Chloroplast Localized FIBRILLIN11 Is Involved in the Osmotic Stress Response during Arabidopsis Seed Germination
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Protein Sequence and Phylogenetic Tree Analysis
2.2. Subcellular Localization
2.3. Plant Material and Growth Conditions
2.4. T-DNA Insertion Homozygous Mutant Selection
2.5. Seed Germination Test
2.6. Primer Design and Construction
2.7. Abiotic Stress Treatments
2.8. Measurements of Root Growth
2.9. RNA Isolation
2.10. cDNA Synthesis and Quantitative Polymerase Chain Reaction Analyses
3. Results
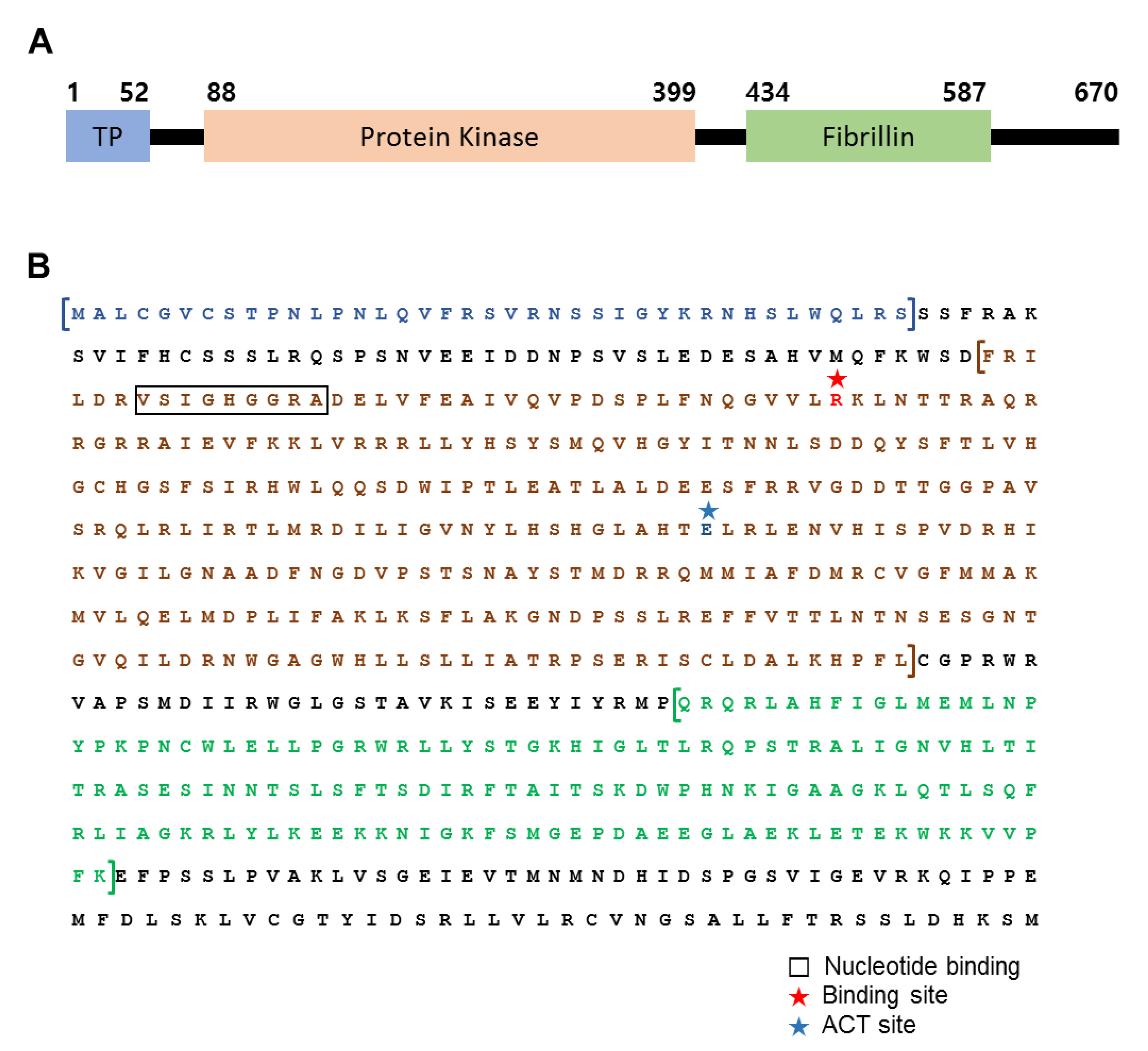
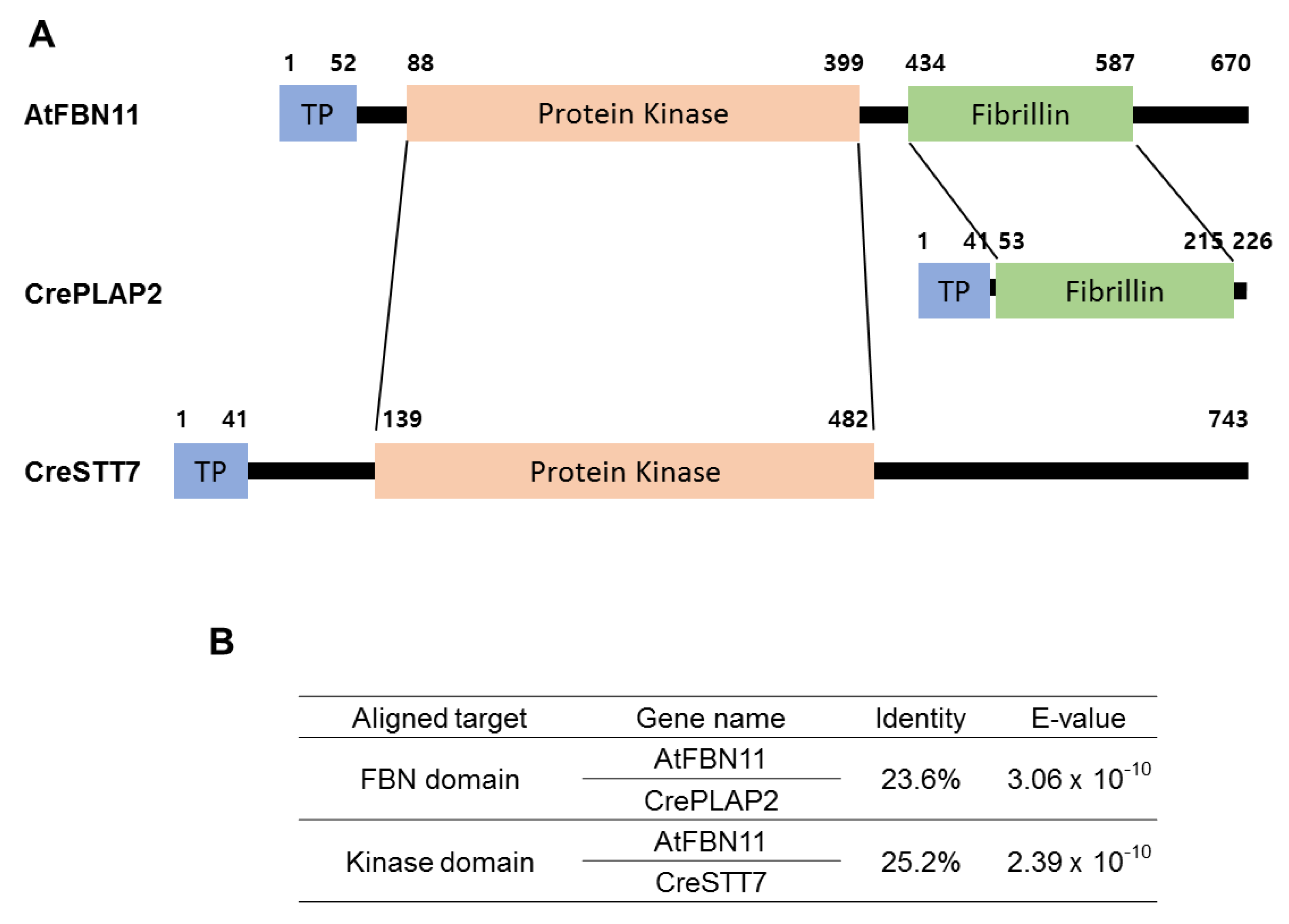
3.1. Structural Features of FBN11
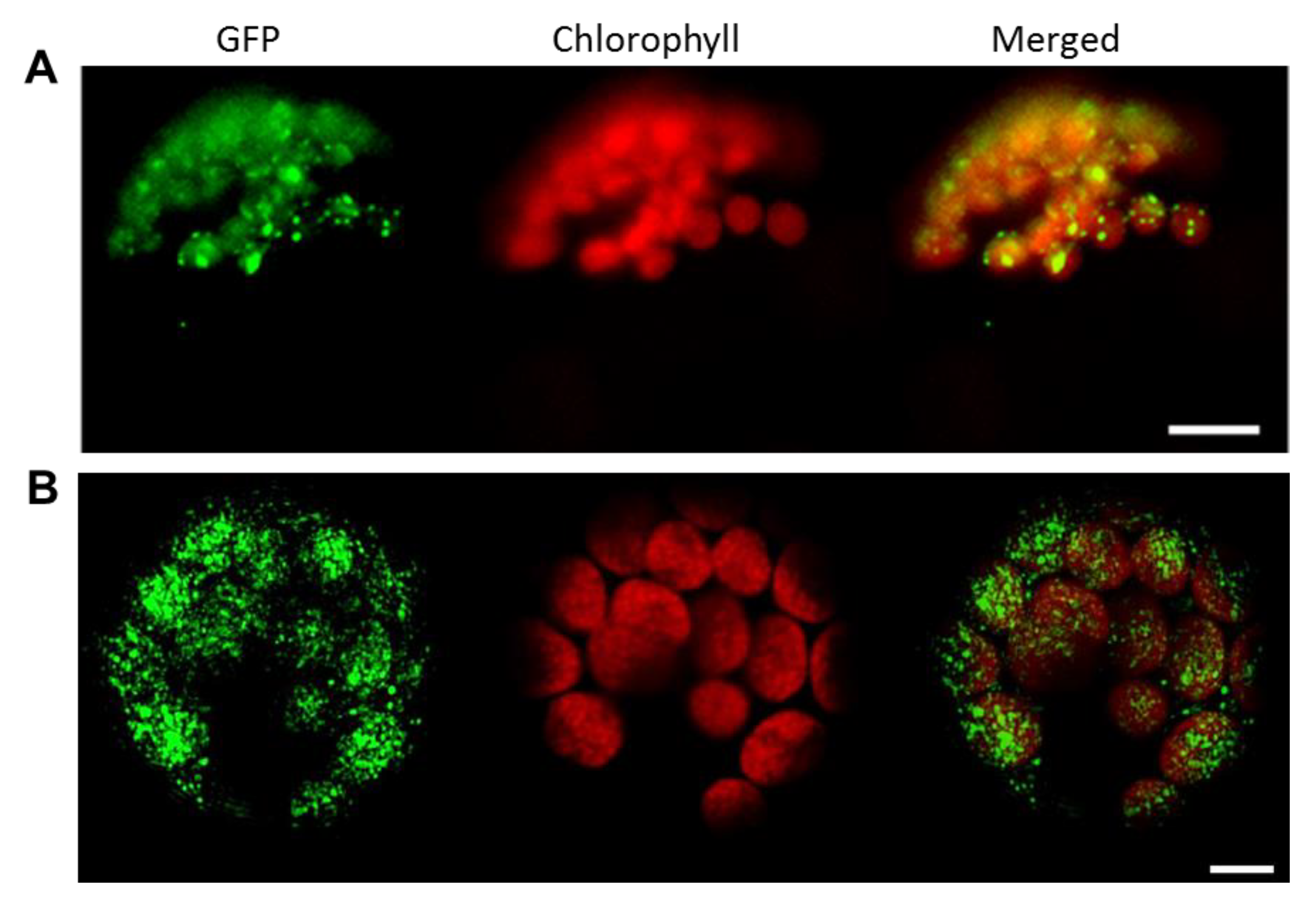
3.2. FBN11 Subcellular Location
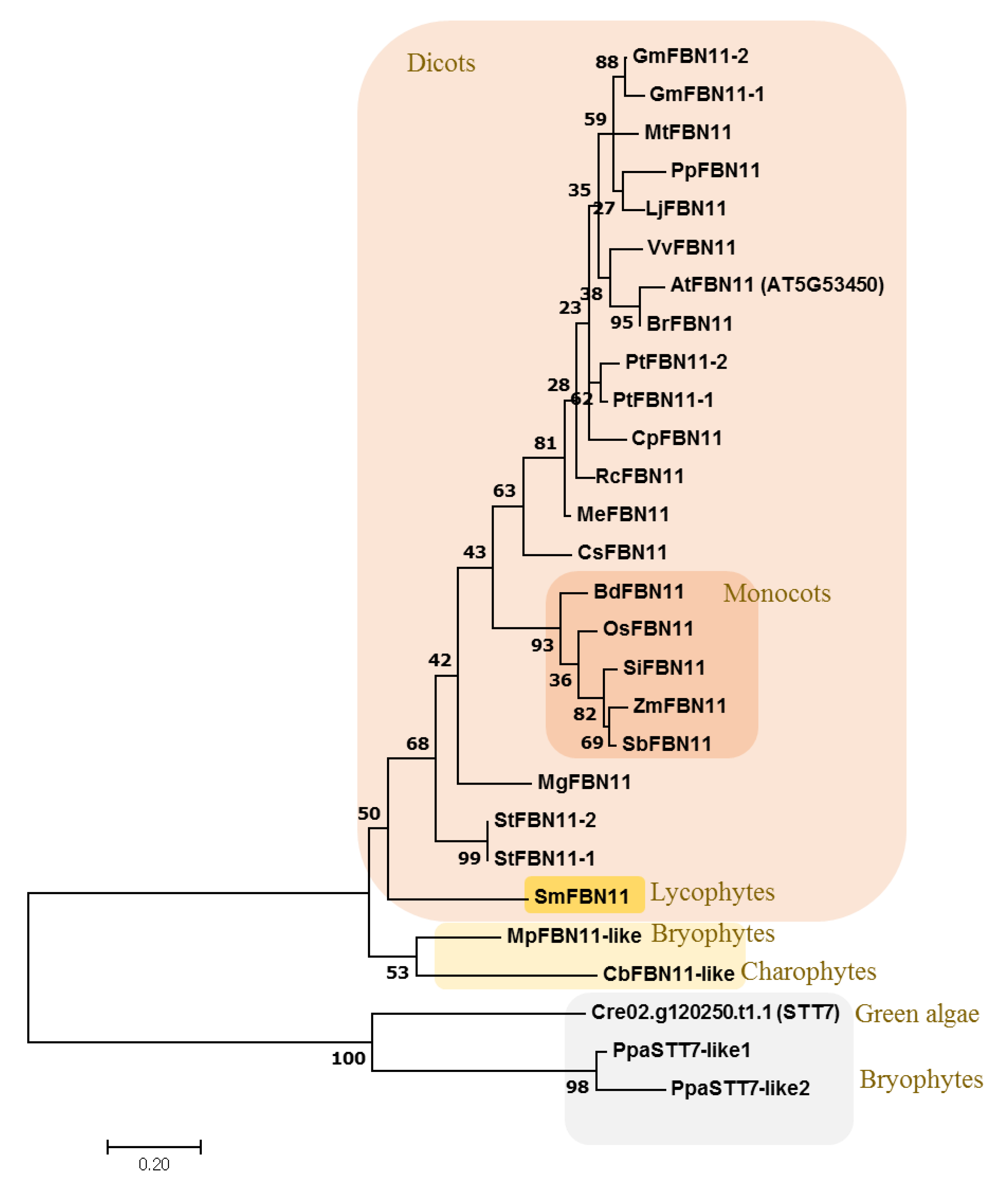
3.3. Evolution of FBN11
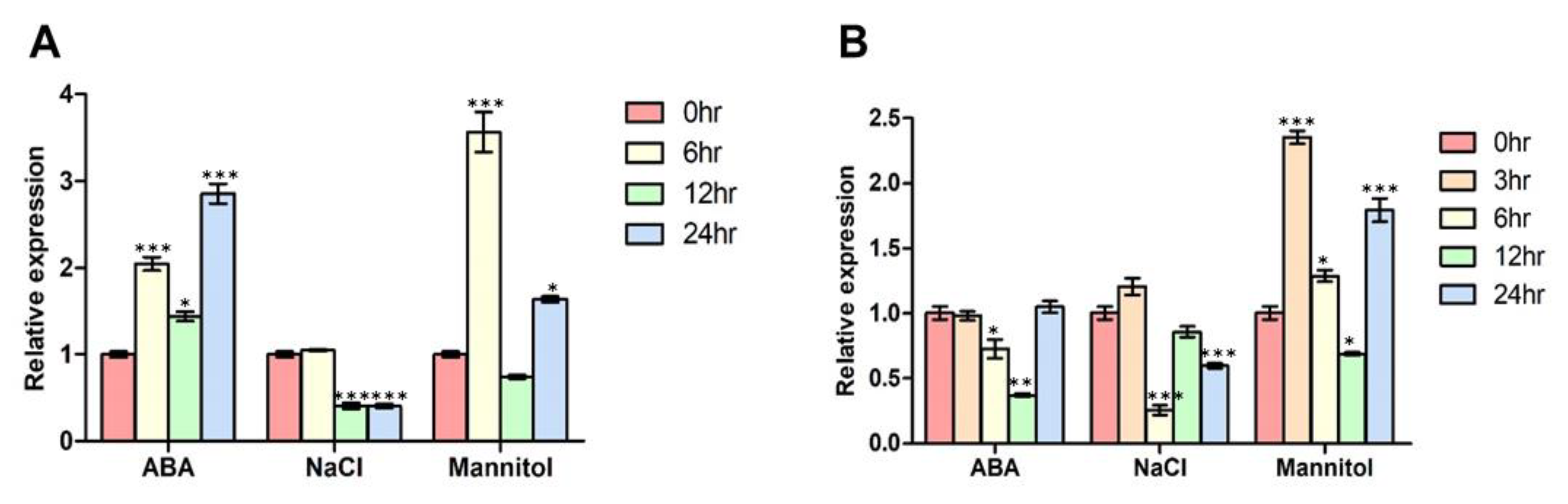
3.4. FBN11 Expression Profiles in Developing Tissue ± ABA, NaCl, or Mannitol Exposure
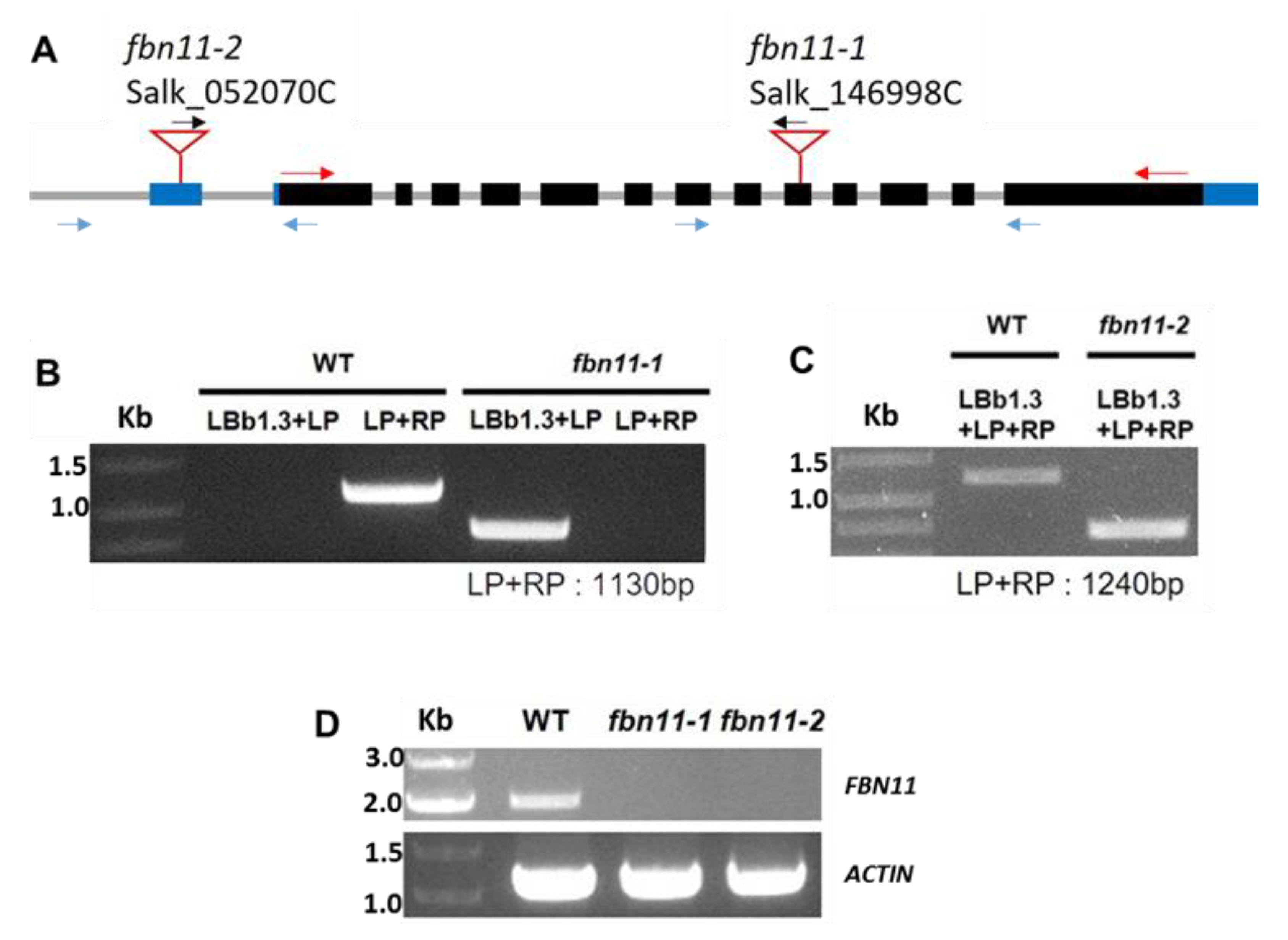
3.5. Characterization of fbn11 Mutants
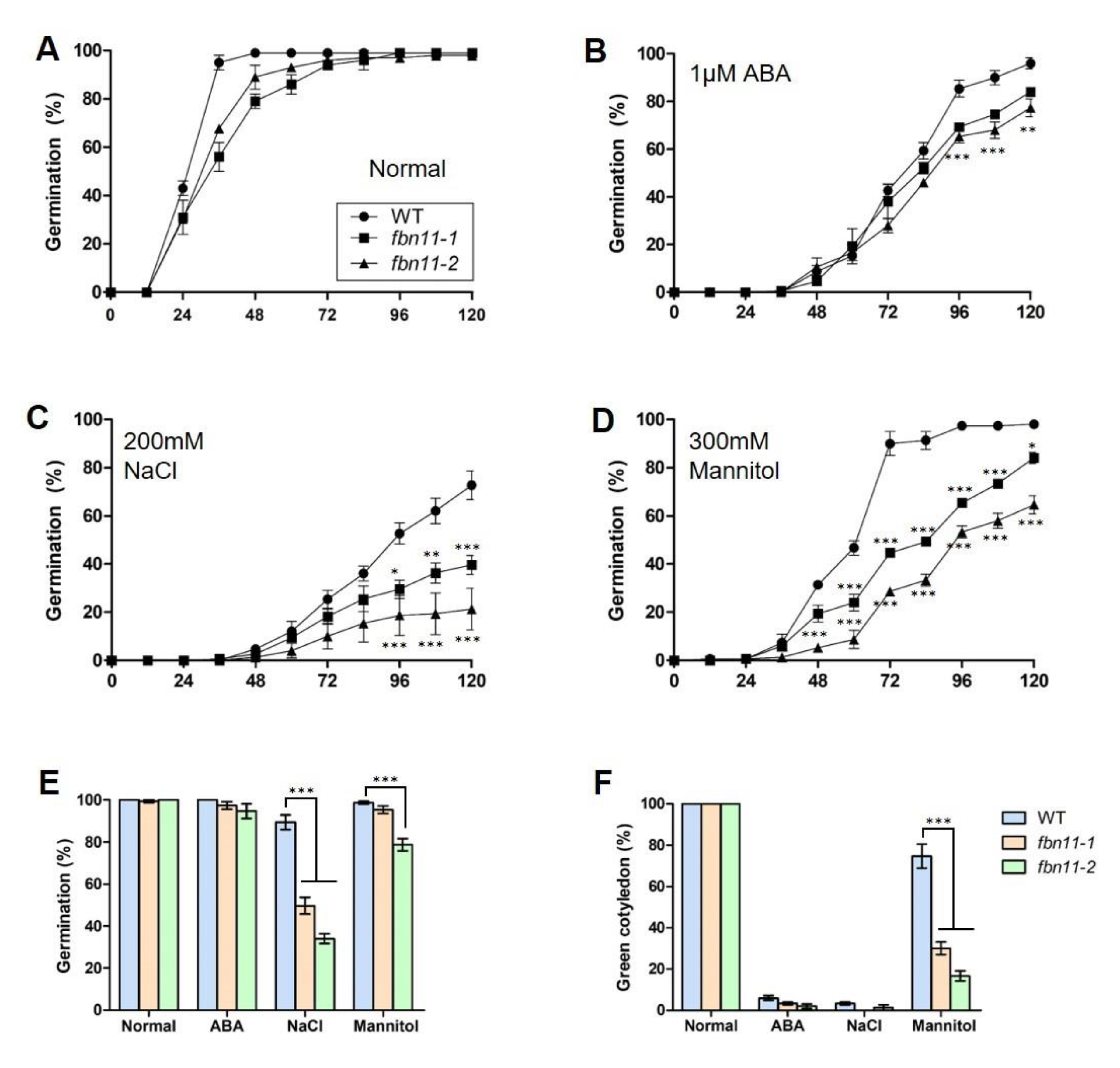
3.6. Phenotype Comparison between Wild Type and fbn11 Mutants Exposed to Abiotic Stresses
3.7. Analysis of fbn11 Effect on Seed Germination Pattern under Different Stress Conditions
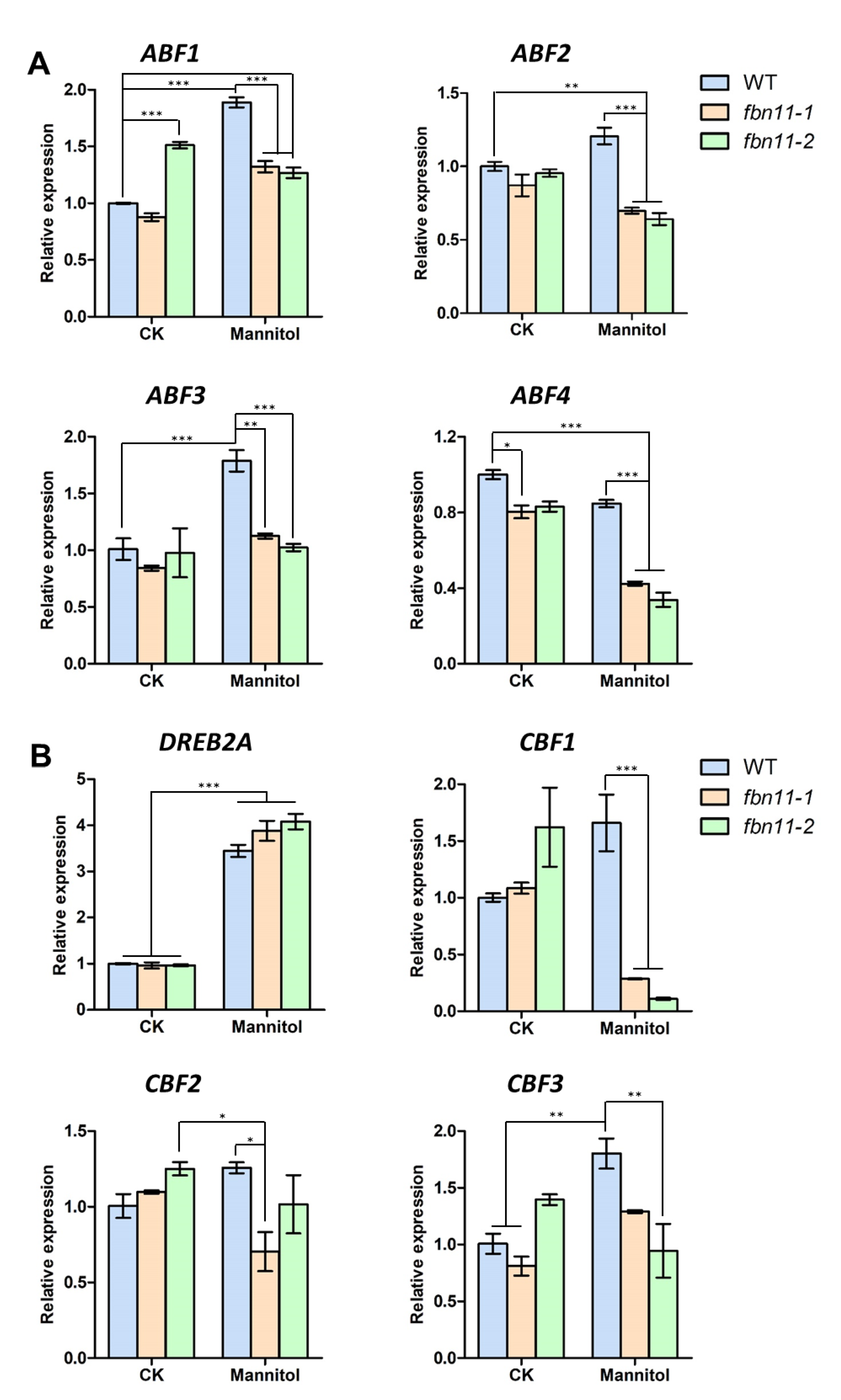
3.8. Expression of Abiotic Stress-Marker Transcription Factors in fbn11 during Seed Germination under Osmotic Stress
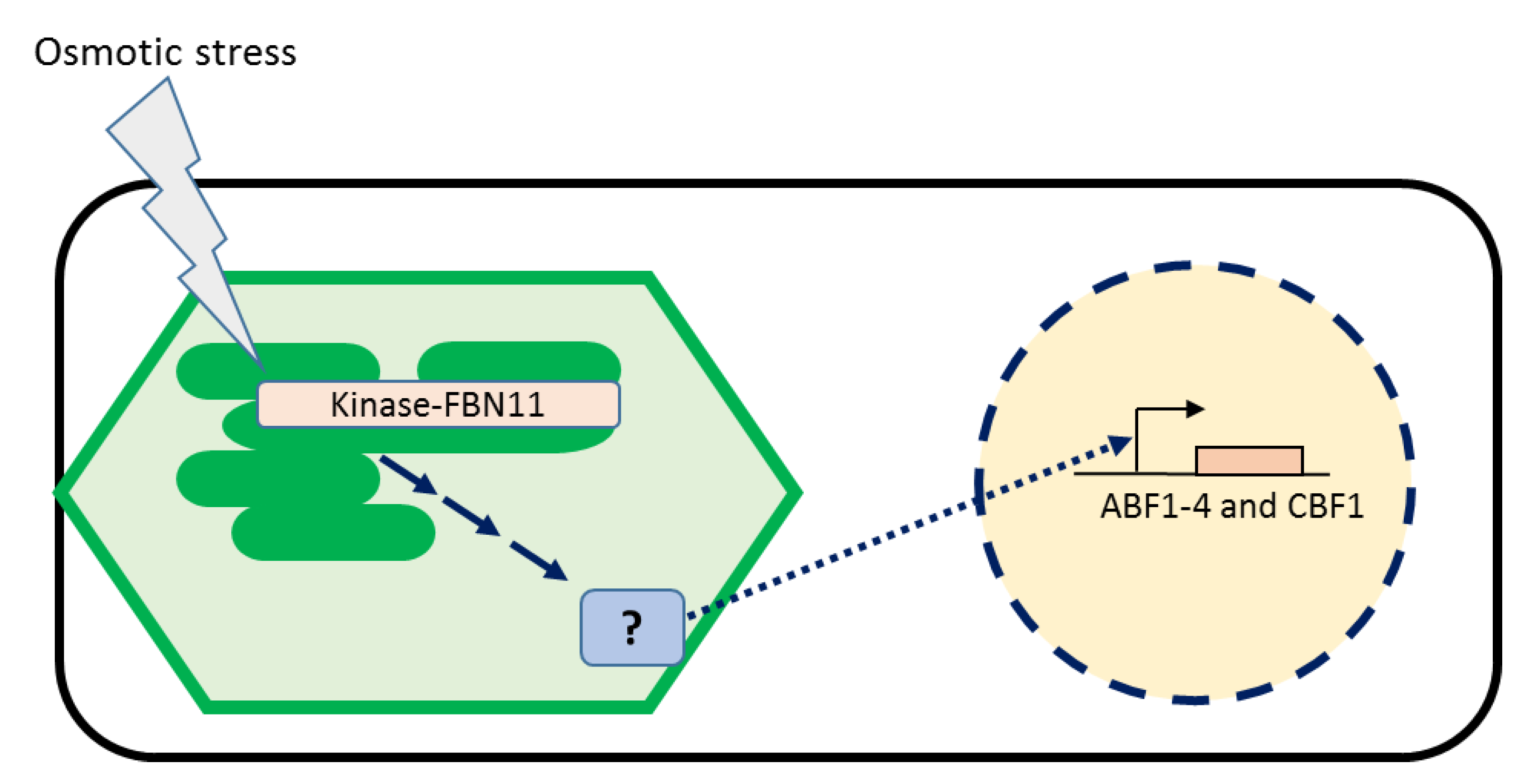
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| FBN | FIBRILLIN |
| PG | Plastoglobule |
| ABA | Abscisic acid |
| DREB | Dehydration responsive element binding protein |
| AREB | Abscisic acid-responsive element binding protein |
| TAIR | The Arabidopsis Information Resource |
| NCBI | National Center for Biotechnology Information |
| ABRC | Arabidopsis Biological Resource Center |
| sGFP | synthetic green fluorescence protein |
| CaMV | Cauliflower mosaic virus |
References
- Austin, J.R., 2nd; Frost, E.; Vidi, P.A.; Kessler, F.; Staehelin, L.A. Plastoglobules are lipoprotein subcompartments of the chloroplast that are permanently coupled to thylakoid membranes and contain biosynthetic enzymes. Plant Cell 2006, 18, 1693–1703. [Google Scholar] [CrossRef] [PubMed]
- Deruere, J.; Romer, S.; d’Harlingue, A.; Backhaus, R.A.; Kuntz, M.; Camara, B. Fibril assembly and carotenoid overaccumulation in chromoplasts: A model for supramolecular lipoprotein structures. Plant Cell 1994, 6, 119–133. [Google Scholar]
- Pozueta-Romero, J.; Rafia, F.; Houlne, G.; Cheniclet, C.; Carde, J.P.; Schantz, M.L.; Schantz, R. A ubiquitous plant housekeeping gene, PAP, encodes a major protein component of bell pepper chromoplasts. Plant Physiol. 1997, 115, 1185–1194. [Google Scholar] [CrossRef]
- Lohscheider, J.N.; Rio Bartulos, C. Plastoglobules in algae: A comprehensive comparative study of the presence of major structural and functional components in complex plastids. Mar. Genom. 2016, 28, 127–136. [Google Scholar] [CrossRef]
- Langenkamper, G.; Manac’h, N.; Broin, M.; Cuine, S.; Becuwe, N.; Kuntz, M.; Rey, P. Accumulation of plastid lipid-associated proteins (fibrillin/CDSP34) upon oxidative stress, ageing and biotic stress in Solanaceae and in response to drought in other species. J. Exp. Bot. 2001, 52, 1545–1554. [Google Scholar] [CrossRef]
- Yang, Y.; Sulpice, R.; Himmelbach, A.; Meinhard, M.; Christmann, A.; Grill, E. Fibrillin expression is regulated by abscisic acid response regulators and is involved in abscisic acid-mediated photoprotection. Proc. Natl. Acad. Sci. USA 2006, 103, 6061–6066. [Google Scholar] [CrossRef]
- Ytterberg, A.J.; Peltier, J.B.; van Wijk, K.J. Protein profiling of plastoglobules in chloroplasts and chromoplasts. A surprising site for differential accumulation of metabolic enzymes. Plant Physiol. 2006, 140, 984–997. [Google Scholar] [CrossRef]
- Brehelin, C.; Kessler, F.; van Wijk, K.J. Plastoglobules: Versatile lipoprotein particles in plastids. Trends Plant Sci. 2007, 12, 260–266. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.K.; Maximova, S.N.; Jensen, P.J.; Lehman, B.L.; Ngugi, H.K.; McNellis, T.W. FIBRILLIN4 is required for plastoglobule development and stress resistance in apple and Arabidopsis. Plant Physiol. 2010, 154, 1281–1293. [Google Scholar] [CrossRef] [PubMed]
- Youssef, A.; Laizet, Y.; Block, M.A.; Marechal, E.; Alcaraz, J.P.; Larson, T.R.; Pontier, D.; Gaffe, J.; Kuntz, M. Plant lipid-associated fibrillin proteins condition jasmonate production under photosynthetic stress. Plant J. 2010, 61, 436–445. [Google Scholar] [CrossRef]
- Singh, D.K.; McNellis, T.W. Fibrillin protein function: The tip of the iceberg? Trends Plant Sci. 2011, 16, 432–441. [Google Scholar] [CrossRef]
- Gamez-Arjona, F.M.; de la Concepcion, J.C.; Raynaud, S.; Merida, A. Arabidopsis thaliana plastoglobule-associated fibrillin 1a interacts with fibrillin 1b in vivo. FEBS Lett. 2014, 588, 2800–2804. [Google Scholar] [CrossRef]
- Li, J.; Yang, J.; Zhu, B.; Xie, G. Overexpressing OsFBN1 enhances plastoglobule formation, reduces grain-filling percent and jasmonate levels under heat stress in rice. Plant Sci. 2019, 285, 230–238. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Hu, H.; Ma, Y.; Zhou, J. Genome-wide identification and characterization of the fibrillin gene family in Triticum aestivum. PeerJ 2020, 8, e9225. [Google Scholar] [CrossRef] [PubMed]
- Lundquist, P.K.; Poliakov, A.; Bhuiyan, N.H.; Zybailov, B.; Sun, Q.; van Wijk, K.J. The functional network of the Arabidopsis plastoglobule proteome based on quantitative proteomics and genome-wide coexpression analysis. Plant Physiol. 2012, 158, 1172–1192. [Google Scholar] [CrossRef]
- Laizet, Y.; Pontier, D.; Mache, R.; Kuntz, M. Subfamily organization and phylogenetic origin of genes encoding plastid lipid-associated proteins of the fibrillin type. J. Genome Sci. Technol. 2004, 3, 19–28. [Google Scholar] [CrossRef]
- Gillet, B.; Beyly, A.; Peltier, G.; Rey, P. Molecular characterization of CDSP 34, a chloroplastic protein induced by water deficit in Solanum tuberosum L. plants, and regulation of CDSP 34 expression by ABA and high illumination. Plant J. 1998, 16, 257–262. [Google Scholar] [CrossRef]
- Otsubo, M.; Ikoma, C.; Ueda, M.; Ishii, Y.; Tamura, N. Functional Role of Fibrillin5 in Acclimation to Photooxidative Stress. Plant Cell Physiol. 2018, 59, 1670–1682. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Li, X.; Khatab, A.A.; Xie, G. Phylogeny, structural diversity and genome-wide expression analysis of fibrillin family genes in rice. Phytochemistry 2020, 175, 112377. [Google Scholar] [CrossRef]
- Kumar, M.; Kesawat, M.S.; Ali, A.; Lee, S.C.; Gill, S.S.; Kim, H.U. Integration of Abscisic Acid Signaling with Other Signaling Pathways in Plant Stress Responses and Development. Plants 2019, 8, 592. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Li, G.J.; Bressan, R.A.; Song, C.P.; Zhu, J.K.; Zhao, Y. Abscisic acid dynamics, signaling, and functions in plants. J Integr. Plant Biol. 2020, 62, 25–54. [Google Scholar] [CrossRef] [PubMed]
- Tuteja, N. Abscisic Acid and abiotic stress signaling. Plant Signal Behav. 2007, 2, 135–138. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, T.; Mogami, J.; Yamaguchi-Shinozaki, K. ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr. Opin. Plant Biol. 2014, 21, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Kumar, K.; Kumar, M.; Kim, S.R.; Ryu, H.; Cho, Y.G. Insights into genomics of salt stress response in rice. Rice (N. Y.) 2013, 6, 27. [Google Scholar] [CrossRef] [PubMed]
- Park, H.J.; Kim, W.Y.; Yun, D.J. A New Insight of Salt Stress Signaling in Plant. Mol. Cells 2016, 39, 447–459. [Google Scholar] [CrossRef] [PubMed]
- Soma, F.; Takahashi, F.; Suzuki, T.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Plant Raf-like kinases regulate the mRNA population upstream of ABA-unresponsive SnRK2 kinases under drought stress. Nat. Commun. 2020, 11, 1373. [Google Scholar] [CrossRef]
- Yoshida, T.; Fujita, Y.; Maruyama, K.; Mogami, J.; Todaka, D.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Four Arabidopsis AREB/ABF transcription factors function predominantly in gene expression downstream of SnRK2 kinases in abscisic acid signalling in response to osmotic stress. Plant Cell Environ. 2015, 38, 35–49. [Google Scholar] [CrossRef]
- Sakuma, Y.; Maruyama, K.; Osakabe, Y.; Qin, F.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell 2006, 18, 1292–1309. [Google Scholar] [CrossRef]
- Kim, E.H.; Lee, D.W.; Lee, K.R.; Jung, S.J.; Jeon, J.S.; Kim, H.U. Conserved Function of Fibrillin5 in the Plastoquinone-9 Biosynthetic Pathway in Arabidopsis and Rice. Front. Plant Sci. 2017, 8, 1197. [Google Scholar] [CrossRef]
- Shen, J.; Fu, J.; Ma, J.; Wang, X.; Gao, C.; Zhuang, C.; Wan, J.; Jiang, L. Isolation, culture, and transient transformation of plant protoplasts. Curr. Protoc. Cell Biol. 2014, 63, 1–17. [Google Scholar] [CrossRef]
- Li, X.; Liang, Y.; Gao, B.; Mijiti, M.; Bozorov, T.A.; Yang, H.; Zhang, D.; Wood, A.J. ScDREB10, an A-5c type of DREB Gene of the Desert Moss Syntrichia caninervis, Confers Osmotic and Salt Tolerances to Arabidopsis. Genes 2019, 10, 146. [Google Scholar] [CrossRef]
- Lee, K.; Lee, H.G.; Yoon, S.; Kim, H.U.; Seo, P.J. The Arabidopsis MYB96 Transcription Factor Is a Positive Regulator of ABSCISIC ACID-INSENSITIVE4 in the Control of Seed Germination. Plant Physiol. 2015, 168, 677–689. [Google Scholar] [CrossRef]
- Kessler, F.; Schnell, D.; Blobel, G. Identification of proteins associated with plastoglobules isolated from pea (Pisum sativum L.) chloroplasts. Planta 1999, 208, 107–113. [Google Scholar] [CrossRef] [PubMed]
- Baek, D.; Chun, H.J.; Kang, S.; Shin, G.; Park, S.J.; Hong, H.; Kim, C.; Kim, D.H.; Lee, S.Y.; Kim, M.C.; et al. A Role for Arabidopsis miR399f in Salt, Drought, and ABA Signaling. Mol. Cells 2016, 39, 111–118. [Google Scholar] [PubMed]
- Lin, L.; Wu, J.; Jiang, M.; Wang, Y. Plant Mitogen-Activated Protein Kinase Cascades in Environmental Stresses. Int. J. Mol. Sci. 2021, 22, 1543. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.H.; Lee, Y.; Kim, H.U. Fibrillin 5 Is Essential for Plastoquinone-9 Biosynthesis by Binding to Solanesyl Diphosphate Synthases in Arabidopsis. Plant. Cell 2015, 27, 2956–2971. [Google Scholar] [CrossRef]
- Uno, Y.; Furihata, T.; Abe, H.; Yoshida, R.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc. Natl. Acad. Sci. USA 2000, 97, 11632–11637. [Google Scholar] [CrossRef]
- Furihata, T.; Maruyama, K.; Fujita, Y.; Umezawa, T.; Yoshida, R.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc. Natl. Acad. Sci. USA 2006, 103, 1988–1993. [Google Scholar] [CrossRef]
- Fujii, H.; Verslues, P.E.; Zhu, J.K. Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant. Cell 2007, 19, 485–494. [Google Scholar] [CrossRef]
- Sakuma, Y.; Maruyama, K.; Qin, F.; Osakabe, Y.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Dual function of an Arabidopsis transcription factor DREB2A in water-stress-responsive and heat-stress-responsive gene expression. Proc. Natl. Acad. Sci. USA 2006, 103, 18822–28827. [Google Scholar] [CrossRef]
- Hsu, C.L.; Prasad, R.; Blackman, C.; Ng, D.T. Endoplasmic reticulum stress regulation of the Kar2p/BiP chaperone alleviates proteotoxicity via dual degradation pathways. Mol. Biol. Cell 2012, 23, 630–641. [Google Scholar] [CrossRef]
- Wagner, D.; Przybyla, D.; Op den Camp, R.; Kim, C.; Landgraf, F.; Lee, K.P.; Wursch, M.; Laloi, C.; Nater, M.; Hideg, E.; et al. The genetic basis of singlet oxygen-induced stress responses of Arabidopsis thaliana. Science 2004, 306, 1183–1185. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.; Meskauskiene, R.; Zhang, S.; Lee, K.P.; Lakshmanan Ashok, M.; Blajecka, K.; Herrfurth, C.; Feussner, I.; Apel, K. Chloroplasts of Arabidopsis are the source and a primary target of a plant-specific programmed cell death signaling pathway. Plant Cell 2012, 24, 3026–3039. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.K. Abiotic Stress Signaling and Responses in Plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.G.; Foley, R.C.; Onate-Sanchez, L.; Lin, C.; Singh, K.B. Target genes for OBP3, a Dof transcription factor, include novel basic helix-loop-helix domain proteins inducible by salicylic acid. Plant J. 2003, 35, 362–372. [Google Scholar] [CrossRef] [PubMed]
- Fernando, V.C.D.; Al Khateeb, W.; Belmonte, M.F.; Schroeder, D.F. Role of Arabidopsis ABF1/3/4 during det1 germination in salt and osmotic stress conditions. Plant Mol. Biol. 2018, 97, 149–163. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Li, Y.J.; Zhang, F.J.; Zhang, G.Z.; Jiang, X.Y.; Yu, H.M.; Hou, B.K. The Arabidopsis UDP-glycosyltransferases UGT79B2 and UGT79B3, contribute to cold, salt and drought stress tolerance via modulating anthocyanin accumulation. Plant. J. 2017, 89, 85–103. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Choi, Y.-R.; Kim, I.; Kumar, M.; Shim, J.; Kim, H.-U. Chloroplast Localized FIBRILLIN11 Is Involved in the Osmotic Stress Response during Arabidopsis Seed Germination. Biology 2021, 10, 368. https://doi.org/10.3390/biology10050368
Choi Y-R, Kim I, Kumar M, Shim J, Kim H-U. Chloroplast Localized FIBRILLIN11 Is Involved in the Osmotic Stress Response during Arabidopsis Seed Germination. Biology. 2021; 10(5):368. https://doi.org/10.3390/biology10050368
Chicago/Turabian StyleChoi, Yu-Ri, Inyoung Kim, Manu Kumar, Jaekyung Shim, and Hyun-Uk Kim. 2021. "Chloroplast Localized FIBRILLIN11 Is Involved in the Osmotic Stress Response during Arabidopsis Seed Germination" Biology 10, no. 5: 368. https://doi.org/10.3390/biology10050368
APA StyleChoi, Y.-R., Kim, I., Kumar, M., Shim, J., & Kim, H.-U. (2021). Chloroplast Localized FIBRILLIN11 Is Involved in the Osmotic Stress Response during Arabidopsis Seed Germination. Biology, 10(5), 368. https://doi.org/10.3390/biology10050368








