Laser Microdissection of Specific Stem-Base Tissue Types from Olive Microcuttings for Isolation of High-Quality RNA
Simple Summary
Abstract
1. Introduction
2. Methods
2.1. Plant Material and Growth Conditions
2.2. Sample Preparation
2.3. Cryosectioning
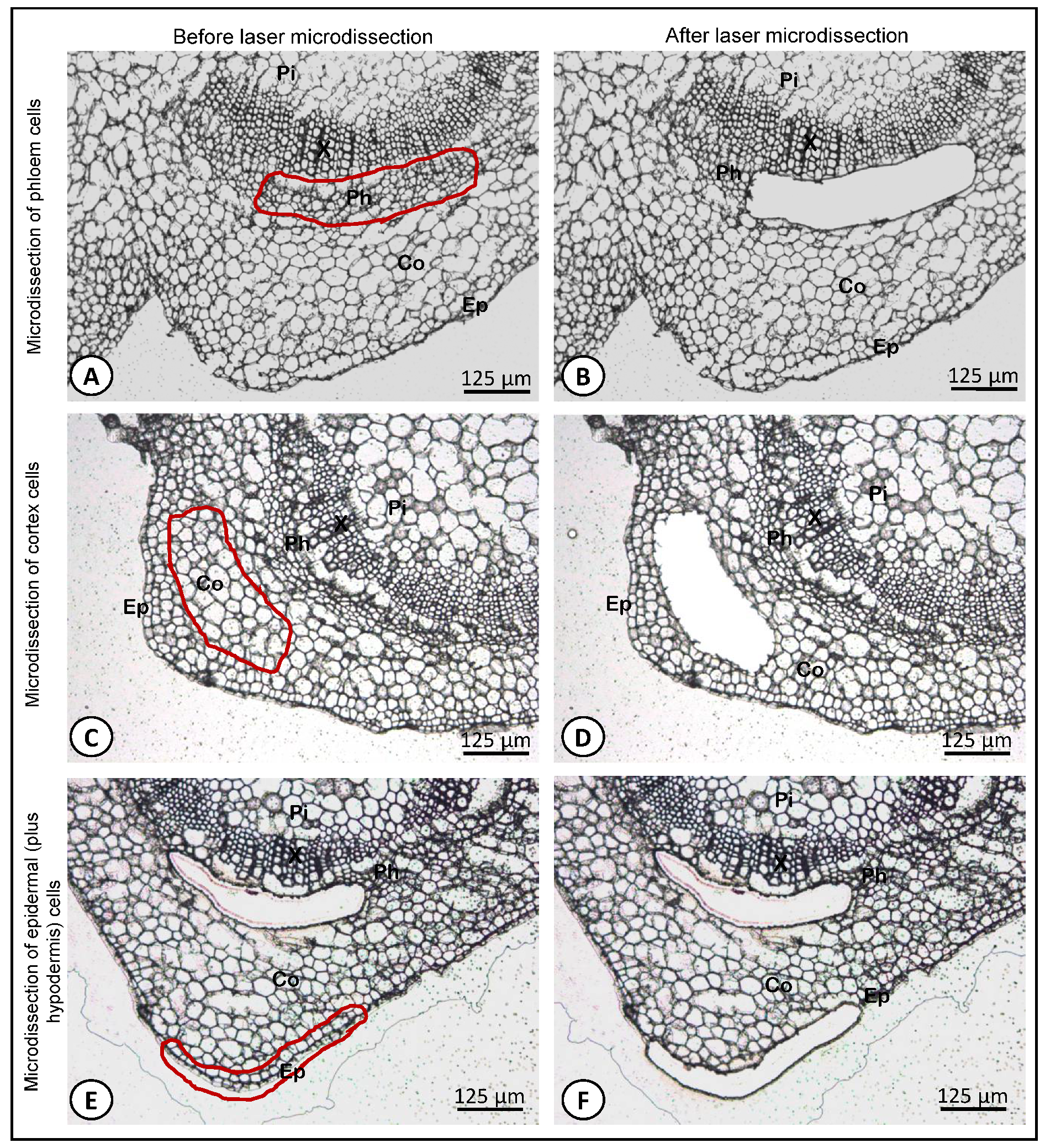
2.4. Laser Microdissection (LM)
2.5. RNA Isolation and Integrity Analysis
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brandt, S.P. Microgenomics: Gene expression analysis at the tissue-specific and single-cell levels. J. Exp. Bot. 2005, 56, 495–505. [Google Scholar] [CrossRef] [PubMed]
- Martin, C.; Bhatt, K.; Baumann, K. Shaping in plant cells. Curr. Opin. Plant Biol. 2001, 4, 540–549. [Google Scholar] [CrossRef]
- Rogers, E.D.; Jackson, T.; Moussaieff, A.; Aharoni, A.; Benfey, P.N. Cell type-specific transcriptional profiling: Implications for metabolite profiling. Plant J. 2012, 70, 5–17. [Google Scholar] [CrossRef] [PubMed]
- Shulse, C.N.; Cole, B.J.; Ciobanu, D.; Lin, J.; Yoshinaga, Y.; Gouran, M.; Turco, G.M.; Zhu, Y.; O’Malley, R.C.; Brady, S.M.; et al. High-Throughput Single-Cell Transcriptome Profiling of Plant Cell Types. Cell Rep. 2019, 27, 2241–2247.e4. [Google Scholar] [CrossRef] [PubMed]
- Shahan, R. The future is now: Gene expression dynamics at single cell resolution. Plant Cell. 2019, 31, 933–934. [Google Scholar] [CrossRef]
- Ryu, K.H.; Huang, L.; Kang, H.M.; Schiefelbein, J. Single-cell RNA sequencing resolves molecular relationships among individual plant cells. Plant Physiol. 2019, 179, 1444–1456. [Google Scholar] [CrossRef] [PubMed]
- Kortz, A.; Hochholdinger, F.; Yu, P. Cell Type-Specific Transcriptomics of Lateral Root Formation and Plasticity. Front. Plant Sci. 2019, 10, 1–7. [Google Scholar] [CrossRef]
- Efroni, I.; Birnbaum, K.D. The potential of single-cell profiling in plants. Genome Biol. 2016, 17, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Libault, M.; Pingault, L.; Zogli, P.; Schiefelbein, J. Plant Systems Biology at the Single-Cell Level. Trends Plant Sci. 2017, 22, 949–960. [Google Scholar] [CrossRef]
- Matas, A.J.; Agustí, J.; Tadeo, F.R.; Talón, M.; Rose, J.K.C. Tissue-specific transcriptome profiling of the citrus fruit epidermis and subepidermis using laser capture microdissection. J. Exp. Bot. 2010, 61, 3321–3330. [Google Scholar] [CrossRef]
- Nelson, T.; Tausta, S.L.; Gandotra, N.; Liu, T. Laser Microdissection of Plant Tissue: What You See Is What You Get. Annu. Rev. Plant Biol. 2006, 57, 181–201. [Google Scholar] [CrossRef]
- Emmert-Buck, M.R.; Bonner, R.F.; Smith, P.D.; Chuaqui, R.F.; Zhuang, Z.; Goldstein, S.R.; Weiss, R.A.; Liotta, L.A. Laser Capture Microdissection. Science 1996, 274, 998–1001. [Google Scholar] [CrossRef]
- Asano, T.; Masumura, T.; Kusano, H.; Kikuchi, S.; Kurita, A.; Shimada, H.; Kadowaki, K.-I. Construction of a specialized cDNA library from plant cells isolated by laser capture microdissection: Toward comprehensive analysis of the genes expressed in the rice phloem. Plant J. 2002, 32, 401–408. [Google Scholar] [CrossRef] [PubMed]
- Blokhina, O.; Valerio, C.; Sokołowska, K.; Zhao, L.; Kärkönen, A.; Niittylä, T.; Fagerstedt, K. Laser capture microdissection protocol for xylem tissues of woody plants. Front Plant Sci. 2017, 7, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Kerk, N.M.; Ceserani, T.; Tausta, S.L.; Sussex, I.M.; Nelson, T.M. Laser Capture Microdissection of Cells from Plant Tissues. Plant Physiol. 2003, 132, 27–35. [Google Scholar] [CrossRef]
- Nakazono, M.; Qiu, F.; Borsuk, L.A.; Schnable, P.S.; Nelissen, H.; Clarke, J.H.; De Block, M.; De Block, S.; Vanderhaeghen, R.; Zielinski, R.E.; et al. Laser-Capture Microdissection, a Tool for the Global Analysis of Gene Expression in Specific Plant Cell Types: Identification of Genes Expressed Differentially in Epidermal Cells or Vascular Tissues of Maize. Plant Cell 2003, 15, 583–596. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, R.T.; Pereira, H. Laser Microdissection applied to plants. In Microscopy: Science, Technology, Applications and Education; Formatex Research Center: Badajoz, Spain, 2010; pp. 986–992. [Google Scholar]
- Teixeira, R.T.; Fortes, A.M.; Bai, H.; Pinheiro, C.; Pereira, H. Transcriptional profiling of cork oak phellogenic cells isolated by laser microdissection. Planta 2017, 247, 317–338. [Google Scholar] [CrossRef]
- Celedon, J.M.; Yuen, M.M.; Chiang, A.; Henderson, H.; Reid, K.E.; Bohlmann, J. Cell-type- and tissue-specific transcriptomes of the white spruce (Picea glauca) bark unmask fine-scale spatial patterns of constitutive and induced conifer defense. Plant J. 2017, 92, 710–726. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Li, H.; Bhatti, S.; Zhou, S.; Yang, Y.; Fish, T.; Thannhauser, T.W. Development of a laser capture microscope-based single-cell-type proteomics tool for studying proteomes of individual cell layers of plant roots. Hortic. Res. 2016, 3, 16026. [Google Scholar] [CrossRef] [PubMed]
- Lenzi, L.; Caruso, C.; Bianchedi, P.L.; Pertot, I.; Perazzolli, M. Laser Microdissection of Grapevine Leaves Reveals Site-Specific Regulation of Transcriptional Response toPlasmopara viticola. Plant Cell Physiol. 2016, 57, 69–81. [Google Scholar] [CrossRef] [PubMed]
- Rajhi, I.; Yamauchi, T.; Takahashi, H.; Nishiuchi, S.; Shiono, K.; Watanabe, R.; Mliki, A.; Nagamura, Y.; Tsutsumi, N.; Nishizawa, N.K.; et al. Identification of genes expressed in maize root cortical cells during lysigenous aerenchyma formation using laser microdissection and microarray analyses. New Phytol. 2010, 190, 351–368. [Google Scholar] [CrossRef]
- Muñoz-Sanhueza, L.G.; Lee, Y.; Tillmann, M.; Cohen, J.D.; Hvoslef-Eide, A.K. Auxin analysis using laser microdissected plant tissues sections. BMC Plant Biol. 2018, 18, 133. [Google Scholar] [CrossRef] [PubMed]
- Harrop, T.W.R.; Din, I.U.; Gregis, V.; Osnato, M.; Jouannic, S.; Adam, H.; Kater, M.M. Gene expression profiling of reproductive meristem types in early rice inflorescences by laser microdissection. Plant J. 2016, 86, 75–88. [Google Scholar] [CrossRef]
- Shinozaki, Y.; Nicolas, P.; Fernandez-Pozo, N.; Ma, Q.; Evanich, D.J.; Shi, Y.; Xu, Y.; Zheng, Y.; Snyder, S.I.; Martin, L.B.B.; et al. High-resolution spatiotemporal transcriptome mapping of tomato fruit development and ripening. Nat. Commun. 2018, 9, 1–13. [Google Scholar] [CrossRef]
- Casson, S.; Spencer, M.; Walker, K.; Lindsey, K. Laser capture microdissection for the analysis of gene expression during embryogenesis of Arabidopsis. Plant J. 2005, 42, 111–123. [Google Scholar] [CrossRef] [PubMed]
- Woll, K.; Borsuk, L.A.; Stransky, H.; Nettleton, D.; Schnable, P.S.; Hochholdinger, F. Isolation, Characterization, and Pericycle-Specific Transcriptome Analyses of the Novel Maize Lateral and Seminal Root Initiation Mutant rum1. Plant Physiol. 2005, 139, 1255–1267. [Google Scholar] [CrossRef] [PubMed]
- Yu, P.; Eggert, K.; von Wirén, N.; Li, C.; Hochholdinger, F. Cell type-specific gene expression analyses by RNA sequencing reveal local high nitrate-triggered lateral root initiation in shoot-borne roots of maize by modulating auxin-related cell cycle regulation. Plant Physiol. 2015, 169, 690–704. [Google Scholar] [CrossRef] [PubMed]
- Macedo, E.; Vieira, C.; Carrizo, D.; Porfirio, S.; Hegewald, H.; Arnholdt-Schmitt, B.; Calado, M.L.; Peixe, A. Adventitious root formation in olive (Olea europaea L.) microshoots: Anatomical evaluation and associated biochemical changes in peroxidase and polyphenoloxidase activities. J. Hortic. Sci. Biotechnol. 2013, 88, 53–59. [Google Scholar] [CrossRef]
- Porfirio, S.; Calado, M.L.; Noceda, C.; Cabrita, M.J.; Da Silva, M.G.; Azadi, P.; Peixe, A. Tracking biochemical changes during adventitious root formation in olive (Olea europaea L.). Sci. Hortic. 2016, 204, 41–53. [Google Scholar] [CrossRef]
- Velada, I.; Grzebelus, D.; Lousa, D.; Soares, C.M.; Macedo, E.S.; Peixe, A.; Arnholdt-Schmitt, B.; Cardoso, H.G. AOX1-Subfamily Gene Members in Olea europaea cv. “Galega Vulgar”—Gene Characterization and Expression of Transcripts during IBA-Induced in Vitro Adventitious Rooting. Int. J. Mol. Sci. 2018, 19, 597. [Google Scholar] [CrossRef] [PubMed]
- Velada, I.; Cardoso, H.; Porfirio, S.; Peixe, A. Expression Profile of PIN-Formed Auxin Efflux Carrier Genes during IBA-Induced In Vitro Adventitious Rooting in Olea europaea L. Plants 2020, 9, 185. [Google Scholar] [CrossRef] [PubMed]
- Santos Macedo, E.; Cardoso, H.G.; Hernández, A.; Peixe, A.A.; Polidoros, A.; Ferreira, A.; Cordeiro, A.; Arnholdt-Schmitt, B. Physiologic responses and gene diversity indicate olive alternative oxidase as a potential source for markers involved in efficient adventitious root induction. Physiol Plant. 2009, 137, 532–552. [Google Scholar] [CrossRef] [PubMed]
- Santos Macedo, E.; Sircar, D.; Cardoso, H.G.; Peixe, A.; Arnholdt-Schmitt, B. Involvement of alternative oxidase (AOX) in adventitious rooting of Olea europaea L. microshoots is linked to adaptive phenylpropanoid and lignin metabolism. Plant Cell Rep. 2012, 31, 1581–1590. [Google Scholar] [CrossRef]
- Li, S.-W.; Xue, L.; Xu, S.; Feng, H.; An, L. Mediators, Genes and Signaling in Adventitious Rooting. Bot. Rev. 2009, 75, 230–247. [Google Scholar] [CrossRef]
- Wang, Z.; Hua, J.; Yin, Y.; Gu, C.; Yu, C.; Shi, Q.; Guo, J.; Xuan, L.; Yu, F. An Integrated Transcriptome and Proteome Analysis Reveals Putative Regulators of Adventitious Root Formation in Taxodium ‘Zhongshanshan’. Int. J. Mol. Sci. 2019, 20, 1225. [Google Scholar] [CrossRef]
- Shang, C.; Yang, H.; Ma, S.; Shen, Q.; Liu, L.; Hou, C.; Cao, X.; Cheng, J. Physiological and Transcriptomic Changes during the Early Phases of Adventitious Root Formation in Mulberry Stem Hardwood Cuttings. Int. J. Mol. Sci. 2019, 20, 3707. [Google Scholar] [CrossRef]
- Quan, J.; Meng, S.; Guo, E.; Zhang, S.; Zhao, Z.; Yang, X. De novo sequencing and comparative transcriptome analysis of adventitious root development induced by exogenous indole-3-butyric acid in cuttings of tetraploid black locust. BMC Genom. 2017, 18, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; Zhang, X.; Nie, H.; Liu, M.; Cheng, J.L. Transcript profiling analysis reveals crucial genes regulating main metabolism during adventitious root formation in cuttings of Morus alba L. Plant Growth Regul. 2015, 79, 251–262. [Google Scholar] [CrossRef]
- De Almeida, M.R.; de Bastiani, D.; Gaeta, M.L.; de Araújo Mariath, J.E.; de Costa, F.; Retallick, J.; Nolan, L.; Tai, H.H.; Strömvik, M.V.; Fett-Neto, A.G. Comparative transcriptional analysis provides new insights into the molecular basis of adventitious rooting recalcitrance in Eucalyptus. Plant Sci. 2015, 239, 155–165. [Google Scholar] [CrossRef]
- Stevens, M.E.; Woeste, K.E.; Pijut, P.M. Localized gene expression changes during adventitious root formation in black walnut (Juglans nigra L.). Tree Physiol. 2018, 38, 877–894. [Google Scholar] [CrossRef]
- Peixe, A.; Raposo, A.; Lourenço, R.; Cardoso, H.; Macedo, E. Coconut water and BAP successfully replaced zeatin in olive (Olea europaea L.) micropropagation. Sci. Hortic. 2007, 113, 1–7. [Google Scholar] [CrossRef]
- Bevilacqua, C.; Makhzami, S.; Helbling, J.-C.; Defrenaix, P.; Martin, P. Maintaining RNA integrity in a homogeneous population of mammary epithelial cells isolated by Laser Capture Microdissection. BMC Cell Biol. 2010, 11, 95. [Google Scholar] [CrossRef] [PubMed]
- Olsen, S.; Krause, K. A rapid preparation procedure for laser microdissection-mediated harvest of plant tissues for gene expression analysis. Plant Methods 2019, 15, 1–10. [Google Scholar] [CrossRef]
- Anjam, M.S.; Ludwig, Y.; Hochholdinger, F.; Miyaura, C.; Inada, M.; Siddique, S.; Grundler, F.M.W. An improved procedure for isolation of high-quality RNA from nematode-infected Arabidopsis roots through laser capture microdissection. Plant Methods 2016, 12, 25. [Google Scholar] [CrossRef] [PubMed]
- Canas, R.A.; Canales, J.; Maldonado, J.G.; Avila, C.; Canovas, F.M. Transcriptome analysis in maritime pine using laser capture microdissection and 454 pyrosequencing. Tree Physiol. 2014, 34, 1278–1288. [Google Scholar] [CrossRef] [PubMed]
- Abbott, E.; Hall, D.; Hamberger, B.; Bohlmann, J. Laser microdissection of conifer stem tissues: Isolation and analysis of high quality RNA, terpene synthase enzyme activity and terpenoid metabolites from resin ducts and cambial zone tissue of white spruce (Picea glauca). BMC Plant Biol. 2010, 10, 106. [Google Scholar] [CrossRef]
- Day, R.C.; Grossniklaus, U.; Macknight, R.C. Be more specific! Laser-assisted microdissection of plant cells. Trends Plant Sci. 2005, 10, 397–406. [Google Scholar] [CrossRef]
- Goldsworthy, S.M.; Stockton, P.S.; Trempus, C.S.; Foley, J.F.; Maronpot, R.R. Effects of fixation on RNA extraction and amplification from laser capture microdissected tissue. Mol. Carcinog. 1999, 25, 86–91. [Google Scholar] [CrossRef]
- Gillespie, J.W.; Best, C.J.; Bichsel, V.E.; Cole, K.A.; Greenhut, S.F.; Hewitt, S.M.; Ahram, M.; Gathright, Y.B.; Merino, M.J.; Strausberg, R.L.; et al. Evaluation of Non-Formalin Tissue Fixation for Molecular Profiling Studies. Am. J. Pathol. 2002, 160, 449–457. [Google Scholar] [CrossRef]
- Gautam, V.; Sarkar, A.K. Laser Assisted Microdissection, an Efficient Technique to Understand Tissue Specific Gene Expression Patterns and Functional Genomics in Plants. Mol. Biotechnol. 2015, 57, 299–308. [Google Scholar] [CrossRef]
- Martin, L.B.B.; Nicolas, P.; Matas, A.J.; Shinozaki, Y.; Catalá, C.; Rose, J.K.C. Laser microdissection of tomato fruit cell and tissue types for transcriptome profiling. Nat. Protoc. 2016, 11, 2376–2388. [Google Scholar] [CrossRef]
- Schad, M.; Mungur, R.; Fiehn, O.; Kehr, J. Metabolic profiling of laser microdissected vascular bundles of Arabidopsis thaliana. Plant Methods 2005, 1, 2. [Google Scholar] [CrossRef]
- Barcala, M.; Fenoll, C.; Escobar, C. Laser Microdissection of Cells and Isolation of High-Quality RNA After Cryosectioning. Adv. Struct. Saf. Stud. 2012, 883, 87–95. [Google Scholar]
- Bova, G.S.; Eltoum, I.A.; Kiernan, J.A.; Siegal, G.P.; Frost, A.R.; Best, C.J.M.; Gillespie, J.W.; Emmert-Buck, M.R.; Su, G.H. Optimal Molecular Profiling of Tissue and Tissue Components: Defining the Best Processing and Microdissection Methods for Biomedical Applications. Pancreat. Cancer 2004, 103, 15–66. [Google Scholar]
- Ofusori, D.A.; Ayoka, A.O.; Adeeyo, O.A.; Adewole, S.O. Mezcla de kerosene y xileno: Una contribución a agentes de aclaramiento. Int. J. Morphol. 2009, 27, 211–218. [Google Scholar]
- Jensen, K.H.; Zwieniecki, M.A. Physical Limits to Leaf Size in Tall Trees. Phys. Rev. Lett. 2013, 110, 018104. [Google Scholar] [CrossRef] [PubMed]
- Chandran, D.; Scanlon, M.J.; Ohtsu, K.; Timmermans, M.C.P.; Schnable, P.S.; Wildermuth, M.C. Laser microdissection-mediated isolation and in vitro transcriptional amplification of plant RNA. Curr. Protoc. Mol. Biol. 2015, 112, 25A-3. [Google Scholar] [CrossRef]
- Sakai, K.; Taconnat, L.; Borrega, N.; Yansouni, J.; Brunaud, V.; Roux, C.P.-L.; Delannoy, E.; Magniette, M.-L.M.; Lepiniec, L.; Faure, J.D.; et al. Combining laser-assisted microdissection (LAM) and RNA-seq allows to perform a comprehensive transcriptomic analysis of epidermal cells of Arabidopsis embryo. Plant Methods 2018, 14, 10. [Google Scholar] [CrossRef]
- Imbeaud, S.; Graudens, E.; Boulanger, V.; Barlet, X.; Zaborski, P.; Eveno, E.; Mueller, O.; Schroeder, A.; Auffray, C. Towards standardization of RNA quality assessment using user-independent classifiers of microcapillary electrophoresis traces. Nucleic Acids Res. 2005, 33, e56. [Google Scholar] [CrossRef]


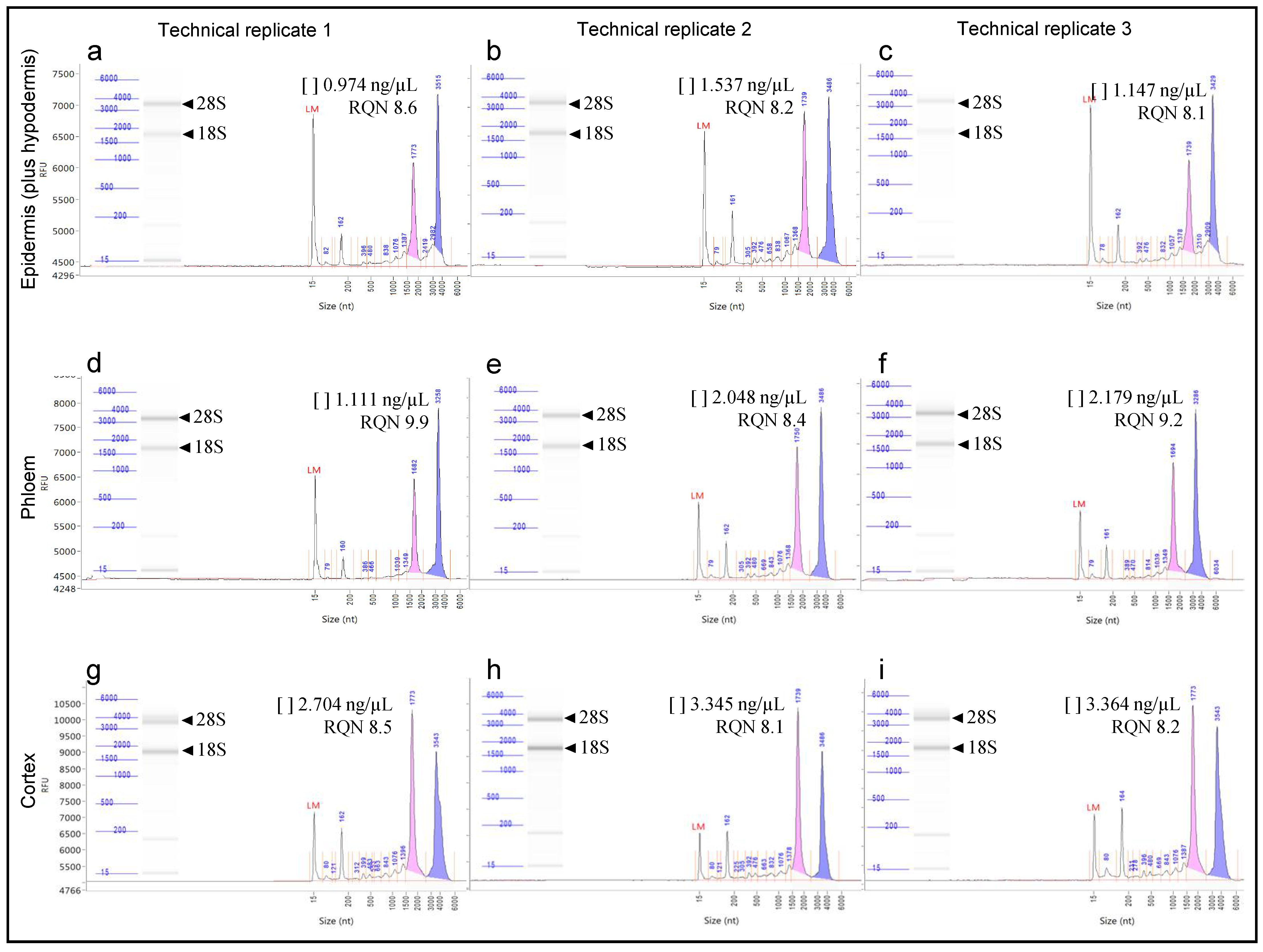
| Tissue Type | Technical Replicate | Microdissected Area (μm2) | RNA Concentration (ng/μL) | RQN 1 |
|---|---|---|---|---|
| Epidermis (plus hypodermis) | 1 | 411,020 | 0.974 | 8.6 |
| 2 | 573,546 | 1.537 | 8.2 | |
| 3 | 513,634 | 1.147 | 8.1 | |
| Phloem | 1 | 651,364 | 1.111 | 9.9 |
| 2 | 648,912 | 2.048 | 8.4 | |
| 3 | 797,119 | 2.179 | 9.2 | |
| Cortex | 1 | 669,198 | 2.704 | 8.5 |
| 2 | 769,165 | 3.345 | 8.1 | |
| 3 | 781,354 | 3.364 | 8.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Velada, I.; Menéndez, E.; Teixeira, R.T.; Cardoso, H.; Peixe, A. Laser Microdissection of Specific Stem-Base Tissue Types from Olive Microcuttings for Isolation of High-Quality RNA. Biology 2021, 10, 209. https://doi.org/10.3390/biology10030209
Velada I, Menéndez E, Teixeira RT, Cardoso H, Peixe A. Laser Microdissection of Specific Stem-Base Tissue Types from Olive Microcuttings for Isolation of High-Quality RNA. Biology. 2021; 10(3):209. https://doi.org/10.3390/biology10030209
Chicago/Turabian StyleVelada, Isabel, Esther Menéndez, Rita Teresa Teixeira, Hélia Cardoso, and Augusto Peixe. 2021. "Laser Microdissection of Specific Stem-Base Tissue Types from Olive Microcuttings for Isolation of High-Quality RNA" Biology 10, no. 3: 209. https://doi.org/10.3390/biology10030209
APA StyleVelada, I., Menéndez, E., Teixeira, R. T., Cardoso, H., & Peixe, A. (2021). Laser Microdissection of Specific Stem-Base Tissue Types from Olive Microcuttings for Isolation of High-Quality RNA. Biology, 10(3), 209. https://doi.org/10.3390/biology10030209










