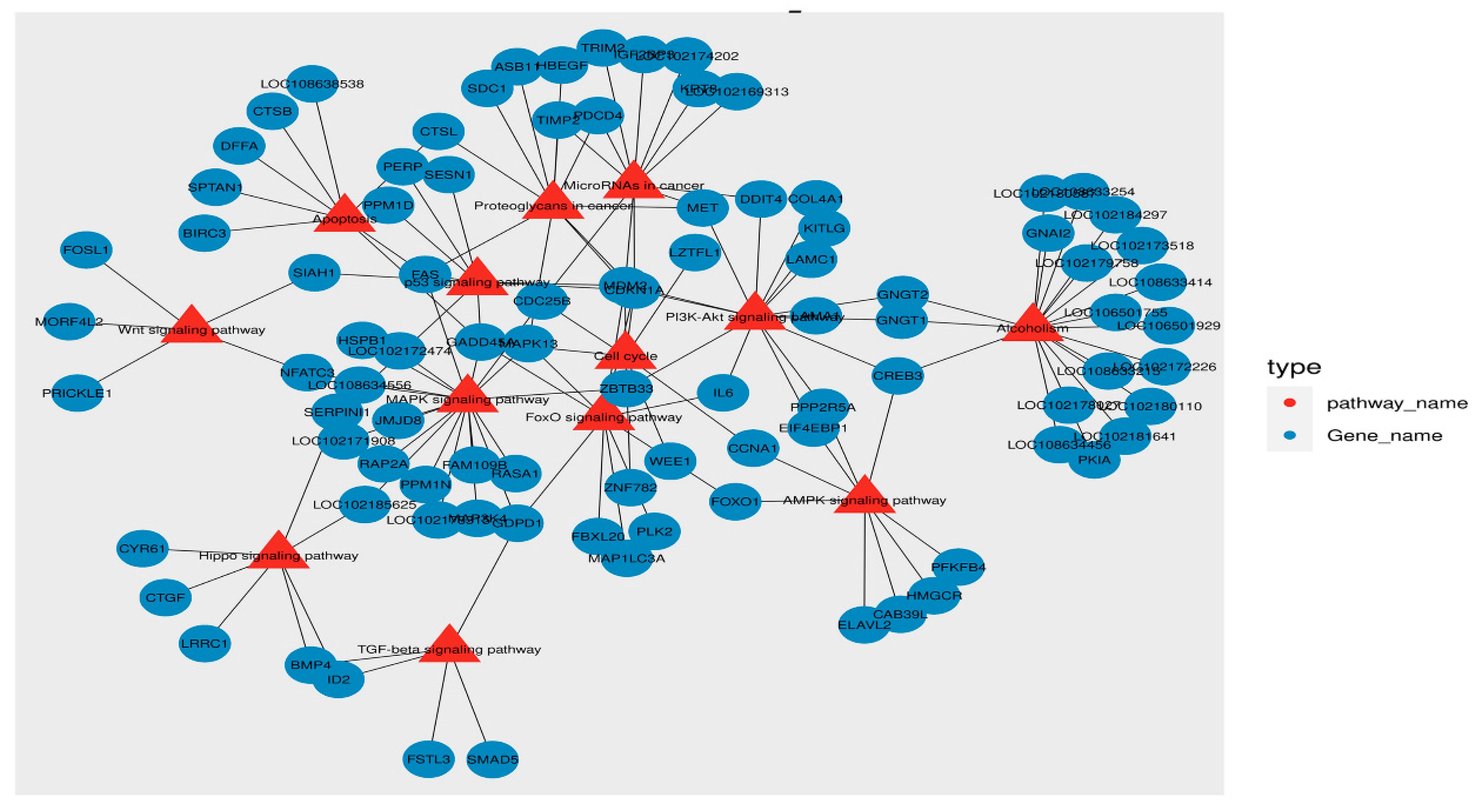
3.6. Analysis of the Protein–Protein Interaction (PPI) and Pathway–Gene Interaction Networks of DEGs
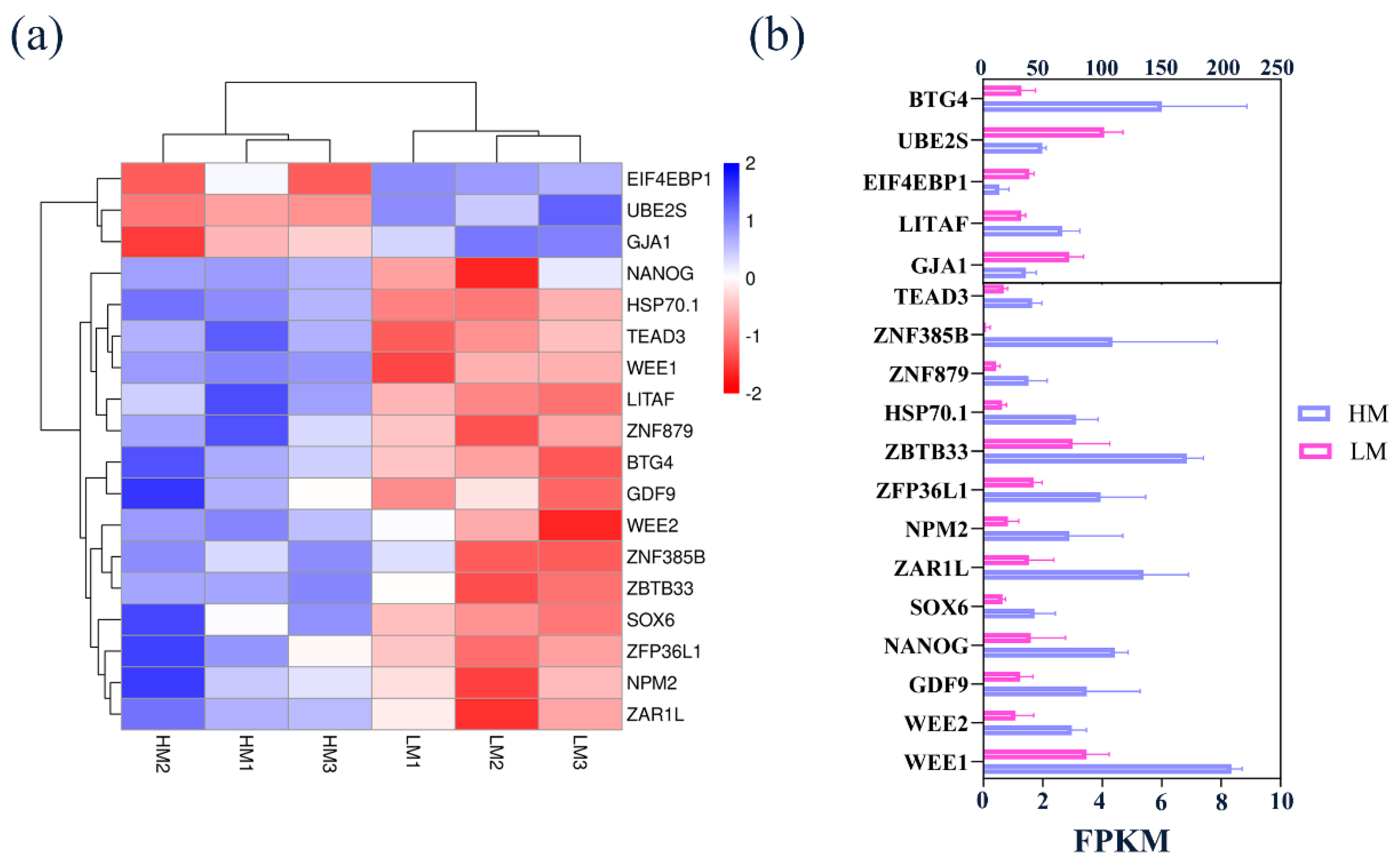
3.7. Key Genes Associated with Oxidative Stress Affecting Embryonic Development and Validation of Sequencing Data
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Amoah, E.A.; Gelaye, S. Biotechnological advances in goat reproduction. J. Anim. Sci. 1997, 75, 578–585. [Google Scholar] [CrossRef]
- Kussano, N.R.; Leme, L.O.; Guimarães, A.L.; Franco, M.M.; Dode, M.A. Molecular markers for oocyte competence in bovine cumulus cells. Theriogenology 2016, 85, 1167–1176. [Google Scholar] [CrossRef]
- Maj, T.; Wang, W.; Crespo, J.; Zhang, H.; Wang, W.; Wei, S.; Zhao, L.; Vatan, L.; Shao, I.; Szeliga, W.; et al. Oxidative stress controls regulatory T cell apoptosis and suppressor activity and PD-L1-blockade resistance in tumor. Nat. Immunol. 2017, 18, 1332–1341. [Google Scholar] [CrossRef]
- Kesavardhana, S.; Kanneganti, T.D. Stressed-out ROS take a silent death route. Nat. Immunol. 2018, 19, 103–105. [Google Scholar] [CrossRef] [PubMed]
- Sies, H. Oxidative stress: A concept in redox biology and medicine. Redox Biol. 2015, 4, 180–183. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, M. Oxidative stress and redox regulation on in vitro development of mammalian embryos. J. Reprod. Dev. 2012, 58, 1–9. [Google Scholar] [CrossRef]
- Fabian, D.; Koppel, J.; Maddox-Hyttel, P. Apoptotic processes during mammalian preimplantation development. Theriogenology 2005, 64, 221–231. [Google Scholar] [CrossRef] [PubMed]
- Jurisicova, A.; Antenos, M.; Varmuza, S.; Tilly, J.L.; Casper, R.F. Expression of apoptosis-related genes during human preimplantation embryo development: Potential roles for the Harakiri gene product and Caspase-3 in blastomere fragmentation. Mol. Hum. Reprod. 2003, 9, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Jurisicova, A.; Latham, K.E.; Casper, R.F.; Casper, R.F.; Varmuza, S.L. Expression and regulation of genes associated with cell death during murine preimplantation embryo development. Mol. Reprod. Dev. 1998, 51, 243–253. [Google Scholar] [CrossRef]
- Byrne, A.T.; Southgate, J.; Brison, D.R.; Leese, H.J. Analysis of apoptosis in the preimplantation bovine embryo using TUNEL. J. Reprod. Fertil. 1999, 117, 97–105. [Google Scholar] [CrossRef]
- Rinaudo, P.F.; Giritharan, G.; Talbi, S.; Dobson, A.T.; Schultz, R.M. Effects of oxygen tension on gene expression in preimplantation mouse embryos. Fertil. Steril. 2006, 86, 1252–1265. [Google Scholar] [CrossRef]
- Alahmar, A.T. Role of Oxidative Stress in Male Infertility: An Updated Review. J. Hum. Reprod. Sci. 2019, 12, 4–18. [Google Scholar] [CrossRef]
- Thompson, J.G.; Partridge, R.J.; Houghton, F.D.; Cox, C.I.; Leese, H.J. Oxygen uptake and carbohydrate metabolism by in vitro derived bovine embryos. J. Reprod. Fertil. 1996, 106, 299–306. [Google Scholar] [CrossRef]
- Fischer, B.; Bavister, B.D. Oxygen tension in the oviduct and uterus of rhesus monkeys, hamsters and rabbits. J. Reprod. Fertil. 1993, 99, 673–679. [Google Scholar] [CrossRef]
- Kasterstein, E.; Strassburger, D.; Komarovsky, D.; Bern, O.; Komsky, A.; Raziel, A.; Friedler, S.; Ron-El, R. The effect of two distinct levels of oxygen concentration on embryo development in a sibling oocyte study. J. Assist. Reprod. Genet. 2013, 30, 1073–1079. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.W.; Hwang, K.J.; Kwon, H.C.; Kim, H.S.; Choi, K.W.; Oh, K.S. Detection of reactive oxygen species (ROS) and apoptosis in human fragmented embryos. Hum. Reprod. 1998, 13, 998–1002. [Google Scholar] [CrossRef] [PubMed]
- Van Soom, A.; Yuan, Y.Q.; Peelman, L.J.; de Matos, D.G.; Dewulf, J.; Laevens, H.; de Kruif, A. Prevalence of apoptosis and inner cell allocation in bovine embryos cultured under different oxygen tensions with or without cysteine addition. Theriogenology 2002, 57, 1453–1465. [Google Scholar] [CrossRef]
- Karagenc, L.; Sertkaya, Z.; Ciray, N.; Ulug, U.; Bahceci, M. Impact of oxygen concentration on embryonic development of mouse zygotes. Reprod. Biomed. Online 2004, 9, 409–417. [Google Scholar] [CrossRef]
- Kitagawa, Y.; Suzuki, K.; Yoneda, A.; Watanabe, T. Effects of oxygen concentration and antioxidants on the in vitro developmental ability, production of reactive oxygen species (ROS), and DNA fragmentation in porcine embryos. Theriogenology 2004, 62, 1186–1197. [Google Scholar] [CrossRef]
- Lee, S.C.; Seo, H.C.; Lee, J.; Jun, J.H.; Choi, K.W. Effects of dynamic oxygen concentrations on the development of mouse pre- and peri-implantation embryos using a double-channel gas supply incubator system. Clin. Exp. Reprod. Med. 2019, 46, 189–196. [Google Scholar] [CrossRef] [PubMed]
- Deng, M.; Liu, Z.; Ren, C.; Zhang, G.; Pang, J.; Zhang, Y.; Wang, F.; Wan, Y. Long noncoding RNAs exchange during zygotic genome activation in goat. Biol. Reprod. 2018, 99, 707–717. [Google Scholar] [CrossRef] [PubMed]
- Picelli, S.; Faridani, O.R.; Björklund, A.K.; Winberg, G.; Sagasser, S.; Sandberg, R. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 2014, 9, 171–181. [Google Scholar] [CrossRef] [PubMed]
- Deng, M.; Chen, B.; Liu, Z.; Cai, Y.; Wan, Y.; Zhang, G.; Fan, Y.; Zhang, Y.; Wang, F. YTHDF2 Regulates Maternal Transcriptome Degradation and Embryo Development in Goat. Front. Cell Dev. Biol. 2020, 8, 580367. [Google Scholar] [CrossRef] [PubMed]
- Wan, Y.; Deng, M.; Zhang, G.; Ren, C.; Zhang, H.; Zhang, Y.; Wang, L.; Wang, F. Abnormal expression of DNA methyltransferases and genomic imprinting in cloned goat fibroblasts. Cell. Biol. Int. 2016, 40, 74–82. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Zhang, G.; Deng, M.; Yang, H.; Pang, J.; Cai, Y.; Wan, Y.; Wang, F. Inhibition of lysine-specific histone demethylase 1A results in meiotic aberration during oocyte maturation in vitro in goats. Theriogenology 2020, 143, 168–178. [Google Scholar] [CrossRef] [PubMed]
- Schultz, R.M.; Stein, P.; Svoboda, P. The oocyte-to-embryo transition in mouse: Past, present, and future. Biol. Reprod. 2018, 99, 160–174. [Google Scholar] [CrossRef] [PubMed]
- Telford, N.A.; Watson, A.J.; Schultz, G.A. Transition from maternal to embryonic control in early mammalian development: A comparison of several species. Mol. Reprod. Dev. 1990, 26, 90–100. [Google Scholar] [CrossRef]
- Lee, M.T.; Bonneau, A.R.; Giraldez, A.J. Zygotic genome activation during the maternal-to-zygotic transition. Annu. Rev. Cell Dev. Biol. 2014, 30, 581–613. [Google Scholar] [CrossRef] [PubMed]
- Tominaga, Y.; Li, C.; Wang, R.H.; Deng, C.X. Murine Wee1 plays a critical role in cell cycle regulation and pre-implantation stages of embryonic development. Int. J. Biol. Sci. 2006, 2, 161–170. [Google Scholar] [CrossRef]
- Lee, G.S.; Kim, H.S.; Hwang, W.S.; Hyun, S.H. Characterization of porcine growth differentiation factor-9 and its expression in oocyte maturation. Mol. Reprod. Dev. 2008, 75, 707–714. [Google Scholar] [CrossRef] [PubMed]
- Bebbere, D.; Bogliolo, L.; Ariu, F.; Fois, S.; Leoni, G.G.; Tore, S.; Succu, S.; Berlinguer, F.; Naitana, S.; Ledda, S. Expression pattern of zygote arrest 1 (ZAR1), maternal antigen that embryo requires (MATER), growth differentiation factor 9 (GDF9) and bone morphogenetic protein 15 (BMP15) genes in ovine oocytes and in vitro-produced preimplantation embryos. Reprod. Fertil. Dev. 2008, 20, 908–915. [Google Scholar] [CrossRef]
- Yu, C.; Ji, S.Y.; Sha, Q.Q.; Dang, Y.; Zhou, J.J.; Zhang, Y.L.; Liu, Y.; Wang, Z.W.; Hu, B.; Sun, Q.Y.; et al. BTG4 is a meiotic cell cycle-coupled maternal-zygotic-transition licensing factor in oocytes. Nat. Struct. Mol. Biol. 2016, 23, 387–394. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhang, Y.; Hou, J.; Qian, X.; Zhang, H.; Zhang, Z.; Li, M.; Wang, R.; Liao, K.; Wang, Y.; et al. Ube2s regulates Sox2 stability and mouse ES cell maintenance. Cell Death Differ. 2016, 23, 393–404. [Google Scholar] [CrossRef] [PubMed]
- Burns, K.H.; Viveiros, M.M.; Ren, Y.; Wang, P.; DeMayo, F.J.; Frail, D.E.; Eppig, J.J.; Matzuk, M.M. Roles of NPM2 in chromatin and nucleolar organization in oocytes and embryos. Science 2003, 300, 633–636. [Google Scholar] [CrossRef]
- Habibi, R.; Hosseini, S.M.; Zadegan, F.G.; Hajian, M.; Ostadhosseini, S.; Vash, N.T.; Naddafpour, A.; Nasr Esfahani, M.H. Functional characterization of NANOG in goat pre-implantation embryonic development. Theriogenology 2018, 120, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Favetta, L.A.; St John, E.J.; King, W.A.; Betts, D.H. High levels of p66shc and intracellular ROS in permanently arrested early embryos. Free. Radic. Biol. Med. 2007, 42, 1201–1210. [Google Scholar] [CrossRef]
- Gaspar, R.C.; Arnold, D.R.; Corrêa, C.A.; da Rocha, C.V., Jr.; Penteado, J.C.; Del Collado, M.; Vantini, R.; Garcia, J.M.; Lopes, F.L. Oxygen tension affects histone remodeling of in vitro-produced embryos in a bovine model. Theriogenology 2015, 83, 1408–1415. [Google Scholar] [CrossRef]
- Agarwal, A.; Saleh, R.A.; Bedaiwy, M.A. Role of reactive oxygen species in the pathophysiology of human reproduction. Fertil. Steril. 2003, 79, 829–843. [Google Scholar] [CrossRef]
- Adam, A.A.; Takahashi, Y.; Katagiri, S.; Nagano, M. Effects of oxygen tension in the gas atmosphere during in vitro maturation, in vitro fertilization and in vitro culture on the efficiency of in vitro production of mouse embryos. Jpn. J. Vet. Res. 2004, 52, 77–84. [Google Scholar]
- Yuan, Y.Q.; Van Soom, A.; Coopman, F.O.; Mintiens, K.; Boerjan, M.L.; Van Zeveren, A.; de Kruif, A.; Peelman, L.J. Influence of oxygen tension on apoptosis and hatching in bovine embryos cultured in vitro. Theriogenology 2003, 59, 1585–1596. [Google Scholar] [CrossRef]
- Mishra, A.; Reddy, I.J.; Gupta, P.S.; Mondal, S. L-carnitine Mediated Reduction in Oxidative Stress and Alteration in Transcript Level of Antioxidant Enzymes in Sheep Embryos Produced In Vitro. Reprod. Domest. Anim. 2016, 51, 311–321. [Google Scholar] [CrossRef] [PubMed]
- Nilson, K.A.; Lawson, C.K.; Mullen, N.J.; Ball, C.B.; Spector, B.M.; Meier, J.L.; Price, D.H. Oxidative stress rapidly stabilizes promoter-proximal paused Pol II across the human genome. Nucleic Acids Res. 2017, 45, 11088–11105. [Google Scholar] [CrossRef] [PubMed]
- Lettieri-Barbato, D.; Ioannilli, L.; Aquilano, K.; Ciccarone, F.; Rosina, M.; Ciriolo, M.R. FoxO1 localizes to mitochondria of adipose tissue and is affected by nutrient stress. Metabolism 2019, 95, 84–92. [Google Scholar] [CrossRef]
- Xu, X.; He, L.; Zhang, A.; Li, Q.; Hu, W.; Chen, H.; Du, J.; Shen, J. Toxoplasma gondii isolate with genotype Chinese 1 triggers trophoblast apoptosis through oxidative stress and mitochondrial dysfunction in mice. Exp. Parasitol. 2015, 154, 51–61. [Google Scholar] [CrossRef]
- Hu, D.B.; Li, Z.S.; Ali, I.; Xu, L.J.; Fang, N.Z. Effect of potential role of p53 on embryo development arrest induced by H(2)O(2) in mouse. In Vitro Cell. Dev. Biol. Anim. 2017, 53, 344–353. [Google Scholar] [CrossRef]
- Wang, H.; Ye, Y.; Yu, Z.L. Proteomic and functional analyses demonstrate the involvement of oxidative stress in the anticancer activities of oridonin in HepG2 cells. Oncol. Rep. 2014, 31, 2165–2172. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.H.; Song, G.X.; Liu, Y.Q.; Sun, W.; Zhou, L.J.; Liu, H.L.; Yang, R.; Sheng, Y.H.; Qian, L.M.; Kong, X.Q. Silencing of FABP3 promotes apoptosis and induces mitochondrion impairment in embryonic carcinoma cells. J. Bioenerg. Biomembr. 2012, 44, 317–323. [Google Scholar] [CrossRef] [PubMed]
- Bonnet, F.; Molina, A.; Roussat, M.; Azais, M.; Bel-Vialar, S.; Gautrais, J.; Pituello, F.; Agius, E. Neurogenic decisions require a cell cycle independent function of the CDC25B phosphatase. Elife 2018, 7, e32937. [Google Scholar] [CrossRef]
- Cao, J.; Dong, J.; Wang, Y.; Chen, Y. The expressions of DNA methyltransferase 1 (DNMT1) and cyclin A1 (CCNA1) in cervical carcinogenesis. Int. J. Clin. Exp. Pathol. 2019, 12, 40–49. [Google Scholar]
- Moiseeva, T.N.; Qian, C.; Sugitani, N.; Osmanbeyoglu, H.U.; Bakkenist, C.J. WEE1 kinase inhibitor AZD1775 induces CDK1 kinase-dependent origin firing in unperturbed G1- and S-phase cells. Proc. Natl. Acad. Sci. USA 2019, 116, 23891–23893. [Google Scholar] [CrossRef]
- Martinsson-Ahlzén, H.S.; Liberal, V.; Grünenfelder, B.; Chaves, S.R.; Spruck, C.H.; Reed, S.I. Cyclin-dependent kinase-associated proteins Cks1 and Cks2 are essential during early embryogenesis and for cell cycle progression in somatic cells. Mol. Cell. Biol. 2008, 28, 5698–5709. [Google Scholar] [CrossRef]
- Ma, S.; Charron, J.; Erikson, R.L. Role of Plk2 (Snk) in mouse development and cell proliferation. Mol. Cell. Biol. 2003, 23, 6936–6943. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Min, J.N.; Masters, S.; Mivechi, N.F.; Moskophidis, D. Insights into function and regulation of small heat shock protein 25 (HSPB1) in a mouse model with targeted gene disruption. Genesis 2007, 45, 487–501. [Google Scholar] [CrossRef] [PubMed]
- Ruzov, A.; Dunican, D.S.; Prokhortchouk, A.; Pennings, S.; Stancheva, I.; Prokhortchouk, E.; Meehan, R.R. Kaiso is a genome-wide repressor of transcription that is essential for amphibian development. Development 2004, 131, 6185–6194. [Google Scholar] [CrossRef] [PubMed]
- Li, H.B.; Jin, X.Q.; Jin, X.; Guo, Z.H.; Ding, X.H.; Wang, Q.; Liu, R.Z. BMP4 knockdown of NCSCs leads to aganglionosis in the middle embryonic stage. Mol. Med. Rep. 2018, 17, 5423–5427. [Google Scholar] [CrossRef] [PubMed]
- Tallquist, M.D.; Weismann, K.E.; Hellström, M.; Soriano, P. Early myotome specification regulates PDGFA expression and axial skeleton development. Development 2000, 127, 5059–5070. [Google Scholar] [CrossRef] [PubMed]
- Park, H.; Bourla, A.B.; Kastner, D.L.; Colbert, R.A.; Siegel, R.M. Lighting the fires within: The cell biology of autoinflammatory diseases. Nat. Rev. Immunol. 2012, 12, 570–580. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; Li, Q.; Cao, Q.; Wang, S.; Liu, H.; Li, Q. Integrated Analysis of miRNA-mRNA Interaction Network in Porcine Granulosa Cells Undergoing Oxidative Stress. Oxid. Med. Cell. Longev. 2019, 2019, 1041583. [Google Scholar] [CrossRef]
- Ebrahimi, K.B.; Cano, M.; Rhee, J.; Datta, S.; Wang, L.; Handa, J.T. Oxidative Stress Induces an Interactive Decline in Wnt and Nrf2 Signaling in Degenerating Retinal Pigment Epithelium. Antioxid. Redox Signal. 2018, 29, 389–407. [Google Scholar] [CrossRef] [PubMed]
- Shen, M.; Jiang, Y.; Guan, Z.; Cao, Y.; Li, L.; Liu, H.; Sun, S.C. Protective mechanism of FSH against oxidative damage in mouse ovarian granulosa cells by repressing autophagy. Autophagy 2017, 13, 1364–1385. [Google Scholar] [CrossRef]
- Wang, Y.; Li, J.; Gao, Y.; Luo, Y.; Luo, H.; Wang, L.; Yi, Y.; Yuan, Z.; Jim Xiao, Z.X. Hippo kinases regulate cell junctions to inhibit tumor metastasis in response to oxidative stress. Redox Biol. 2019, 26, 101233. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Anjomani-Virmouni, S.; Koundouros, N.; Dimitriadi, M.; Choo-Wing, R.; Valle, A.; Zheng, Y.; Chiu, Y.H.; Agnihotri, S.; Zadeh, G.; et al. PARK2 Depletion Connects Energy and Oxidative Stress to PI3K/Akt Activation via PTEN S-Nitrosylation. Mol. Cell 2017, 65, 999–1013. [Google Scholar] [CrossRef] [PubMed]
- Kang, K.A.; Wang, Z.H.; Zhang, R.; Piao, M.J.; Kim, K.C.; Kang, S.S.; Kim, Y.W.; Lee, J.; Park, D.; Hyun, J.W. Myricetin protects cells against oxidative stress-induced apoptosis via regulation of PI3K/Akt and MAPK signaling pathways. Int. J. Mol. Sci. 2010, 11, 4348–4360. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wan, Y.; Li, D.; Deng, M.; Liu, Z.; Liu, L.; Wang, F. Comprehensive Transcriptome Analysis of mRNA Expression Patterns of Early Embryo Development in Goat under Hypoxic and Normoxic Conditions. Biology 2021, 10, 381. https://doi.org/10.3390/biology10050381
Wan Y, Li D, Deng M, Liu Z, Liu L, Wang F. Comprehensive Transcriptome Analysis of mRNA Expression Patterns of Early Embryo Development in Goat under Hypoxic and Normoxic Conditions. Biology. 2021; 10(5):381. https://doi.org/10.3390/biology10050381
Chicago/Turabian StyleWan, Yongjie, Dongxu Li, Mingtian Deng, Zifei Liu, Liang Liu, and Feng Wang. 2021. "Comprehensive Transcriptome Analysis of mRNA Expression Patterns of Early Embryo Development in Goat under Hypoxic and Normoxic Conditions" Biology 10, no. 5: 381. https://doi.org/10.3390/biology10050381
APA StyleWan, Y., Li, D., Deng, M., Liu, Z., Liu, L., & Wang, F. (2021). Comprehensive Transcriptome Analysis of mRNA Expression Patterns of Early Embryo Development in Goat under Hypoxic and Normoxic Conditions. Biology, 10(5), 381. https://doi.org/10.3390/biology10050381





