Determination of the Concentration of IgG against the Spike Receptor-Binding Domain That Predicts the Viral Neutralizing Activity of Convalescent Plasma and Serum against SARS-CoV-2
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Expression and Purification of RBD
2.2. Human Samples
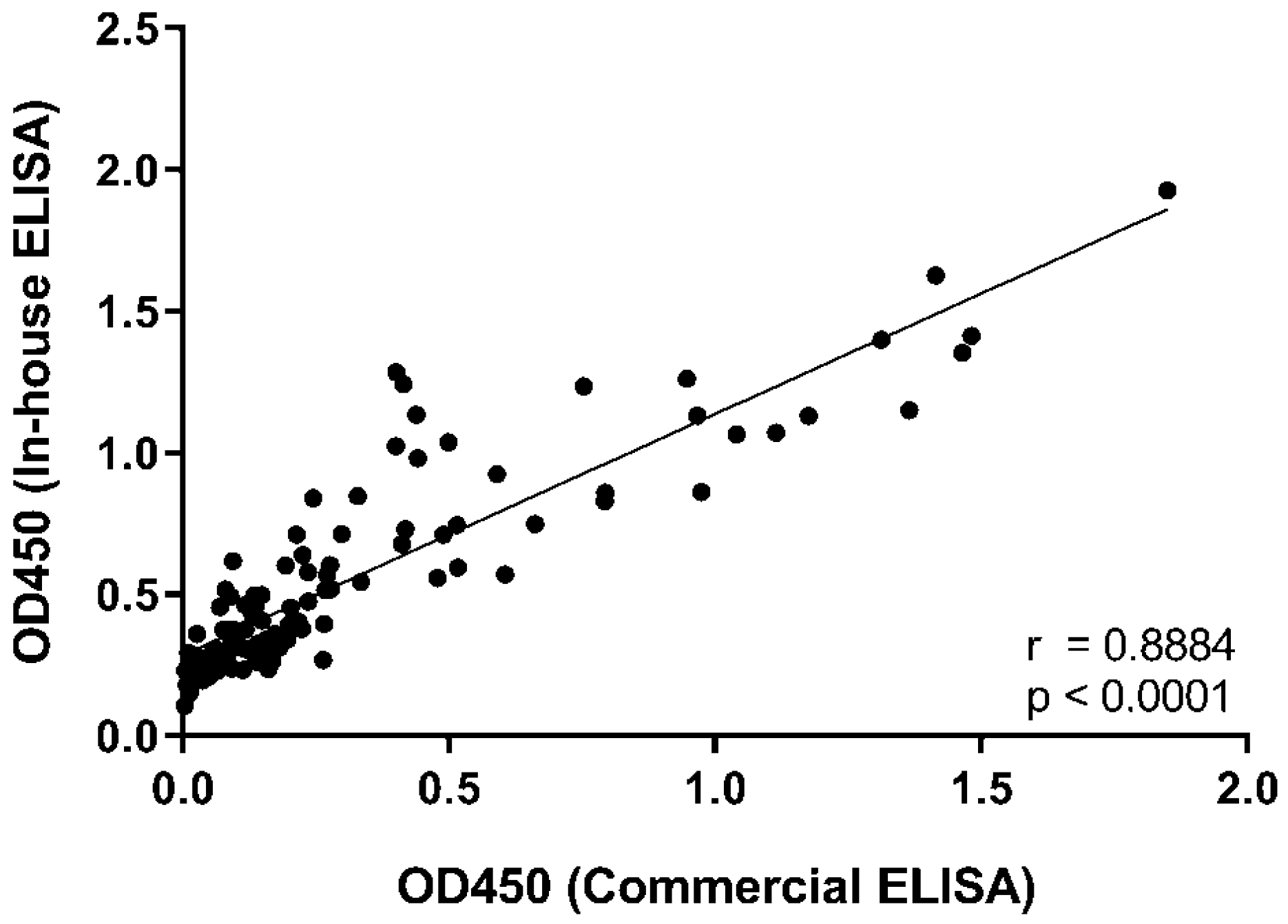
2.3. Determination of Anti-RBD IgG Levels by ELISA
2.4. Virus Isolation and Expansion in Cell Culture
2.5. SARS-CoV-2 Titration
2.6. Micro-Neutralization Assay
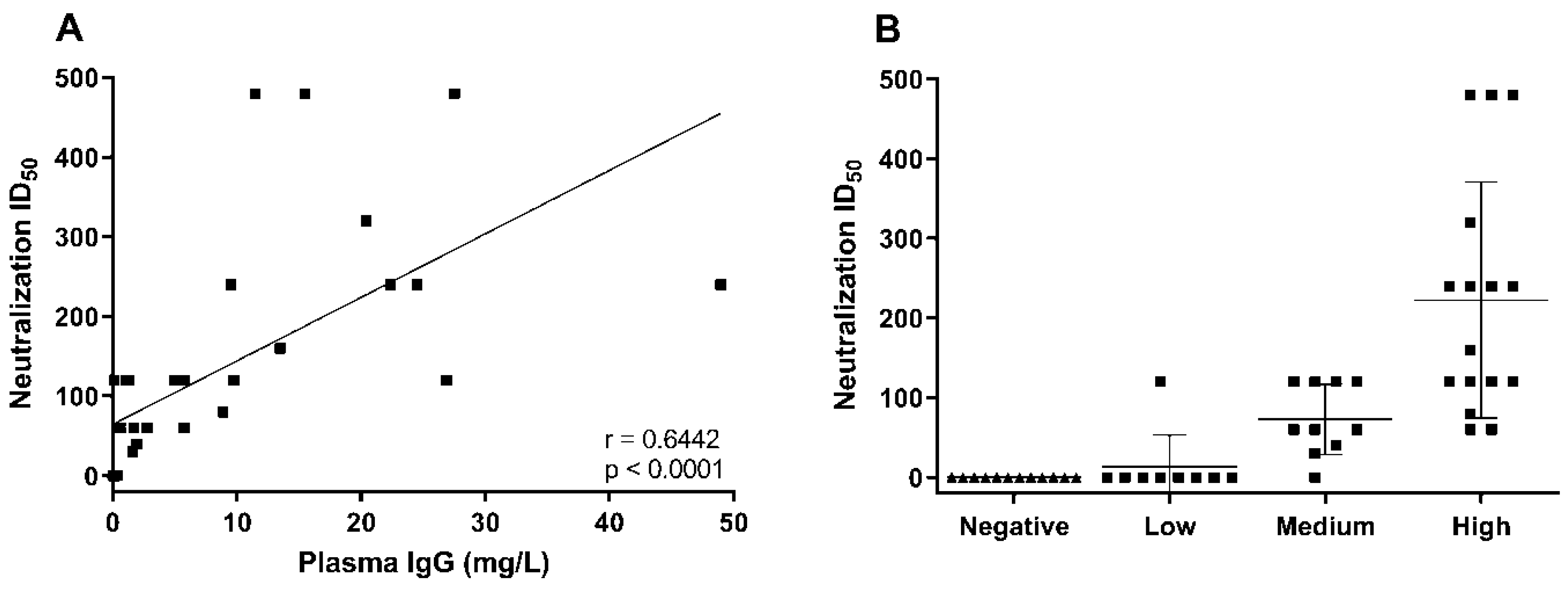
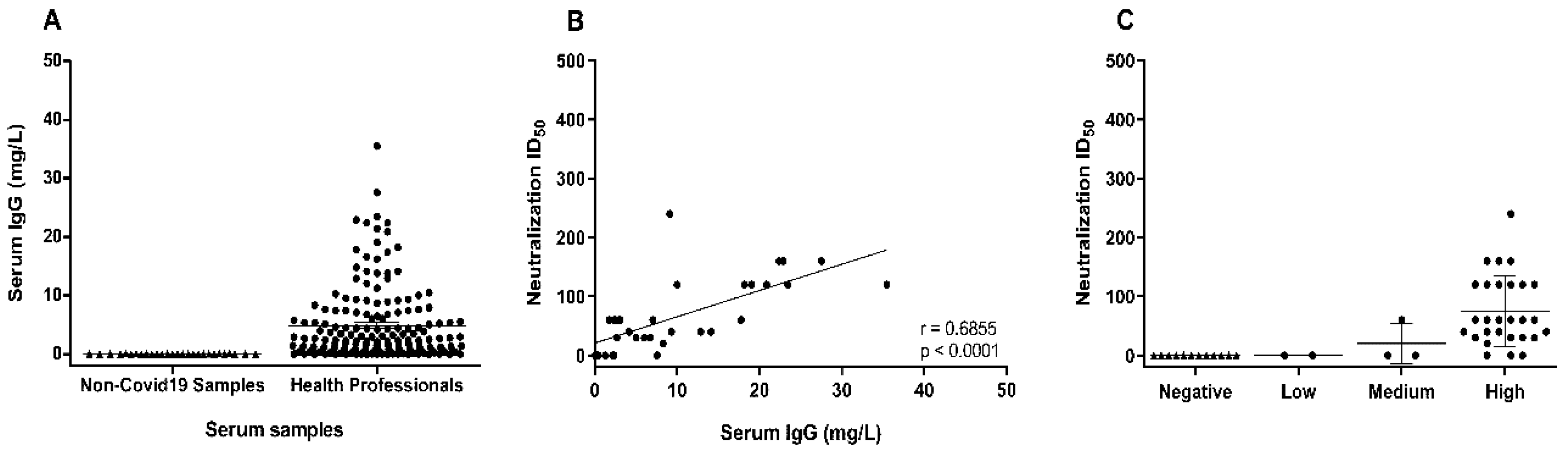
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.M.; Wang, W.; Song, Z.G.; Hu, Y.; Tao, Z.W.; Tian, J.H.; Pei, Y.Y.; et al. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef]
- Peeri, N.C.; Shrestha, N.; Rahman, M.S.; Zaki, R.; Tan, Z.; Bibi, S.; Baghbanzadeh, M.; Aghamohammadi, N.; Zhang, W.; Haque, U. The SARS, MERS and novel coronavirus (COVID-19) epidemics, the newest and biggest global health threats: What lessons have we learned? Int. J. Epidemiol. 2020, 49, 717–726. [Google Scholar] [CrossRef]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef]
- Xu, Z.; Shi, L.; Wang, Y.; Zhang, J.; Huang, L.; Zhang, C.; Liu, S.; Zhao, P.; Liu, H.; Zhu, L.; et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020, 8, 420–422. [Google Scholar] [CrossRef]
- Grifoni, A.; Weiskopf, D.; Ramirez, S.I.; Mateus, J.; Dan, J.M.; Moderbacher, C.R.; Rawlings, S.A.; Sutherland, A.; Premkumar, L.; Jadi, R.S.; et al. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell 2020, 181, 1489–1501.e15. [Google Scholar] [CrossRef] [PubMed]
- Weiskopf, D.; Schmitz, K.S.; Raadsen, M.P.; Grifoni, A.; Okba, N.M.A.; Endeman, H.; van den Akker, J.P.C.; Molenkamp, R.; Koopmans, M.P.G.; van Gorp, E.C.M.; et al. Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci. Immunol. 2020, 5. [Google Scholar] [CrossRef]
- Braun, J.; Loyal, L.; Frentsch, M.; Wendisch, D.; Georg, P.; Kurth, F.; Hippenstiel, S.; Dingeldey, M.; Kruse, B.; Fauchere, F.; et al. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature 2020. [Google Scholar] [CrossRef]
- Zhao, J.; Yuan, Q.; Wang, H.; Liu, W.; Liao, X.; Su, Y.; Wang, X.; Yuan, J.; Li, T.; Li, J.; et al. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2020. [Google Scholar] [CrossRef]
- Xiao, A.T.; Gao, C.; Zhang, S. Profile of specific antibodies to SARS-CoV-2: The first report. J. Infect. 2020, 81, 147–178. [Google Scholar] [CrossRef]
- Robbiani, D.F.; Gaebler, C.; Muecksch, F.; Lorenzi, J.C.C.; Wang, Z.; Cho, A.; Agudelo, M.; Barnes, C.O.; Gazumyan, A.; Finkin, S.; et al. Convergent Antibody Responses to SARS-CoV-2 Infection in Convalescent Individuals. Nature 2020. [Google Scholar] [CrossRef]
- Odak, I.; Barros-Martins, J.; Bosnjak, B.; Stahl, K.; David, S.; Wiesner, O.; Busch, M.; Hoeper, M.M.; Pink, I.; Welte, T.; et al. Reappearance of effector T cells is associated with recovery from COVID-19. EBioMedicine 2020, 57, 102885. [Google Scholar] [CrossRef]
- Liu, J.; Li, S.; Liu, J.; Liang, B.; Wang, X.; Wang, H.; Li, W.; Tong, Q.; Yi, J.; Zhao, L.; et al. Longitudinal characteristics of lymphocyte responses and cytokine profiles in the peripheral blood of SARS-CoV-2 infected patients. EBioMedicine 2020, 55, 102763. [Google Scholar] [CrossRef] [PubMed]
- Qin, C.; Zhou, L.; Hu, Z.; Zhang, S.; Yang, S.; Tao, Y.; Xie, C.; Ma, K.; Shang, K.; Wang, W.; et al. Dysregulation of Immune Response in Patients with Coronavirus 2019 (COVID-19) in Wuhan, China. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2020, 71, 762–768. [Google Scholar] [CrossRef]
- Zheng, M.; Gao, Y.; Wang, G.; Song, G.; Liu, S.; Sun, D.; Xu, Y.; Tian, Z. Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cell. Mol. Immunol. 2020, 17, 533–535. [Google Scholar] [CrossRef] [PubMed]
- McCreary, E.K.; Pogue, J.M. Coronavirus Disease 2019 Treatment: A Review of Early and Emerging Options. Open Forum Infect. Dis. 2020, 7, ofaa105. [Google Scholar] [CrossRef]
- Fischer, J.C.; Zanker, K.; van Griensven, M.; Schneider, M.; Kindgen-Milles, D.; Knoefel, W.T.; Lichtenberg, A.; Tamaskovics, B.; Djiepmo-Njanang, F.J.; Budach, W.; et al. The role of passive immunization in the age of SARS-CoV-2: An update. Eur. J. Med. Res. 2020, 25, 16. [Google Scholar] [CrossRef]
- Kumar, S.; Sharma, V.; Priya, K. Battle against COVID-19: Efficacy of Convalescent Plasma as an emergency therapy. Am. J. Emerg. Med. 2020. [Google Scholar] [CrossRef]
- Perotti, C.; Baldanti, F.; Bruno, R.; Del Fante, C.; Seminari, E.; Casari, S.; Percivalle, E.; Glingani, C.; Musella, V.; Belliato, M.; et al. Mortality reduction in 46 severe Covid-19 patients treated with hyperimmune plasma. A proof of concept single arm multicenter trial. Haematologica 2020. [Google Scholar] [CrossRef]
- Duan, K.; Liu, B.; Li, C.; Zhang, H.; Yu, T.; Qu, J.; Zhou, M.; Chen, L.; Meng, S.; Hu, Y.; et al. Effectiveness of convalescent plasma therapy in severe COVID-19 patients. Proc. Natl. Acad. Sci. USA 2020, 117, 9490–9496. [Google Scholar] [CrossRef]
- Liu, S.T.H.; Lin, H.M.; Baine, I.; Wajnberg, A.; Gumprecht, J.P.; Rahman, F.; Rodriguez, D.; Tandon, P.; Bassily-Marcus, A.; Bander, J.; et al. Convalescent plasma treatment of severe COVID-19: A propensity score-matched control study. Nat. Med. 2020, 26, 1708–1713. [Google Scholar] [CrossRef]
- Simonovich, V.A.; Burgos Pratx, L.D.; Scibona, P.; Beruto, M.V.; Vallone, M.G.; Vazquez, C.; Savoy, N.; Giunta, D.H.; Perez, L.G.; Sanchez, M.D.L.; et al. A Randomized Trial of Convalescent Plasma in Covid-19 Severe Pneumonia. N. Engl. J. Med. 2021, 384, 619–629. [Google Scholar] [CrossRef] [PubMed]
- Marano, G.; Vaglio, S.; Pupella, S.; Facco, G.; Catalano, L.; Liumbruno, G.M.; Grazzini, G. Convalescent plasma: New evidence for an old therapeutic tool? Blood Transfus. 2016, 14, 152–157. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Joyner, M.J.; Senefeld, J.W.; Klassen, S.A.; Mills, J.R.; Johnson, P.W.; Theel, E.S.; Wiggins, C.C.; Bruno, K.A.; Klompas, A.M.; Lesser, E.R.; et al. Effect of Convalescent Plasma on Mortality among Hospitalized Patients with COVID-19: Initial Three-Month Experience. medRxiv Prepr. Serv. Health Sci. 2020. [Google Scholar] [CrossRef]
- Li, L.; Zhang, W.; Hu, Y.; Tong, X.; Zheng, S.; Yang, J.; Kong, Y.; Ren, L.; Wei, Q.; Mei, H.; et al. Effect of Convalescent Plasma Therapy on Time to Clinical Improvement in Patients with Severe and Life-threatening COVID-19: A Randomized Clinical Trial. JAMA J. Am. Med Assoc. 2020, 324, 460–470. [Google Scholar] [CrossRef]
- Gharbharan, A.; Jordans, C.C.E.; GeurtsvanKessel, C.; den Hollander, J.G.; Karim, F.; Mollema, F.P.N.; Stalenhoef, J.E.; Dofferhoff, A.; Ludwig, I.; Koster, A.; et al. Convalescent Plasma for COVID-19. A randomized clinical trial. medRxiv Prepr. Serv. Health Sci. 2020. [Google Scholar] [CrossRef]
- Avendano-Sola, C.; Ramos-Martinez, A.; Munez-Rubio, E.; Ruiz-Antoran, B.; Malo de Molina, R.; Torres, F.; Fernandez-Cruz, A.; Callejas-Diaz, A.; Calderon, J.; Payares-Herrera, C.; et al. Convalescent Plasma for COVID-19: A multicenter, randomized clinical trial. medRxiv Prepr. Serv. Health Sci. 2020. [Google Scholar] [CrossRef]
- Agarwal, A.; Mukherjee, A.; Kumar, G.; Chatterjee, P.; Bhatnagar, T.; Malhotra, P.; Collaborators, P.T. Convalescent plasma in the management of moderate covid-19 in adults in India: Open label phase II multicentre randomised controlled trial (PLACID Trial). Bmj 2020, 371, m3939. [Google Scholar] [CrossRef]
- Wu, L.P.; Wang, N.C.; Chang, Y.H.; Tian, X.Y.; Na, D.Y.; Zhang, L.Y.; Zheng, L.; Lan, T.; Wang, L.F.; Liang, G.D. Duration of antibody responses after severe acute respiratory syndrome. Emerg. Infect. Dis. 2007, 13, 1562–1564. [Google Scholar] [CrossRef] [PubMed]
- Payne, D.C.; Iblan, I.; Rha, B.; Alqasrawi, S.; Haddadin, A.; Al Nsour, M.; Alsanouri, T.; Ali, S.S.; Harcourt, J.; Miao, C.; et al. Persistence of Antibodies against Middle East Respiratory Syndrome Coronavirus. Emerg. Infect. Dis. 2016, 22, 1824–1826. [Google Scholar] [CrossRef]
- Wajnberg, A.; Amanat, F.; Firpo, A.; Altman, D.R.; Bailey, M.J.; Mansour, M.; McMahon, M.; Meade, P.; Mendu, D.R.; Muellers, K.; et al. Robust neutralizing antibodies to SARS-CoV-2 infection persist for months. Science. 2020. [Google Scholar] [CrossRef] [PubMed]
- Dan, J.M.; Mateus, J.; Kato, Y.; Hastie, K.M.; Faliti, C.E.; Ramirez, S.I.; Frazier, A.; Yu, E.D.; Grifoni, A.; Rawlings, S.A.; et al. Immunological memory to SARS-CoV-2 assessed for greater than six months after infection. bioRxiv Prepr. Serv. Biol. 2020. [Google Scholar] [CrossRef]
- Premkumar, L.; Segovia-Chumbez, B.; Jadi, R.; Martinez, D.R.; Raut, R.; Markmann, A.; Cornaby, C.; Bartelt, L.; Weiss, S.; Park, Y.; et al. The receptor binding domain of the viral spike protein is an immunodominant and highly specific target of antibodies in SARS-CoV-2 patients. Sci. Immunol. 2020, 5. [Google Scholar] [CrossRef]
- Liu, L.; Wang, P.; Nair, M.S.; Yu, J.; Rapp, M.; Wang, Q.; Luo, Y.; Chan, J.F.; Sahi, V.; Figueroa, A.; et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature 2020, 584, 450–456. [Google Scholar] [CrossRef]
- Amanat, F.; Stadlbauer, D.; Strohmeier, S.; Nguyen, T.H.O.; Chromikova, V.; McMahon, M.; Jiang, K.; Arunkumar, G.A.; Jurczyszak, D.; Polanco, J.; et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med. 2020, 26, 1033–1036. [Google Scholar] [CrossRef]
- Garcia-Garcia, A.; Ceballos-Laita, L.; Serna, S.; Artschwager, R.; Reichardt, N.C.; Corzana, F.; Hurtado-Guerrero, R. Structural basis for substrate specificity and catalysis of alpha1,6-fucosyltransferase. Nat. Commun. 2020, 11, 973. [Google Scholar] [CrossRef]
- Rambaut, A.; Holmes, E.C.; O’Toole, A.; Hill, V.; McCrone, J.T.; Ruis, C.; du Plessis, L.; Pybus, O.G. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishnan, M.A. Determination of 50% endpoint titer using a simple formula. World J. Virol. 2016, 5, 85–86. [Google Scholar] [CrossRef] [PubMed]
- Hou, H.; Wang, T.; Zhang, B.; Luo, Y.; Mao, L.; Wang, F.; Wu, S.; Sun, Z. Detection of IgM and IgG antibodies in patients with coronavirus disease 2019. Clin. Transl. Immunol. 2020, 9, e01136. [Google Scholar] [CrossRef]
- Liu, X.; Wang, J.; Xu, X.; Liao, G.; Chen, Y.; Hu, C.H. Patterns of IgG and IgM antibody response in COVID-19 patients. Emerg. Microbes Infect. 2020, 9, 1269–1274. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Tong, X.; Chen, H.; He, R.; Lv, Q.; Yang, R.; Zhao, L.; Wang, J.; Xu, H.; Liu, C.; et al. Characteristics and serological patterns of COVID-19 convalescent plasma donors: Optimal donors and timing of donation. Transfusion 2020. [Google Scholar] [CrossRef]
- Salazar, E.; Kuchipudi, S.V.; Christensen, P.A.; Eagar, T.N.; Yi, X.; Zhao, P.; Jin, Z.; Long, S.W.; Olsen, R.J.; Chen, J.; et al. Relationship between Anti-Spike Protein Antibody Titers and SARS-CoV-2 In Vitro Virus Neutralization in Convalescent Plasma. bioRxiv Prepr. Serv. Biol. 2020. [Google Scholar] [CrossRef]
- Manenti, A.; Maggetti, M.; Casa, E.; Martinuzzi, D.; Torelli, A.; Trombetta, C.M.; Marchi, S.; Montomoli, E. Evaluation of SARS-CoV-2 neutralizing antibodies using a CPE-based colorimetric live virus micro-neutralization assay in human serum samples. J. Med Virol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Morrison, B.J.; Martin, N.J.; Rehman, T.; Ewing, D.; Dewar, R.L.; Metcalf, J.; Sun, P.; Beigel, J.; Luke, T.C.; Raviprakash, K. Influence of sample collection tube method, anticoagulant-containing plasma versus serum, on influenza virus hemagglutination inhibition titer and microneutralization titer serological assays. BMC Health Serv. Res. 2018, 18, 651. [Google Scholar] [CrossRef] [PubMed]
- Suthar, M.S.; Zimmerman, M.; Kauffman, R.; Mantus, G.; Linderman, S.; Vanderheiden, A.; Nyhoff, L.; Davis, C.; Adekunle, S.; Affer, M.; et al. Rapid generation of neutralizing antibody responses in COVID-19 patients. medRxiv Prepr. Serv. Health Sci. 2020. [Google Scholar] [CrossRef]
- Harvala, H.; Robb, M.; Watkins, N.; Ijaz, S.; Dicks, S.; Patel, M.; Supasa, P.; Dejnirattisai, W.; Liu, C.; Mongkolsapaya, J.; et al. Convalescent plasma therapy for the treatment of patients with COVID-19: Assessment of methods available for antibody detection and their correlation with neutralising antibody levels. medRxiv Prepr. Serv. Health Sci. 2020. [Google Scholar] [CrossRef]
- Collier, D.A.; De Marco, A.; Ferreira, I.A.T.M.; Meng, B.; Datir, R.; Walls, A.C.; Kemp, S.S.A.; Bassi, J.; Pinto, D.; Fregni, C.S.; et al. SARS-CoV-2 B.1.1.7 escape from mRNA vaccine-elicited neutralizing antibodies. medRxiv Prepr. Serv. Health Sci. 2021. [Google Scholar] [CrossRef]
- Wang, Z.; Schmidt, F.; Weisblum, Y.; Muecksch, F.; Barnes, C.O.; Finkin, S.; Schaefer-Babajew, D.; Cipolla, M.; Gaebler, C.; Lieberman, J.A.; et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. bioRxiv Prepr. Serv. Biol. 2021. [Google Scholar] [CrossRef]
- Wu, K.; Werner, A.P.; Moliva, J.I.; Koch, M.; Choi, A.; Stewart-Jones, G.B.E.; Bennett, H.; Boyoglu-Barnum, S.; Shi, W.; Graham, B.S.; et al. mRNA-1273 vaccine induces neutralizing antibodies against spike mutants from global SARS-CoV-2 variants. bioRxiv Prepr. Serv. Biol. 2021. [Google Scholar] [CrossRef]
- Andreano, E.; Piccini, G.; Licastro, D.; Casalino, L.; Johnson, N.V.; Paciello, I.; Monego, S.D.; Pantano, E.; Manganaro, N.; Manenti, A.; et al. SARS-CoV-2 escape in vitro from a highly neutralizing COVID-19 convalescent plasma. bioRxiv Prepr. Serv. Biol. 2020. [Google Scholar] [CrossRef]
- Greaney, A.J.; Starr, T.N.; Gilchuk, P.; Zost, S.J.; Binshtein, E.; Loes, A.N.; Hilton, S.K.; Huddleston, J.; Eguia, R.; Crawford, K.H.D.; et al. Complete mapping of mutations to the SARS-CoV-2 spike receptor-binding domain that escape antibody recognition. bioRxiv Prepr. Serv. Biol. 2020. [Google Scholar] [CrossRef]
- Wibmer, C.K.; Ayres, F.; Hermanus, T.; Madzivhandila, M.; Kgagudi, P.; Lambson, B.E.; Vermeulen, M.; van den Berg, K.; Rossouw, T.; Boswell, M.; et al. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. bioRxiv Prepr. Serv. Biol. 2021. [Google Scholar] [CrossRef]
- Abolghasemi, H.; Eshghi, P.; Cheraghali, A.M.; Imani Fooladi, A.A.; Bolouki Moghaddam, F.; Imanizadeh, S.; Moeini Maleki, M.; Ranjkesh, M.; Rezapour, M.; Bahramifar, A.; et al. Clinical efficacy of convalescent plasma for treatment of COVID-19 infections: Results of a multicenter clinical study. Transfus. Apher. Sci. Off. J. World Apher. Assoc. Off. J. Eur. Soc. Haemapheresis 2020, 102875. [Google Scholar] [CrossRef] [PubMed]
- Piechotta, V.; Chai, K.L.; Valk, S.J.; Doree, C.; Monsef, I.; Wood, E.M.; Lamikanra, A.; Kimber, C.; McQuilten, Z.; So-Osman, C.; et al. Convalescent plasma or hyperimmune immunoglobulin for people with COVID-19: A living systematic review. Cochrane Database Syst. Rev. 2020, 7, CD013600. [Google Scholar] [CrossRef] [PubMed]
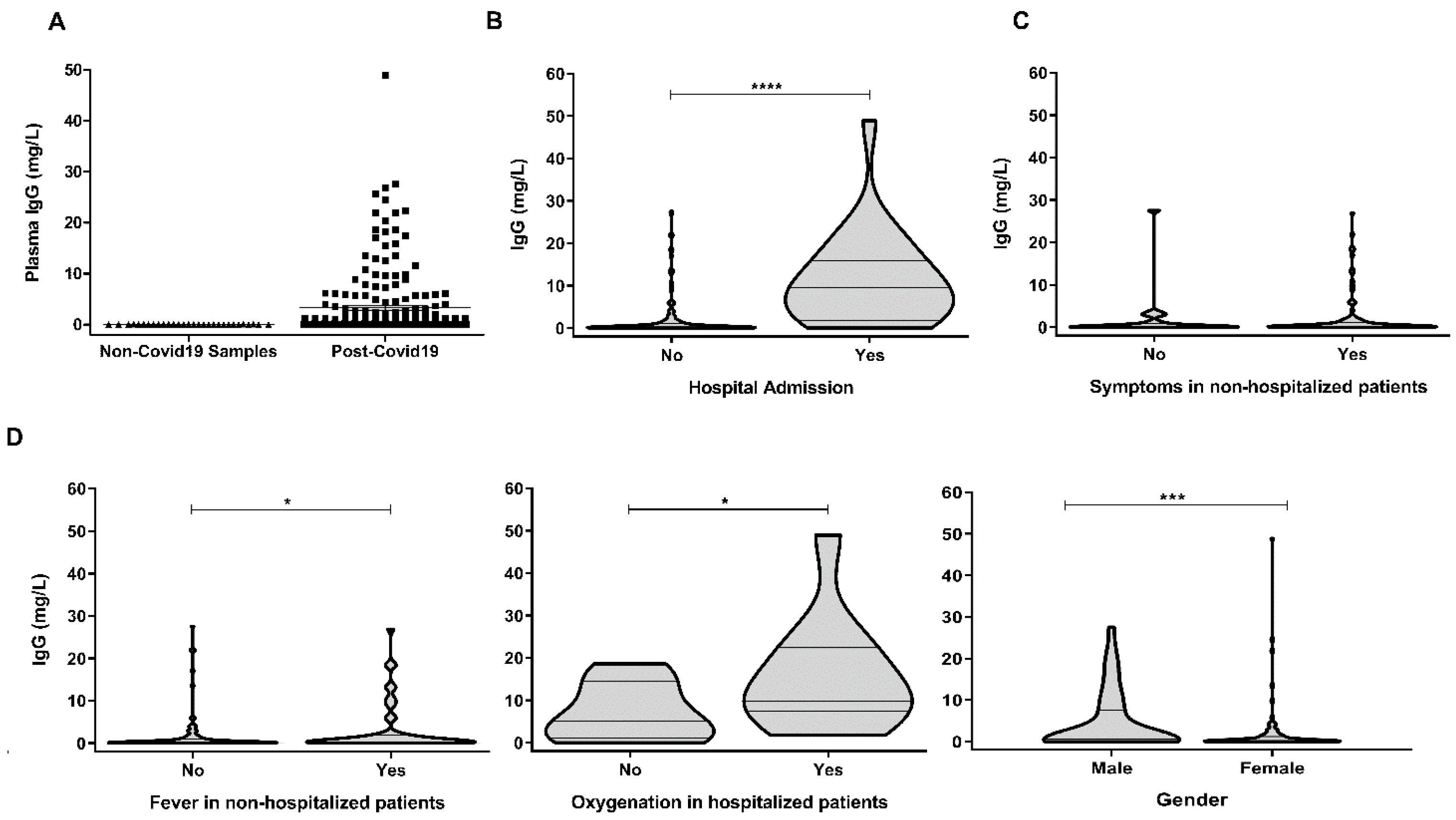
| Age (Median; Range) | 44 (20–64) |
|---|---|
| Gender (female) | 92/177 (52%) |
| Asymptomatic | 22/177 (12%) |
| Time from symptom onset (median; range) | 45 (22–111) |
| Moderate/severe COVID-19 (hospitalization) | 24/177 (13.6%) |
| Fever, non-hospitalized | 47/153 (30.7%) |
| Oxygenation, hospitalized | 11/24 (45.8%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santiago, L.; Uranga-Murillo, I.; Arias, M.; González-Ramírez, A.M.; Macías-León, J.; Moreo, E.; Redrado, S.; García-García, A.; Taleb, V.; Lira-Navarrete, E.; et al. Determination of the Concentration of IgG against the Spike Receptor-Binding Domain That Predicts the Viral Neutralizing Activity of Convalescent Plasma and Serum against SARS-CoV-2. Biology 2021, 10, 208. https://doi.org/10.3390/biology10030208
Santiago L, Uranga-Murillo I, Arias M, González-Ramírez AM, Macías-León J, Moreo E, Redrado S, García-García A, Taleb V, Lira-Navarrete E, et al. Determination of the Concentration of IgG against the Spike Receptor-Binding Domain That Predicts the Viral Neutralizing Activity of Convalescent Plasma and Serum against SARS-CoV-2. Biology. 2021; 10(3):208. https://doi.org/10.3390/biology10030208
Chicago/Turabian StyleSantiago, Llipsy, Iratxe Uranga-Murillo, Maykel Arias, Andrés Manuel González-Ramírez, Javier Macías-León, Eduardo Moreo, Sergio Redrado, Ana García-García, Víctor Taleb, Erandi Lira-Navarrete, and et al. 2021. "Determination of the Concentration of IgG against the Spike Receptor-Binding Domain That Predicts the Viral Neutralizing Activity of Convalescent Plasma and Serum against SARS-CoV-2" Biology 10, no. 3: 208. https://doi.org/10.3390/biology10030208
APA StyleSantiago, L., Uranga-Murillo, I., Arias, M., González-Ramírez, A. M., Macías-León, J., Moreo, E., Redrado, S., García-García, A., Taleb, V., Lira-Navarrete, E., Hurtado-Guerrero, R., Aguilo, N., del Mar Encabo-Berzosa, M., Hidalgo, S., Galvez, E. M., Ramirez-Labrada, A., de Miguel, D., Benito, R., Miranda, P., ... Pardo, J. (2021). Determination of the Concentration of IgG against the Spike Receptor-Binding Domain That Predicts the Viral Neutralizing Activity of Convalescent Plasma and Serum against SARS-CoV-2. Biology, 10(3), 208. https://doi.org/10.3390/biology10030208







