Mucociliary Respiratory Epithelium Integrity in Molecular Defense and Susceptibility to Pulmonary Viral Infections
Abstract
Simple Summary
Abstract
1. Introduction
2. Organization and Components of the Human Respiratory System
3. Structure and Composition of the MCC Apparatus in Humans
3.1. Functional Role of Cilia in MCC
3.2. Components of the Propeller Machinery of the MCC Apparatus
3.3. Molecular Network of the Lung Cilia and MCC Machinery Regulating Mucociliary Clearance in Humans: A Protein Network Analysis
3.4. Physiological Importance of the MCC Apparatus and Associated Disease
3.5. Ciliopathies and Their Relevance to MCC
3.6. Mucociliary Dysfunctions upon Polymicrobial Infections
4. Coronavirus Disease Manifestation in the Respiratory System
4.1. Coronavirus-Induced Dysfunctions of the Lung Cilia
4.2. SARS-CoV-2-Manifested Implications on the Molecular Machinery Regulating Mucociliary Clearance
4.3. Modulation of Gene Expression of the MCC System upon SARS-CoV-2 Infection
5. Molecular Basis of Repurposing of Drugs for Mitigating SARS-CoV-2-Induced Lung Cilia and MCC Dysfunctions
6. Concluding Remarks
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Whitsett, J.A. Airway epithelial differentiation and mucociliary clearance. Ann. Am. Thorac. Soc. 2018, 15, S143–S148. [Google Scholar] [CrossRef] [PubMed]
- Munkholm, M.; Mortensen, J. Mucociliary clearance: Pathophysiological aspects. Clin. Physiol. Funct. Imaging 2014, 34, 171–177. [Google Scholar] [CrossRef] [PubMed]
- Crystal, R.G.; Randell, S.H.; Engelhardt, J.F.; Voynow, J.; Sunday, M.E. Airway epithelial cells: Current concepts and challenges. Proc. Am. Thorac. Soc. 2008, 5, 772–777. [Google Scholar] [CrossRef] [PubMed]
- Jeffrey, P.K. The development of large and small airways. Am. J. Respir. Crit. Care Med. 1998, 157, S174–S180. [Google Scholar] [CrossRef]
- Tilley, A.E.; Walters, M.S.; Shaykhiev, R.; Crystal, R.G. Cilia dysfunction in lung disease. Annu. Rev. Physiol. 2015, 77, 379–406. [Google Scholar] [CrossRef]
- Wisnivesky, J.P.; De-Torres, J.P. The global burden of pulmonary diseases: Most prevalent problems and opportunities for improvement. Ann. Glob. Health 2019, 85, 85. [Google Scholar] [CrossRef]
- Bustamante-Marin, X.M.; Ostrowski, L.E. Cilia and mucociliary clearance. Cold Spring Harb. Perspect. Biol. 2017, 9, a028241. [Google Scholar] [CrossRef]
- Sahin-Yilmaz, A.; Naclerio, R.M. Anatomy and physiology of the upper airway. Proc. Am. Thorac. Soc. 2011, 8, 31–39. [Google Scholar] [CrossRef]
- Thurlbeck, W.M. Postnatal growth and development of the lung. Am. Rev. Respir. Dis. 1975, 111, 803–844. [Google Scholar]
- Wine, J.J.; Joo, N.S. Submucosal glands and airway defense. Proc. Am. Thorac. Soc. 2004, 1, 47–53. [Google Scholar] [CrossRef]
- Rawlins, E.L.; Hogan, B.L.M. Ciliated epithelial cell lifespan in the mouse trachea and lung. Am. J. Physiol. Lung Cell. Mol. 2008, 295, L231–L234. [Google Scholar] [CrossRef] [PubMed]
- Mercer, R.R.; Russell, M.L.; Roggli, V.L.; Crapo, J.D. Cell number and distribution in human and rat airways. Am. J. Respir. Cell Mol. Biol. 1994, 10, 613–624. [Google Scholar] [CrossRef] [PubMed]
- Knight, D.A.; Holgate, S.T. The airway epithelium: Structural and functional properties in health and disease. Respirology 2003, 8, 432–446. [Google Scholar] [CrossRef]
- Kikkawa, M. Big steps toward understanding dynein. J. Cell Biol. 2013, 202, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Knowles, M.R.; Boucher, R.C. Mucus clearance as a primary innate defense mechanism for mammalian airways. J. Clin. Investig. 2002, 109, 571–577. [Google Scholar] [CrossRef]
- Okubo, T.; Knoepfler, P.S.; Eisenman, R.N.; Hogan, B.L. Nmyc plays an essential role during lung development as a dosage-sensitive regulator of progenitor cell proliferation and differentiation. Development 2005, 132, 1363–1374. [Google Scholar] [CrossRef]
- Zhou, F.; Narasimhan, V.; Shboul, M.; Chong, Y.L.; Reversade, B.; Roy, S. Gmnc is a master regulator of the multiciliated cell differentiation program. Curr. Biol. 2015, 25, 3267–3273. [Google Scholar] [CrossRef]
- Stubbs, J.L.; Vladar, E.K.; Axelrod, J.D.; Kintner, C. Multicilin promotes centriole assembly and ciliogenesis during multiciliate cell differentiation. Nat. Cell Biol. 2012, 14, 140–147. [Google Scholar] [CrossRef]
- Ma, L.; Quigley, I.; Omran, H.; Kintner, C. Multicilin drives centriole biogenesis via E2f proteins. Genes Dev. 2014, 28, 1461–1471. [Google Scholar] [CrossRef]
- Vladar, E.K.; Mitchell, B.J. It’s a family act: The geminin triplets take center stage in motile ciliogenesis. EMBO J. 2016, 35, 904–906. [Google Scholar] [CrossRef]
- You, Y.; Huang, T.; Richer, E.J.; Schmidt, J.-E.H.; Zabner, J.; Borok, Z.; Brody, S.L. Role of f-box factor foxj1 in differentiation of ciliated airway epithelial cells. Am. J. Physiol. Lung Cell. Mol. Physiol. 2004, 286, L650–L657. [Google Scholar] [CrossRef]
- Didon, L.; Zwick, R.K.; Chao, I.W.; Walters, M.S.; Wang, R.; Hackett, N.R.; Crystal, R.G. RFX3 modulation of FOXJ1 regulation of cilia genes in the human airway epithelium. Respir. Res. 2013, 14, 70. [Google Scholar] [CrossRef]
- Pan, J.-H.; Adair-Kirk, T.L.; Patel, A.C.; Huang, T.; Yozamp, N.S.; Xu, J.; Reddy, E.P.; Byers, D.E.; Pierce, R.A.; Holtzman, M.J.; et al. Myb permits multilineage airway epithelial cell differentiation. Stem Cells 2014, 32, 3245–3256. [Google Scholar] [CrossRef]
- Rock, J.R.; Gao, X.; Xue, Y.; Randell, S.H.; Kong, Y.-Y.; Hogan, B. Notch-dependent differentiation of adult airway basal stem cells. Cell Stem Cell 2011, 8, 639–648. [Google Scholar] [CrossRef]
- Chen, G.; Korfhagen, T.R.; Xu, Y.; Kitzmiller, J.; Wert, S.E.; Maeda, Y.; Gregorieff, A.; Clevers, H.; Whitsett, J.A. SPDEF is required for mouse pulmonary goblet cell differentiation and regulates a network of genes associated with mucus production. J. Clin. Investig. 2009, 119, 2914–2924. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Korfhagen, T.R.; Karp, C.L.; Impey, S.; Xu, Y.; Randell, S.H.; Kitzmiller, J.; Maeda, Y.; Haitchi, H.M.; Sridharan, A.; et al. Foxa3 Induces Goblet Cell Metaplasia and Inhibits Innate Antiviral Immunity. Am. J. Respir. Crit. Care Med. 2014, 189, 301–313. [Google Scholar] [CrossRef] [PubMed]
- Rajavelu, P.; Chen, G.; Xu, Y.; Kitzmiller, J.A.; Korfhagen, T.R.; Whitsett, J.A. Airway epithelial SPDEF integrates goblet cell differentiation and pulmonary Th2 inflammation. J. Clin. Investig. 2015, 125, 2021–2031. [Google Scholar] [CrossRef] [PubMed]
- Thornton, D.J.; Rousseau, K.; McGuckin, M.A. Structure and function of the polymeric mucins in airways mucus. Annu. Rev. Physiol. 2008, 70, 459–486. [Google Scholar] [CrossRef]
- Rogan, M.P.; Geraghty, P.; Greene, C.; O’Neill, S.; Taggart, C.C.; McElvaney, N.G. Antimicrobial proteins and polypeptides in pulmonary innate defence. Respir. Res. 2006, 7, 29. [Google Scholar] [CrossRef]
- Thai, P.; Loukoianov, A.; Wachi, S.; Wu, R. Regulation of airway mucin gene expression. Annu. Rev. Physiol. 2008, 70, 405–429. [Google Scholar] [CrossRef]
- Rogers, D.F. Airway mucus hypersecretion in asthma: An undervalued pathology? Curr. Opin. Pharmacol. 2004, 4, 241–250. [Google Scholar] [CrossRef] [PubMed]
- Braschi, B.; Denny, P.; Gray, K.A.; Jones, T.E.M.; Seal, R.L.; Tweedie, S.; Yates, B.; Bruford, E.A. Genenames.org: The HGNC and VGNC resources in 2019. Nucleic Acids Res. 2019, 47, D786–D792. [Google Scholar] [CrossRef] [PubMed]
- Hamed, R.; Fiegel, J. Synthetic tracheal mucus with native rheological and surface tension properties. J. Biomed. Mater. Res. Part. A 2014, 102, 1788–1798. [Google Scholar] [CrossRef] [PubMed]
- Kirkham, S.; Sheehan, J.K.; Knight, D.; Richardson, P.S.; Thornton, D.J. Heterogeneity of airways mucus: Variations in the amounts and glycoforms of the major oligomeric mucins MUC5AC and MUC5B. Biochem. J. 2002, 361, 537–546. [Google Scholar] [CrossRef] [PubMed]
- Davies, J.R.; Svitacheva, N.; Lannefors, L.; Kornfalt, R.; Carlstedt, I. Identification of MUC5B, MUC5AC and small amounts of MUC2 mucins in cystic fibrosis airway secretions. Biochem. J. 1999, 344, 321–330. [Google Scholar] [CrossRef]
- Hovenberg, H.W.; Davies, J.R.; Herrmann, A.; Linden, C.J.; Carlstedt, I. MUC5AC, but not MUC2, is a prominent mucin in respiratory secretions. Glycoconj. J. 1996, 13, 839–847. [Google Scholar] [CrossRef]
- Tarran, R.; Button, B.; Picher, M.; Paradiso, A.M.; Ribeiro, C.M.; Lazarowski, E.R.; Zhang, L.; Collins, P.L.; Pickles, R.J.; Fredberg, J.J.; et al. Normal and cystic fibrosis airway surface liquid homeostasis. The effects of phasic shear stress and viral infections. J. Biol. Chem. 2005, 280, 35751–35759. [Google Scholar] [CrossRef]
- Button, B.; Cai, L.-H.; Ehre, C.; Kesimer, M.; Hill, D.B.; Sheehan, J.K.; Boucher, R.C.; Rubinstein, M. A periciliary brush promotes the lung health by separating the mucus layer from airway epithelia. Science 2012, 337, 937–941. [Google Scholar] [CrossRef]
- Hattrup, C.L.; Gendler, S.J. Structure and function of the cell surface (tethered) mucins. Annu. Rev. Physiol. 2008, 70, 431–457. [Google Scholar] [CrossRef]
- Sleigh, M.A.; Blake, J.R.; Liron, N. The propulsion of mucus by cilia. Am. Rev. Respir. Dis. 1988, 137, 726–741. [Google Scholar] [CrossRef]
- Jain, R.; Ray, J.M.; Pan, J.-H.; Brody, S.L. Sex hormone–dependent regulation of cilia beat frequency in airway epithelium. Am. J. Respir. Cell Mol. Biol. 2012, 46, 446–453. [Google Scholar] [CrossRef] [PubMed]
- Jiao, J.; Wang, H.; Lou, W.; Jin, S.; Fan, E.; Li, Y.; Han, D.-M.; Zhang, L. Regulation of ciliary beat frequency by the nitric oxide signaling pathway in mouse nasal and tracheal epithelial cells. Exp. Cell Res. 2011, 317, 2548–2553. [Google Scholar] [CrossRef] [PubMed]
- Salathe, M. Regulation of mammalian ciliary beating. Annu. Rev. Physiol. 2007, 69, 401–422. [Google Scholar] [CrossRef]
- Schmid, A.; Salathe, M. Ciliary beat co-ordination by calcium. Biol. Cell 2011, 103, 159–169. [Google Scholar] [CrossRef] [PubMed]
- Button, B.; Boucher, R.C. Role of mechanical stress in regulating airway surface hydration and mucus clearance rates. Respir. Physiol. Neurobiol. 2008, 163, 189–201. [Google Scholar] [CrossRef]
- Fahy, J.V.; Dickey, B.F. Airway mucus function and dysfunction. N. Engl. J. Med. 2010, 363, 2233–2247. [Google Scholar] [CrossRef]
- Agius, A.M.; Smallman, L.A.; Pahor, A.L. Age, smoking and nasal ciliary beat frequency. Clin. Otolaryngol. Allied Sci. 1998, 23, 227–230. [Google Scholar] [CrossRef]
- Muns, G.; Singer, P.; Wolf, F.; Rubinstein, I. Impaired nasal mucociliary clearance in long-distance runners. Int. J. Sports Med. 1995, 16, 209–213. [Google Scholar] [CrossRef]
- Yaghi, A.; Zaman, A.; Cox, G.; Dolovich, M. Ciliary beating is depressed in nasal cilia from chronic obstructive pulmonary disease subjects. Respir. Med. 2012, 106, 1139–1147. [Google Scholar] [CrossRef]
- Ho, J.C.; Chan, K.N.; Hu, W.H.; Lam, W.K.; Zheng, L.; Tipoe, G.L.; Sun, J.; Leung, R.; Tsang, K.W. The effect of aging on nasal mucociliary clearance, beat frequency, and ultrastructure of respiratory cilia. Am. J. Respir. Crit. Care Med. 2001, 163, 983–988. [Google Scholar] [CrossRef]
- Shah, A.S.; Ben-Shahar, Y.; Moninger, T.O.; Kline, J.N.; Welsh, M.J. Motile cilia of human airway epithelia are chemosensory. Science 2009, 325, 1131–1134. [Google Scholar] [CrossRef] [PubMed]
- Avidor-Reiss, T.; Maer, A.M.; Koundakjian, E.; Polyanovsky, A.; Keil, T.; Subramaniam, S.; Zuker, C.S. Decoding cilia function: Defining specialized genes required for compartmentalized cilia biogenesis. Cell 2004, 117, 527–539. [Google Scholar] [CrossRef]
- Taschner, M.; Lorentzen, E. The intraflagellar transport machinery. Cold Spring Harb. Perspect. Biol. 2016, 8, a028092. [Google Scholar] [CrossRef] [PubMed]
- Bisgrove, B.W.; Yost, H.J. The roles of cilia in developmental disorders and disease. Development 2006, 133, 4131–4143. [Google Scholar] [CrossRef]
- Singla, V.; Reiter, J.F. The primary cilium as the cell’s antenna: Signaling at a sensory organelle. Science 2006, 313, 629–633. [Google Scholar] [CrossRef]
- Ishikawa, H.; Marshall, W.F. Ciliogenesis: Building the cell’s antenna. Nat. Rev. Mol. Cell Biol. 2011, 12, 222–234. [Google Scholar] [CrossRef]
- Roberts, A.J.; Kon, T.; Knight, P.J.; Sutoh, K.; Burgess, S.A. Functions and mechanics of dynein motor proteins. Nat. Rev. Mol. Cell Biol. 2013, 14, 713–726. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017: Quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Brandes, U. A faster algorithm for betweenness centrality*. J. Math. Sociol. 2001, 25, 163–177. [Google Scholar] [CrossRef]
- Enright, A.J.; Van Dongen, S.; Ouzounis, C.A. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002, 30, 1575–1584. [Google Scholar] [CrossRef] [PubMed]
- Siller, S.S.; Burke, M.C.; Li, F.-Q.; Takemaru, K.-I. Chibby functions to preserve normal ciliary morphology through the regulation of intraflagellar transport in airway ciliated cells. Cell Cycle 2015, 14, 3163–3172. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Burke, M.C.; Li, F.-Q.; Cyge, B.; Arashiro, T.; Brechbuhl, H.M.; Chen, X.; Siller, S.S.; Weiss, M.A.; O’Connell, C.B.; Love, D.; et al. Chibby promotes ciliary vesicle formation and basal body docking during airway cell differentiation. J. Cell Biol. 2014, 207, 123–137. [Google Scholar] [CrossRef] [PubMed]
- Siller, S.S.; Sharma, H.; Li, S.; Yang, J.; Zhang, Y.; Holtzman, M.J.; Winuthayanon, W.; Colognato, H.; Holdener, B.C.; Li, F.-Q.; et al. Conditional knockout mice for the distal appendage protein CEP164 reveal its essential roles in airway multiciliated cell differentiation. PLoS Genet. 2017, 13, e1007128. [Google Scholar] [CrossRef]
- Bonnefoy, S.; Watson, C.M.; Kernohan, K.D.; Lemos, M.; Hutchinson, S.; Poulter, J.A.; Crinnion, L.A.; Berry, I.; Simmonds, J.; Vasudevan, P.; et al. Biallelic mutations in LRRC56, encoding a protein associated with intraflagellar transport, cause mucociliary clearance and laterality defects. Am. J. Hum. Genet. 2018, 103, 727–739. [Google Scholar] [CrossRef]
- Tsang, W.Y.; Dynlacht, B.D. CP110 and its network of partners coordinately regulate cilia assembly. Cilia 2013, 2, 9. [Google Scholar] [CrossRef]
- Spektor, A.; Tsang, W.Y.; Khoo, D.; Dynlacht, B.D. Cep97 and CP110 suppress a cilia assembly program. Cell 2007, 130, 678–690. [Google Scholar] [CrossRef]
- Lai, Y.; Chen, B.; Shi, J.; Palmer, J.N.; Kennedy, D.W.; Cohen, N.A. Inflammation-mediated upregulation of centrosomal protein 110, a negative modulator of ciliogenesis, in patients with chronic rhinosinusitis. J. Allergy Clin. Immunol. 2011, 128, 1207–1215.e1. [Google Scholar] [CrossRef]
- Cao, J.; Shen, Y.; Zhu, L.; Xu, Y.; Zhou, Y.; Wu, Z.; Li, Y.; Yan, X.; Zhu, X. miR-129-3p controls cilia assembly by regulating CP110 and actin dynamics. Nat. Cell Biol. 2012, 14, 697–706. [Google Scholar] [CrossRef]
- Walentek, P.; Quigley, I.K.; Sun, D.I.; Sajjan, U.K.; Kintner, C.; Harland, R.M. Ciliary transcription factors and miRNAs precisely regulate Cp110 levels required for ciliary adhesions and ciliogenesis. eLife 2016, 5, e17557. [Google Scholar] [CrossRef]
- Choksi, S.P.; Lauter, G.; Swoboda, P.; Roy, S. Switching on cilia: Transcriptional networks regulating ciliogenesis. Development 2014, 141, 1427–1441. [Google Scholar] [CrossRef] [PubMed]
- Chung, M.-I.; Kwon, T.; Tu, F.; Brooks, E.R.; Gupta, R.; Meyer, M.; Baker, J.C.; Marcotte, E.M.; Wallingford, J.B. Coordinated genomic control of ciliogenesis and cell movement by RFX2. eLife 2014, 3, e01439. [Google Scholar] [CrossRef] [PubMed]
- Song, R.; Walentek, P.; Sponer, N.; Klimke, A.; Lee, J.S.; Dixon, G.; Harland, R.; Wan, Y.; Lishko, P.; Lize, M.; et al. miR-34/449 miRNAs are required for motile ciliogenesis by repressing cp110. Nature 2014, 510, 115–120. [Google Scholar] [CrossRef] [PubMed]
- Mercey, O.; Kodjabachian, L.; Barbry, P.; Marcet, B. MicroRNAs as key regulators of GTPase-mediated apical actin reorganization in multiciliated epithelia. Small GTPases 2016, 7, 54–58. [Google Scholar] [CrossRef] [PubMed]
- Boon, M.; Wallmeier, J.; Ma, L.; Loges, N.T.; Jaspers, M.; Olbrich, H.; Dougherty, G.W.; Raidt, J.; Werner, C.; Amirav, I.; et al. MCIDAS mutations result in a mucociliary clearance disorder with reduced generation of multiple motile cilia. Nat. Commun. 2014, 5, 4418. [Google Scholar] [CrossRef] [PubMed]
- Funk, M.C.; Bera, A.N.; Menchen, T.; Kuales, G.; Thriene, K.; Lienkamp, S.S.; Dengjel, J.; Omran, H.; Frank, M.; Arnold, S.J. Cyclin O (Ccno) functions during deuterosome-mediated centriole amplification of multiciliated cells. EMBO J. 2015, 34, 1078–1089. [Google Scholar] [CrossRef] [PubMed]
- Wallmeier, J.; Al-Mutairi, D.A.; Chen, C.-T.; Loges, N.T.; Pennekamp, P.; Menchen, T.; Ma, L.; Shamseldin, H.E.; Olbrich, H.; Dougherty, G.W.; et al. Mutations in CCNO result in congenital mucociliary clearance disorder with reduced generation of multiple motile cilia. Nat. Genet. 2014, 46, 646–651. [Google Scholar] [CrossRef]
- Terré, B.; Piergiovanni, G.; Segura-Bayona, S.; Gil-Gómez, G.; Youssef, S.A.; Attolini, C.S.; Wilsch-Bräuninger, M.; Jung, C.; Rojas, A.M.; Marjanović, M.; et al. GEMC 1 is a critical regulator of multiciliated cell differentiation. EMBO J. 2016, 35, 942–960. [Google Scholar] [CrossRef]
- Arbi, M.; Pefani, D.; Kyrousi, C.; Lalioti, M.; Kalogeropoulou, A.; Papanastasiou, A.D.; Taraviras, S.; Lygerou, Z. GemC1 controls multiciliogenesis in the airway epithelium. EMBO Rep. 2016, 17, 400–413. [Google Scholar] [CrossRef]
- Walz, G. Role of primary cilia in non-dividing and post-mitotic cells. Cell Tissue Res. 2017, 369, 11–25. [Google Scholar] [CrossRef]
- Wang, Z.; Plasschaert, L.W.; Aryal, S.; Renaud, N.A.; Yang, Z.; Choo-Wing, R.; Pessotti, A.D.; Kirkpatrick, N.D.; Cochran, N.R.; Carbone, W.; et al. TRRAP is a central regulator of human multiciliated cell formation. J. Cell Biol. 2018, 217, 1941–1955. [Google Scholar] [CrossRef] [PubMed]
- Herceg, Z.; Hulla, W.; Gell, D.A.; Cuenin, C.; Lleonart, M.E.; Jackson, S.P.; Wang, Z.-Q. Disruption of Trrap causes early embryonic lethality and defects in cell cycle progression. Nat. Genet. 2001, 29, 206–211. [Google Scholar] [CrossRef] [PubMed]
- Borlado, L.R.; Méndez, J. CDC6: From DNA replication to cell cycle checkpoints and oncogenesis. Carcinogenesis 2008, 29, 237–243. [Google Scholar] [CrossRef] [PubMed]
- Bowers, J.L.; Randell, J.C.; Chen, S.; Bell, S.P. ATP Hydrolysis by ORC Catalyzes reiterative Mcm2-7 assembly at a defined origin of replication. Mol. Cell 2004, 16, 967–978. [Google Scholar] [CrossRef] [PubMed]
- Randell, J.C.; Bowers, J.L.; Rodríguez, H.K.; Bell, S.P. Sequential ATP hydrolysis by Cdc6 and ORC directs loading of the Mcm2-7 helicase. Mol. Cell 2006, 21, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Fujita, M. Cdt1 revisited: Complex and tight regulation during the cell cycle and consequences of deregulation in mammalian cells. Cell Div. 2006, 1, 22. [Google Scholar] [CrossRef][Green Version]
- Pavia, D.; Thomson, M.L.; Pocock, S.J.; Pavia, M.L.T.D. Evidence for temporary slowing of mucociliary clearance in the lung caused by tobacco smoking. Nature 1971, 231, 325–326. [Google Scholar] [CrossRef]
- Auerbach, O.; Stout, A.P.; Hammond, E.C.; Garfinkel, L. Changes in bronchial epithelium in relation to sex, age, residence, smoking and pneumonia. N. Engl. J. Med. 1962, 267, 111–119. [Google Scholar] [CrossRef]
- Leopold, P.L.; O’Mahony, M.J.; Lian, X.J.; Tilley, A.E.; Harvey, B.-G.; Crystal, R.G. Smoking Is Associated with Shortened Airway Cilia. PLoS ONE 2009, 4, e8157. [Google Scholar] [CrossRef]
- Shaykhiev, R.; Zuo, W.-L.; Chao, I.; Fukui, T.; Witover, B.; Brekman, A.; Crystal, R.G. EGF shifts human airway basal cell fate toward a smoking-associated airway epithelial phenotype. Proc. Natl. Acad. Sci. USA 2013, 110, 12102–12107. [Google Scholar] [CrossRef]
- Brekman, A.; Walters, M.S.; Tilley, A.E.; Crystal, R.G. FOXJ1 prevents cilia growth inhibition by cigarette smoke in human airway epithelium in vitro. Am. J. Respir. Cell Mol. Biol. 2014, 51, 688–700. [Google Scholar] [CrossRef] [PubMed]
- Elwany, S.; Ibrahim, A.A.; Mandour, Z.; Talaat, I.M. Effect of passive smoking on the ultrastructure of the nasal mucosa in children. Laryngoscope 2012, 122, 965–969. [Google Scholar] [CrossRef] [PubMed]
- Calderon-Garciduenas, L.; Rodriguez-Alcaraz, A.; Villarreal-Calderon, A.; Lyght, O.; Janszen, D.; Morgan, K.T. Nasal epithelium as a sentinel for airborne environmental pollution. Toxicol. Sci. 1998, 46, 352–364. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, M. Ciliary activity and pollution. Lung 1990, 168, 368–376. [Google Scholar] [CrossRef]
- Hessel, J.; Heldrich, J.; Fuller, J.; Staudt, M.R.; Radisch, S.; Hollmann, C.; Harvey, B.-G.; Kaner, R.J.; Salit, J.; Yee-Levin, J.; et al. Intraflagellar transport gene expression associated with short cilia in smoking and COPD. PLoS ONE 2014, 9, e85453. [Google Scholar] [CrossRef]
- Dunnill, M.S. The pathology of asthma, with special reference to changes in the bronchial mucosa. J. Clin. Pathol. 1960, 13, 27–33. [Google Scholar] [CrossRef]
- Cokugras, H.; Akcakaya, N.; Camcioglu, Y.; Sarimurat, N.; Aksoy, F. Ultrastructural examination of bronchial biopsy specimens from children with moderate asthma. Thorax 2001, 56, 25–29. [Google Scholar] [CrossRef]
- Laitinen, L.A.; Heino, M.; Laitinen, A.; Kava, T.; Haahtela, T. Damage of the airway epithelium and bronchial reactivity in patients with asthma. Am. Rev. Respir. Dis. 1985, 131, 599–606. [Google Scholar] [CrossRef]
- Thomas, B.; Rutman, A.; Hirst, R.A.; Haldar, P.; Wardlaw, A.J.; Bankart, J.; Brightling, C.E.; O’Callaghan, C. Ciliary dysfunction and ultrastructural abnormalities are features of severe asthma. J. Allergy Clin. Immunol. 2010, 126, 722–729. [Google Scholar] [CrossRef]
- Rael, E.L.; Lockey, R.F. Interleukin-13 signaling and its role in asthma. World Allergy Organ. J. 2011, 4, 54–64. [Google Scholar] [CrossRef]
- Munitz, A.; Brandt, E.B.; Mingler, M.; Finkelman, F.D.; Rothenberg, M.E. Distinct roles for IL-13 and IL-4 via IL-13 receptor 1 and the type II IL-4 receptor in asthma pathogenesis. Proc. Natl. Acad. Sci. USA 2008, 105, 7240–7245. [Google Scholar] [CrossRef] [PubMed]
- Laoukili, J.; Perret, E.; Willems, T.; Minty, A.; Parthoens, E.; Houcine, O.; Coste, A.; Jorissen, M.; Marano, F.; Caput, D.; et al. IL-13 alters mucociliary differentiation and ciliary beating of human respiratory epithelial cells. J. Clin. Investig. 2001, 108, 1817–1824. [Google Scholar] [CrossRef] [PubMed]
- Gomperts, B.N.; Kim, L.J.; Flaherty, S.A.; Hackett, B.P. IL-13 regulates cilia loss and foxj1 expression in human airway epithelium. Am. J. Respir. Cell Mol. Biol. 2007, 37, 339–346. [Google Scholar] [CrossRef] [PubMed]
- Kovacic, M.B.; Biagini, J.M.; Wang, N.; Martin, L.J.; Lindsey, M.; Ericksen, M.B.; He, H.; Patterson, T.L.; Baye, T.M.; Torgerson, D.; et al. Identification of KIF3A as a novel candidate gene for childhood asthma using RNA expression and population allelic frequencies differences. PLoS ONE 2011, 6, e23714. [Google Scholar] [CrossRef]
- Götz, M.; Stockinger, L. Aplasia of respiratory tract cilia. Lancet 1983, 1, 1283. [Google Scholar] [CrossRef]
- DeBoeck, K.; Jorissen, M.; Wouters, K.; Van Der Schueren, B.; Eyssen, M.; Casteels-Vandaele, M.; Corbeel, L. Aplasia of respiratory tract cilia. Pediatr. Pulmonol. 1992, 13, 259–265. [Google Scholar] [CrossRef] [PubMed]
- Ehre, C.; Ridley, C.; Thornton, D.J. Cystic fibrosis: An inherited disease affecting mucin-producing organs. Int. J. Biochem. Cell Biol. 2014, 52, 136–145. [Google Scholar] [CrossRef]
- Vankeerberghen, A.; Cuppens, H.; Cassiman, J.-J. The cystic fibrosis transmembrane conductance regulator: An intriguing protein with pleiotropic functions. J. Cyst. Fibros. 2002, 1, 13–29. [Google Scholar] [CrossRef]
- Sosnay, P.R.; Siklosi, K.R.; Van Goor, F.; Kaniecki, K.; Yu, H.; Sharma, N.; Ramalho, A.S.; Amaral, M.D.; Dorfman, R.; Zielenski, J.; et al. Defining the disease liability of variants in the cystic fibrosis transmembrane conductance regulator gene. Nat. Genet. 2013, 45, 1160–1167. [Google Scholar] [CrossRef]
- Lewis, H.A.; Zhao, X.; Wang, C.; Sauder, J.M.; Rooney, I.; Noland, B.W.; Lorimer, D.; Kearins, M.C.; Conners, K.; Condon, B.; et al. Impact of the deltaF508 mutation in first nucleotide-binding domain of human cystic fibrosis transmembrane conductance regulator on domain folding and structure. J. Biol. Chem. 2005, 280, 1346–1353. [Google Scholar] [CrossRef]
- Boucher, R.C.; Cotton, C.U.; Gatzy, J.T.; Knowles, M.R.; Yankaskas, J.R. Evidence for reduced Cl- and increased Na+ permeability in cystic fibrosis human primary cell cultures. J. Physiol. 1988, 405, 77–103. [Google Scholar] [CrossRef] [PubMed]
- Bhagirath, A.Y.; Li, Y.; Somayajula, D.; Dadashi, M.; Badr, S.; Duan, K. Cystic fibrosis lung environment and Pseudomonas aeruginosa infection. BMC Pulm. Med. 2016, 16, 174. [Google Scholar] [CrossRef] [PubMed]
- Boucher, R.C. New concepts of the pathogenesis of cystic fibrosis lung disease. Eur. Respir. J. 2004, 23, 146–158. [Google Scholar] [CrossRef] [PubMed]
- Lamblin, G.; DeGroote, S.; Perini, J.-M.; Delmotte, P.; Scharfman, A.; Davril, M.; Lo-Guidice, J.-M.; Houdret, N.; Dumur, V.; Klein, A.; et al. Human airway mucin glycosylation: A combinatory of carbohydrate determinants which vary in cystic fibrosis. Glycoconj. J. 2001, 18, 661–684. [Google Scholar] [CrossRef] [PubMed]
- Alaiwa, M.H.A.; Beer, A.M.; Pezzulo, A.A.; Launspach, J.L.; Horan, R.A.; Stoltz, D.A.; Starner, T.D.; Welsh, M.J.; Zabner, J. Neonates with cystic fibrosis have a reduced nasal liquid pH.; A small pilot study. J. Cyst. Fibros. 2014, 13, 373–377. [Google Scholar] [CrossRef] [PubMed]
- Pezzulo, A.A.; Tang, X.X.; Hoegger, M.J.; Alaiwa, M.H.A.; Ramachandran, S.; Moninger, T.O.; Karp, P.H.; Wohlford-Lenane, C.L.; Haagsman, H.P.; Van Eijk, M.; et al. Reduced airway surface pH impairs bacterial killing in the porcine cystic fibrosis lung. Nature 2012, 487, 109–113. [Google Scholar] [CrossRef]
- Horani, A.; Ferkol, T.W. Advances in the genetics of primary ciliary dyskinesia: Clinical implications. Chest 2018, 154, 645–652. [Google Scholar] [CrossRef]
- Horani, A.; Ferkol, T.W.; Dutcher, S.K.; Brody, S.L. Genetics and biology of primary ciliary dyskinesia. Paediatr. Respir. Rev. 2016, 18, 18–24. [Google Scholar] [CrossRef]
- Knowles, M.R.; Daniels, L.A.; Davis, S.D.; Zariwala, M.A.; Leigh, M.W. Primary ciliary dyskinesia. recent advances in diagnostics, genetics, and characterization of clinical disease. Am. J. Respir. Crit. Care Med. 2013, 188, 913–922. [Google Scholar] [CrossRef]
- Knowles, M.R.; Zariwala, M.; Leigh, M. Primary ciliary dyskinesia. Clin. Chest Med. 2016, 37, 449–461. [Google Scholar] [CrossRef]
- Shapiro, A.J.; Zariwala, M.A.; Ferkol, T.W.; Davis, S.D.; Sagel, S.D.; Dell, S.D.; Rosenfeld, M.; Olivier, K.N.; Milla, C.E.; Daniel, S.J.; et al. Diagnosis, monitoring, and treatment of primary ciliary dyskinesia: PCD foundation consensus recommendations based on state of the art review. Pediatr. Pulmonol. 2016, 51, 115–132. [Google Scholar] [CrossRef] [PubMed]
- Rossman, C.M.; Lee, R.M.; Forrest, J.B.; Newhouse, M.T. Nasal ciliary ultrastructure and function in patients with primary ciliary dyskinesia compared with that in normal subjects and in subjects with various respiratory diseases. Am. Rev. Respir. Dis. 1984, 129, 161–167. [Google Scholar] [PubMed]
- Möller, W.; Häußinger, K.; Ziegler-Heitbrock, L.; Heyder, J. Mucociliary and long-term particle clearance in airways of patients with immotile cilia. Respir. Res. 2006, 7, 10. [Google Scholar] [CrossRef] [PubMed]
- Guichard, C.; Harricane, M.-C.; Lafitte, J.-J.; Godard, P.; Zaegel, M.; Tack, V.; Lalau, G.; Bouvagnet, P. Axonemal dynein intermediate-chain gene (DNAI1) mutations result in situs inversus and primary ciliary dyskinesia (kartagener syndrome). Am. J. Hum. Genet. 2001, 68, 1030–1035. [Google Scholar] [CrossRef] [PubMed]
- Olbrich, H.; Häffner, K.; Kispert, A.; Völkel, A.; Volz, A.; Sasmaz, G.; Reinhardt, R.; Hennig, S.; Lehrach, H.; Konietzko, N.; et al. Mutations in DNAH5 cause primary ciliary dyskinesia and randomization of left–right asymmetry. Nat. Genet. 2002, 30, 143–144. [Google Scholar] [CrossRef] [PubMed]
- Mazor, M.; Alkrinawi, S.; Chalifa-Caspi, V.; Manor, E.; Sheffield, V.C.; Aviram, M.; Parvari, R. Primary ciliary dyskinesia caused by homozygous mutation in DNAL1, encoding dynein light chain 1. Am. J. Hum. Genet. 2011, 88, 599–607. [Google Scholar] [CrossRef][Green Version]
- Loges, N.T.; Olbrich, H.; Fenske, L.; Mussaffi, H.; Horvath, J.; Fliegauf, M.; Kuhl, H.; Baktai, G.; Peterffy, E.; Chodhari, R.; et al. DNAI2 mutations cause primary ciliary dyskinesia with defects in the outer dynein arm. Am. J. Hum. Genet. 2008, 83, 547–558. [Google Scholar] [CrossRef]
- Duriez, B.; Duquesnoy, P.; Escudier, E.; Bridoux, A.-M.; Escalier, D.; Rayet, I.; Marcos, E.; Vojtek, A.-M.; Bercher, J.-F.; Amselem, S. A common variant in combination with a nonsense mutation in a member of the thioredoxin family causes primary ciliary dyskinesia. Proc. Natl. Acad. Sci. USA 2007, 104, 3336–3341. [Google Scholar] [CrossRef]
- Knowles, M.R.; Leigh, M.W.; Carson, J.L.; Davis, S.D.; Dell, S.D.; Ferkol, T.W.; Olivier, K.N.; Sagel, S.D.; Rosenfeld, M.; Burns, K.A.; et al. Mutations ofDNAH11in patients with primary ciliary dyskinesia with normal ciliary ultrastructure. Thorax 2012, 67, 433–441. [Google Scholar] [CrossRef]
- Pifferi, M.; Michelucci, A.; Conidi, M.E.; Cangiotti, A.M.; Simi, P.; Macchia, P.; Boner, A.L. New DNAH11 mutations in primary ciliary dyskinesia with normal axonemal ultrastructure. Eur. Respir. J. 2010, 35, 1413–1416. [Google Scholar] [CrossRef]
- Raidt, J.; Wallmeier, J.; Hjeij, R.; Onnebrink, J.G.; Pennekamp, P.; Loges, N.T.; Olbrich, H.; Häffner, K.; Dougherty, G.W.; Omran, H.; et al. Ciliary beat pattern and frequency in genetic variants of primary ciliary dyskinesia. Eur. Respir. J. 2014, 44, 1579–1588. [Google Scholar] [CrossRef] [PubMed]
- Hjeij, R.; Onoufriadis, A.; Watson, C.M.; Slagle, C.E.; Klena, N.T.; Dougherty, G.W.; Kurkowiak, M.; Loges, N.T.; Diggle, C.P.; Morante, N.F.; et al. CCDC151 mutations cause primary ciliary dyskinesia by disruption of the outer dynein arm docking complex formation. Am. J. Hum. Genet. 2014, 95, 257–274. [Google Scholar] [CrossRef] [PubMed]
- Knowles, M.R.; Leigh, M.W.; Ostrowski, L.E.; Huang, L.; Carson, J.L.; Hazucha, M.J.; Yin, W.; Berg, J.S.; Davis, S.D.; Dell, S.D.; et al. Exome sequencing identifies mutations in CCDC114 as a cause of primary ciliary dyskinesia. Am. J. Hum. Genet. 2013, 92, 99–106. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Merveille, A.-C.; Davis, E.E.; Becker-Heck, A.; Legendre, M.; Amirav, I.; Bataille, G.; Belmont, J.W.; Beydon, N.; Billen, F.; Clément, A.; et al. CCDC39 is required for assembly of inner dynein arms and the dynein regulatory complex and for normal ciliary motility in humans and dogs. Nat. Genet. 2010, 43, 72–78. [Google Scholar] [CrossRef] [PubMed]
- Becker-Heck, A.; Zohn, I.E.; Okabe, N.; Pollock, A.; Lenhart, K.B.; Sullivan-Brown, J.; McSheene, J.; Loges, N.T.; Olbrich, H.; Haeffner, K.; et al. The coiled-coil domain containing protein CCDC40 is essential for motile cilia function and left-right axis formation. Nat. Genet. 2011, 43, 79–84. [Google Scholar] [CrossRef]
- Olbrich, H.; Schmidts, M.; Werner, C.; Onoufriadis, A.; Loges, N.T.; Raidt, J.; Banki, N.F.; Shoemark, A.; Burgoyne, T.; Al Turki, S.; et al. Recessive HYDIN mutations cause primary ciliary dyskinesia without randomization of left-right body asymmetry. Am. J. Hum. Genet. 2012, 91, 672–684. [Google Scholar] [CrossRef]
- Castleman, V.H.; Romio, L.; Chodhari, R.; Hirst, R.A.; De Castro, S.C.; Parker, K.A.; Ybot-Gonzalez, P.; Emes, R.D.; Wilson, S.W.; Wallis, C.; et al. Mutations in radial spoke head protein genes RSPH9 and RSPH4A cause primary ciliary dyskinesia with central-microtubular-pair abnormalities. Am. J. Hum. Genet. 2009, 84, 197–209. [Google Scholar] [CrossRef]
- Kott, E.; Legendre, M.; Copin, B.; Papon, J.-F.; Moal, F.D.-L.; Montantin, G.; Duquesnoy, P.; Piterboth, W.; Amram, D.; Bassinet, L.; et al. Loss-of-function mutations in RSPH1 cause primary ciliary dyskinesia with central-complex and radial-spoke defects. Am. J. Hum. Genet. 2013, 93, 561–570. [Google Scholar] [CrossRef]
- Knowles, M.R.; Ostrowski, L.E.; Leigh, M.W.; Sears, P.R.; Davis, S.D.; Wolf, W.E.; Hazucha, M.J.; Carson, J.L.; Olivier, K.N.; Sagel, S.D.; et al. Mutations in RSPH1 cause primary ciliary dyskinesia with a unique clinical and ciliary phenotype. Am. J. Respir. Crit. Care Med. 2014, 189, 707–717. [Google Scholar] [CrossRef]
- Davis, S.D.; Ferkol, T.W.; Rosenfeld, M.; Lee, H.-S.; Dell, S.D.; Sagel, S.D.; Milla, C.; Zariwala, M.A.; Pittman, J.E.; Shapiro, A.J.; et al. Clinical features of childhood primary ciliary dyskinesia by genotype and ultrastructural phenotype. Am. J. Respir. Crit. Care Med. 2015, 191, 316–324. [Google Scholar] [CrossRef]
- Oda, T.; Yanagisawa, H.; Kamiya, R.; Kikkawa, M. A molecular ruler determines the repeat length in eukaryotic cilia and flagella. Science 2014, 346, 857–860. [Google Scholar] [CrossRef] [PubMed]
- Horani, A.; Brody, S.L.; Ferkol, T.W.; Shoseyov, D.; Wasserman, M.G.; Ta-Shma, A.; Wilson, K.S.; Bayly, P.V.; Amirav, I.; Cohen-Cymberknoh, M.; et al. CCDC65 mutation causes primary ciliary dyskinesia with normal ultrastructure and hyperkinetic cilia. PLoS ONE 2013, 8, e72299. [Google Scholar] [CrossRef] [PubMed]
- Horani, A.; Druley, T.E.; Zariwala, M.A.; Patel, A.C.; Levinson, B.T.; Van Arendonk, L.G.; Thornton, K.C.; Giacalone, J.C.; Albee, A.J.; Wilson, K.S.; et al. Whole-exome capture and sequencing identifies HEATR2 mutation as a cause of primary ciliary dyskinesia. Am. J. Hum. Genet. 2012, 91, 685–693. [Google Scholar] [CrossRef] [PubMed]
- Moore, D.J.; Onoufriadis, A.; Shoemark, A.; Simpson, M.A.; Lage, P.I.Z.; De Castro, S.C.; Bartoloni, L.; Gallone, G.; Petridi, S.; Woollard, W.J.; et al. Mutations in ZMYND10, a gene essential for proper axonemal assembly of inner and outer dynein arms in humans and flies, cause primary ciliary dyskinesia. Am. J. Hum. Genet. 2013, 93, 346–356. [Google Scholar] [CrossRef] [PubMed]
- Knowles, M.R.; Ostrowski, L.E.; Loges, N.T.; Hurd, T.; Leigh, M.W.; Huang, L.; Wolf, W.E.; Carson, J.L.; Hazucha, M.J.; Yin, W.; et al. Mutations in SPAG1 cause primary ciliary dyskinesia associated with defective outer and inner dynein arms. Am. J. Hum. Genet. 2013, 93, 711–720. [Google Scholar] [CrossRef] [PubMed]
- Horani, A.; Ferkol, T.W.; Shoseyov, D.; Wasserman, M.G.; Oren, Y.S.; Kerem, B.; Amirav, I.; Cohen-Cymberknoh, M.; Dutcher, S.K.; Brody, S.L.; et al. LRRC6 mutation causes primary ciliary dyskinesia with dynein arm defects. PLoS ONE 2013, 8, e59436. [Google Scholar] [CrossRef]
- Hjeij, R.; Lindstrand, A.; Francis, R.; Zariwala, M.A.; Liu, X.; Li, Y.; Damerla, R.; Dougherty, G.W.; Abouhamed, M.; Olbrich, H.; et al. ARMC4 mutations cause primary ciliary dyskinesia with randomization of left/right body asymmetry. Am. J. Hum. Genet. 2013, 93, 357–367. [Google Scholar] [CrossRef] [PubMed]
- Loges, N.T.; Olbrich, H.; Becker-Heck, A.; Häffner, K.; Heer, A.; Reinhard, C.; Schmidts, M.; Kispert, A.; Zariwala, M.A.; Leigh, M.W.; et al. Deletions and point mutations of LRRC50 cause primary ciliary dyskinesia due to dynein arm defects. Am. J. Hum. Genet. 2009, 85, 883–889. [Google Scholar] [CrossRef]
- Omran, H.; Kobayashi, D.; Olbrich, H.; Tsukahara, T.; Loges, N.T.; Hagiwara, H.; Zhang, Q.; Leblond, G.; O’Toole, E.; Hara, C.; et al. Ktu/PF13 is required for cytoplasmic pre-assembly of axonemal dyneins. Nature 2008, 456, 611–616. [Google Scholar] [CrossRef]
- Mitchison, H.M.; Schmidts, M.; Loges, N.T.; Freshour, J.; Dritsoula, A.; Hirst, R.A.; O’Callaghan, C.; Blau, H.; Al Dabbagh, M.; Olbrich, H.; et al. Mutations in axonemal dynein assembly factor DNAAF3 cause primary ciliary dyskinesia. Nat. Genet. 2012, 44, 381–389. [Google Scholar] [CrossRef]
- Panizzi, J.R.; Becker-Heck, A.; Castleman, V.H.; Al-Mutairi, D.A.; Liu, Y.; Loges, N.T.; Pathak, N.; Austin-Tse, C.; Sheridan, E.; Schmidts, M.; et al. CCDC103 mutations cause primary ciliary dyskinesia by disrupting assembly of ciliary dynein arms. Nat. Genet. 2012, 44, 714–719. [Google Scholar] [CrossRef] [PubMed]
- Tarkar, A.; Loges, N.T.; Slagle, C.E.; Francis, R.; Dougherty, G.W.; Tamayo, J.V.; Shook, B.; Cantino, M.; Schwartz, D.; Jahnke, C.; et al. DYX1C1 is required for axonemal dynein assembly and ciliary motility. Nat. Genet. 2013, 45, 995–1003. [Google Scholar] [CrossRef] [PubMed]
- Bustamante-Marin, X.M.; Horani, A.; Stoyanova, M.; Charng, W.-L.; Bottier, M.; Sears, P.R.; Yin, W.-N.; Daniels, M.A.; Bowen, H.; Conrad, D.F.; et al. Mutation of CFAP57, a protein required for the asymmetric targeting of a subset of inner dynein arms in Chlamydomonas, causes primary ciliary dyskinesia. PLoS Genet. 2020, 16, e1008691. [Google Scholar] [CrossRef] [PubMed]
- Chivukula, R.R.; Montoro, D.T.; Leung, H.M.; Yang, J.; Shamseldin, H.E.; Taylor, M.S.; Dougherty, G.W.; Zariwala, M.A.; Carson, J.; Daniels, M.A.; et al. A human ciliopathy reveals essential functions for NEK10 in airway mucociliary clearance. Nat. Med. 2020, 26, 244–251. [Google Scholar] [CrossRef] [PubMed]
- Bottier, M.; Thomas, K.A.; Dutcher, S.K.; Bayly, P.V. How does cilium length affect beating? Biophys. J. 2019, 116, 1292–1304. [Google Scholar] [CrossRef]
- Rogers, G.B.; Hart, C.A.; Mason, J.R.; Hughes, M.; Walshaw, M.J.; Bruce, K.D. Bacterial diversity in cases of lung infection in cystic fibrosis patients: 16S ribosomal DNA (rDNA) length heterogeneity PCR and 16S rDNA terminal restriction fragment length polymorphism profiling. J. Clin. Microbiol. 2003, 41, 3548–3558. [Google Scholar] [CrossRef]
- Etherington, C.; Naseer, R.; Conway, S.P.; Whitaker, P.; Denton, M.; Peckham, D. The role of respiratory viruses in adult patients with cystic fibrosis receiving intravenous antibiotics for a pulmonary exacerbation. J. Cyst. Fibros. 2014, 13, 49–55. [Google Scholar] [CrossRef]
- Willger, S.D.; Grim, S.L.; Dolben, E.L.; Shipunova, A.; Hampton, T.H.; Morrison, H.G.; Filkins, L.M.; O’Toole, G.A.; Moulton, L.A.; Ashare, A.; et al. Characterization and quantification of the fungal microbiome in serial samples from individuals with cystic fibrosis. Microbiome 2014, 2, 40. [Google Scholar] [CrossRef]
- Balder, R.; Krunkosky, T.M.; Nguyen, C.Q.; Feezel, L.; Lafontaine, E.R. Hag mediates adherence of moraxella catarrhalis to ciliated human airway cells. Infect. Immun. 2009, 77, 4597–4608. [Google Scholar] [CrossRef]
- Noone, P.G.; Leigh, M.W.; Sannuti, A.; Minnix, S.L.; Carson, J.L.; Hazucha, M.; Zariwala, M.A.; Knowles, M.R. Primary ciliary dyskinesia: Diagnostic and phenotypic features. Am. J. Respir. Crit. Care Med. 2004, 169, 459–467. [Google Scholar] [CrossRef]
- Wijers, C.D.; Chmiel, J.F.; Gaston, B.M. Bacterial infections in patients with primary ciliary dyskinesia: Comparison with cystic fibrosis. Chronic Respir. Dis. 2017, 14, 392–406. [Google Scholar] [CrossRef] [PubMed]
- Maglione, M.; Bush, A.; Nielsen, K.G.; Hogg, C.; Montella, S.; Marthin, J.K.; Di Giorgio, A.; Santamaria, F. Multicenter analysis of body mass index, lung function, and sputum microbiology in primary ciliary dyskinesia. Pediatr. Pulmonol. 2014, 49, 1243–1250. [Google Scholar] [CrossRef] [PubMed]
- Rogers, G.; Carroll, M.P.; Zain, N.M.M.; Bruce, K.D.; Lock, K.; Walker, W.; Jones, G.; Daniels, T.W.V.; Lucas, J.S. Complexity, Temporal stability, and clinical correlates of airway bacterial community composition in primary ciliary dyskinesia. J. Clin. Microbiol. 2013, 51, 4029–4035. [Google Scholar] [CrossRef] [PubMed]
- Alanin, M.C.; Nielsen, K.G.; von Buchwald, C.; Skov, M.; Aanaes, K.; Hoiby, N.; Johansen, H.K. A longitudinal study of lung bacterial pathogens in patients with primary ciliary dyskinesia. Clin. Microbiol. Infect. 2015, 21, 1093.e1–1093.e7. [Google Scholar] [CrossRef] [PubMed]
- Emerson, J.; Rosenfeld, M.; McNamara, S.; Ramsey, B.; Gibson, R.L. Pseudomonas aeruginosa and other predictors of mortality and morbidity in young children with cystic fibrosis. Pediatr. Pulmonol. 2002, 34, 91–100. [Google Scholar] [CrossRef]
- Bhatt, J.M. Treatment of pulmonary exacerbations in cystic fibrosis. Eur. Respir. Rev. 2013, 22, 205–216. [Google Scholar] [CrossRef] [PubMed]
- Goss, C.H.; Burns, J.L. Exacerbations in cystic fibrosis 1: Epidemiology and pathogenesis. Thorax 2007, 62, 360–367. [Google Scholar] [CrossRef]
- Flume, P.A.; Mogayzel, P.J., Jr.; Robinson, K.A.; Goss, C.H.; Rosenblatt, R.L.; Kuhn, R.J.; Marshall, B.C. Cystic fibrosis pulmonary guidelines: Treatment of pulmonary exacerbations. Am. J. Respir. Crit. Care Med. 2009, 180, 802–808. [Google Scholar] [CrossRef]
- Gilligan, P.H. Infections in patients with cystic fibrosis: Diagnostic microbiology update. Clin. Lab. Med. 2014, 34, 197–217. [Google Scholar] [CrossRef]
- Filkins, L.M.; O’Toole, G.A. Cystic fibrosis lung infections: Polymicrobial, complex, and hard to treat. PLoS Pathog. 2015, 11, e1005258. [Google Scholar] [CrossRef]
- Amitani, R.; Wilson, R.; Rutman, A.; Read, R.; Ward, C.; Burnett, D.; Stockley, R.A.; Cole, P.J. Effects of human neutrophil elastase and pseudomonas aeruginosa proteinases on human respiratory epithelium. Am. J. Respir. Cell Mol. Biol. 1991, 4, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Kao, S.S.; Ramezanpour, M.; Bassiouni, A.; Wormald, P.; Psaltis, A.J.; Vreugde, S. The effect of neutrophil serine proteases on human nasal epithelial cell barrier function. Int. Forum Allergy Rhinol. 2019, 9, 1220–1226. [Google Scholar] [CrossRef] [PubMed]
- Kantar, A.; Oggiano, N.; Giorgi, P.L.; Braga, P.C.; Fiorini, R. Polymorphonuclear leukocyte-generated oxygen metabolites decrease beat frequency of human respiratory cilia. Lung 1994, 172, 215–222. [Google Scholar] [CrossRef] [PubMed]
- Van Der Gast, C.J.; Walker, A.W.; Stressmann, F.A.; Rogers, G.B.; Scott, P.J.B.; Daniels, T.W.; Carroll, M.P.; Parkhill, J.; Bruce, K.D. Partitioning core and satellite taxa from within cystic fibrosis lung bacterial communities. ISME J. 2011, 5, 780–791. [Google Scholar] [CrossRef] [PubMed]
- Sibley, C.D.; Parkins, M.D.; Rabin, H.R.; Duan, K.; Norgaard, J.C.; Surette, M.G. A polymicrobial perspective of pulmonary infections exposes an enigmatic pathogen in cystic fibrosis patients. Proc. Natl. Acad. Sci. USA 2008, 105, 15070–15075. [Google Scholar] [CrossRef] [PubMed]
- Rayner, C.F.; Rutman, A.; Dewar, A.; Cole, P.J.; Wilson, R. Ciliary disorientation in patients with chronic upper respiratory tract inflammation. Am. J. Respir. Crit. Care Med. 1995, 151, 800–804. [Google Scholar] [CrossRef] [PubMed]
- Look, D.C.; Walter, M.J.; Williamson, M.R.; Pang, L.; You, Y.; Sreshta, J.N.; Johnson, J.E.; Zander, D.S.; Brody, S.L. Effects of paramyxoviral infection on airway epithelial cell foxj1 expression, ciliogenesis, and mucociliary function. Am. J. Pathol. 2001, 159, 2055–2069. [Google Scholar] [CrossRef]
- Crothers, K.; Huang, L.; Goulet, J.L.; Goetz, M.B.; Brown, S.T.; Rodriguez-Barradas, M.C.; Oursler, K.K.; Rimland, D.; Gibert, C.L.; Butt, A.A.; et al. HIV infection and risk for incident pulmonary diseases in the combination antiretroviral therapy era. Am. J. Respir. Crit. Care Med. 2011, 183, 388–395. [Google Scholar] [CrossRef] [PubMed]
- Chinnapaiyan, S.; Parira, T.; Dutta, R.; Agudelo, M.; Morris, A.; Nair, M.; Unwalla, H.J. HIV infects bronchial epithelium and suppresses components of the mucociliary clearance apparatus. PLoS ONE 2017, 12, e0169161. [Google Scholar] [CrossRef]
- Palm, J.; Lidman, C.; Graf, P.; Alving, K.; Lundberg, J. Nasal nitric oxide is reduced in patients with HIV. Acta Otolaryngol. 2000, 120, 420–423. [Google Scholar] [CrossRef]
- Milgrim, L.M.; Rubin, J.S.; Small, C.B. Mucociliary clearance abnormalities in the HIV-infected patient: A precursor to acute sinusitis. Laryngoscope 1995, 105, 1202–1208. [Google Scholar] [CrossRef] [PubMed]
- Hament, J.M.; Kimpen, J.L.; Fleer, A.; Wolfs, T.F. Respiratory viral infection predisposing for bacterial disease: A concise review. FEMS Immunol. Med. Microbiol. 1999, 26, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Takala, A.K.; Meurman, O.; Kleemola, M.; Kela, E.; Rönnberg, P.-R.; Eskola, J.; Mäkelä, P.H. Preceding respiratory infection predisposing for primary and secondary invasive Haemophilus influenzae type b disease. Pediatr. Infect. Dis. J. 1993, 12, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Glezen, W.P.; Greenberg, S.B.; Atmar, R.L.; Piedra, P.A.; Couch, R.B. Impact of respiratory virus infections on persons with chronic underlying conditions. JAMA 2000, 283, 499–505. [Google Scholar] [CrossRef]
- Jiang, Z.; Nagata, N.; Molina, E.; Bakaletz, L.O.; Hawkins, H.; Patel, J.A. Fimbria-mediated enhanced attachment of nontypeable haemophilus influenzae to respiratory syncytial virus-infected respiratory epithelial cells. Infect. Immun. 1999, 67, 187–192. [Google Scholar] [CrossRef]
- Nicolas de Lamballerie, C.; Pizzorno, A.; Dubois, J.; Julien, T.; Padey, B.; Bouveret, M.; Traversier, A.; Legras-Lachuer, C.; Lina, B.; Boivin, G.; et al. Characterization of cellular transcriptomic signatures induced by different respiratory viruses in human reconstituted airway epithelia. Sci. Rep. 2019, 9, 11493. [Google Scholar] [CrossRef]
- Fehr, A.R.; Perlman, S. Coronaviruses: An overview of their replication and pathogenesis. Methods Mol. Biol. 2015, 1282, 1–23. [Google Scholar]
- Su, S.; Wong, G.; Shi, W.; Liu, J.; Lai, A.C.; Zhou, J.; Liu, W.; Bi, Y.; Gao, G.F. Epidemiology, Genetic Recombination, and Pathogenesis of Coronaviruses. Trends Microbiol. 2016, 24, 490–502. [Google Scholar] [CrossRef]
- Fung, T.S.; Liu, D.X. Human coronavirus: Host-pathogen interaction. Annu. Rev. Microbiol. 2019, 73, 529–557. [Google Scholar] [CrossRef]
- Rabenau, H.F.; Cinatl, J.; Morgenstern, B.; Bauer, G.; Preiser, W.; Doerr, H.W. Stability and inactivation of SARS coronavirus. Med. Microbiol. Immunol. 2004, 194, 1–6. [Google Scholar] [CrossRef]
- Singh, S.K. Middle East Respiratory Syndrome Virus Pathogenesis. Semin. Respir. Crit. Care Med. 2016, 37, 572–577. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Shi, Z.; Yu, M.; Ren, W.; Smith, C.; Epstein, J.H.; Wang, H.; Crameri, G.; Hu, Z.; Zhang, H.; et al. Bats are natural reservoirs of SARS-like coronaviruses. Science 2005, 310, 676–679. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.S.; Zheng, J.P.; Mok, Y.W.; Li, Y.M.; Liu, Y.-N.; Chu, C.M.; Ip, M.S. SARS: Prognosis, outcome and sequelae. Respirology 2003, 8, S36–S40. [Google Scholar] [CrossRef] [PubMed]
- Sung, J.J.Y.; Wu, A.; Joynt, G.M.; Yuen, K.Y.; Lee, N.; Chan, P.K.S.; Cockram, C.S.; Ahuja, A.T.; Yu, L.M.; Wong, V.W.; et al. Severe acute respiratory syndrome: Report of treatment and outcome after a major outbreak. Thorax 2004, 59, 414–420. [Google Scholar] [CrossRef] [PubMed]
- Lo, A.W.I.; Tang, N.L.; To, K.-F. How the SARS coronavirus causes disease: Host or organism? J. Pathol. 2006, 208, 142–151. [Google Scholar] [CrossRef]
- Ngai, J.C.; Ko, F.W.; Ng, S.S.; To, K.-W.; Tong, M.; Hui, D.S. The long-term impact of severe acute respiratory syndrome on pulmonary function, exercise capacity and health status. Respirology 2010, 15, 543–550. [Google Scholar] [CrossRef]
- Chu, W.C.-W.; Li, A.M.; Ng, A.; So, H.-K.; Lam, W.W.M.; Lo, K.L.; Yeung, M.-C.A.; Yau, Y.-S.; Chiu, W.-K.; Leung, C.-W.; et al. Thin-section CT 12 months after the diagnosis of severe acute respiratory syndrome in pediatric patients. AJR Am. J. Roentgenol. 2006, 186, 1707–1714. [Google Scholar] [CrossRef]
- Zhang, P.; Li, J.; Liu, H.; Han, N.; Ju, J.; Kou, Y.; Chen, L.; Jiang, M.; Pan, F.; Zheng, Y.; et al. Long-term bone and lung consequences associated with hospital-acquired severe acute respiratory syndrome: A 15-year follow-up from a prospective cohort study. Bone Res. 2020, 8, 8. [Google Scholar] [CrossRef]
- Peiris, J.S.M.; Lai, S.T.; Poon, L.L.M.; Guan, Y.; Yam, L.Y.C.; Lim, W.; Nicholls, J.; Yee, W.K.S.; Yan, W.W.; Cheung, M.T.; et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 2003, 361, 1319–1325. [Google Scholar] [CrossRef]
- Leung, W.K.; To, K.F.; Chan, P.K.; Chan, H.L.; Wu, A.K.; Lee, N.; Yuen, K.Y.; Sung, J.J. Enteric involvement of severe acute respiratory syndrome-associated coronavirus infection. Gastroenterology 2003, 125, 1011–1017. [Google Scholar] [CrossRef]
- Chan, H.L.-Y.; Leung, W.K.; To, K.-F.; Chan, P.K.; Lee, N.; Wu, A.; Tam, J.S.L.; Sung, J.J. Retrospective analysis of liver function derangement in severe acute respiratory syndrome. Am. J. Med. 2004, 116, 566–567. [Google Scholar] [CrossRef]
- Chu, K.H.; Tsang, W.K.; Tang, C.S.; Lam, M.F.; Lai, F.M.; To, K.F.; Fung, K.S.; Tang, H.L.; Yan, W.W.; Chan, H.W.; et al. Acute renal impairment in coronavirus-associated severe acute respiratory syndrome. Kidney Int. 2005, 67, 698–705. [Google Scholar] [CrossRef] [PubMed]
- Wong, R.S.M.; Wu, A.; To, K.F.; Lee, N.; Lam, C.W.K.; Wong, C.K.; Chan, P.K.S.; Ng, M.H.L.; Yu, L.M.; Hui, D.S.; et al. Haematological manifestations in patients with severe acute respiratory syndrome: Retrospective analysis. BMJ 2003, 326, 1358–1362. [Google Scholar] [CrossRef] [PubMed]
- Hung, E.C.; Chim, S.S.; Chan, P.K.; Tong, Y.K.; Ng, E.K.; Chiu, R.W.; Leung, C.-B.; Sung, J.J.; Tam, J.S.; Lo, Y.D. Detection of SARS coronavirus RNA in the cerebrospinal fluid of a patient with severe acute respiratory syndrome. Clin. Chem. 2003, 49, 2108–2109. [Google Scholar] [CrossRef] [PubMed]
- Ng, K.H.L.; Wu, A.K.L.; Cheng, V.C.C.; Tang, B.S.F.; Chan, C.Y.; Yung, C.Y.; Luk, S.H.; Lee, T.W.; Chow, L.; Yuen, K.Y. Pulmonary artery thrombosis in a patient with severe acute respiratory syndrome. Postgrad. Med. J. 2005, 81, e3. [Google Scholar] [CrossRef] [PubMed]
- Umapathi, T.; Kor, A.C.; Venketasubramanian, N.; Lim, C.C.T.; Pang, B.C.; Yeo, T.T.; Lee, C.C.; Lim, P.L.; Ponnudurai, K.; Chuah, K.L.; et al. Large artery ischaemic stroke in severe acute respiratory syndrome (SARS). J. Neurol. 2004, 251, 1227–1231. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Hon, K.-L.E.; Li, K.; Fok, T.F.; Li, C.-K. The effect of SARS coronavirus on blood system: Its clinical findings and the pathophysiologic hypothesis. Zhongguo Shi Yan Xue Ye Xue Za Zhi 2003, 11, 217–221. [Google Scholar] [PubMed]
- Wong, C.K.; Lam, C.W.K.; Wu, A.K.L.; Ip, W.K.; Lee, N.L.S.; Chan, I.H.S.; Lit, L.C.W.; Hui, D.S.C.; Chan, M.H.M.; Chung, S.S.C.; et al. Plasma inflammatory cytokines and chemokines in severe acute respiratory syndrome. Clin. Exp. Immunol. 2004, 136, 95–103. [Google Scholar] [CrossRef]
- Corman, V.M.; Ithete, N.L.; Richards, L.R.; Schoeman, M.C.; Preiser, W.; Drosten, C.; Drexler, J.F.; Raposo, R.A.S.; Abdel-Mohsen, M.; Deng, X.; et al. Rooting the phylogenetic tree of middle east respiratory syndrome coronavirus by characterization of a conspecific virus from an African bat. J. Virol. 2014, 88, 11297–11303. [Google Scholar] [CrossRef]
- Cui, J.; Li, F.; Shi, Z.-L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019, 17, 181–192. [Google Scholar] [CrossRef]
- Zumla, A.; Hui, D.S.; Perlman, S. Middle East respiratory syndrome. Lancet 2015, 386, 995–1007. [Google Scholar] [CrossRef]
- She, J.; Jiang, J.; Ye, L.; Hu, L.; Bai, C.; Song, Y. 2019 novel coronavirus of pneumonia in Wuhan, China: Emerging attack and management strategies. Clin. Transl. Med. 2020, 9, 19. [Google Scholar] [CrossRef] [PubMed]
- Graham, R.L.; Donaldson, E.F.; Baric, R.S. A decade after SARS: Strategies for controlling emerging coronaviruses. Nat. Rev. Microbiol. 2013, 11, 836–848. [Google Scholar] [CrossRef] [PubMed]
- Assiri, A.; Al-Tawfiq, J.A.; Al-Rabeeah, A.; Al-Rabiah, F.; Al-Hajjar, S.; Al-Barrak, A.; Flemban, H.; Al-Nassir, W.N.; Balkhy, H.H.; Al-Hakeem, R.F.; et al. Epidemiological, demographic, and clinical characteristics of 47 cases of Middle East respiratory syndrome coronavirus disease from Saudi Arabia: A descriptive study. Lancet Infect. Dis. 2013, 13, 752–761. [Google Scholar] [CrossRef]
- Arabi, Y.M.; Balkhy, H.H.; Hayden, F.G.; Bouchama, A.; Luke, T.; Baillie, J.K.; Al-Omari, A.; Hajeer, A.H.; Senga, M.; Denison, M.R.; et al. Middle East Respiratory Syndrome. N. Engl. J. Med. 2017, 376, 584–594. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Yang, X.-L.; Wang, X.-G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.-R.; Zhu, Y.; Li, B.; Huang, C.-L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed]
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Kruger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef]
- Lukassen, S.; Chua, R.L.; Trefzer, T.; Kahn, N.C.; Schneider, M.A.; Muley, T.; Winter, H.; Meister, M.; Veith, C.; Boots, A.W.; et al. SARS -CoV-2 receptor ACE 2 and TMPRSS 2 are primarily expressed in bronchial transient secretory cells. EMBO J. 2020, 39, e105114. [Google Scholar] [CrossRef]
- Sungnak, W.; Huang, N.; Becavin, C.; Berg, M.; Queen, R.; Litvinukova, M.; Talavera-Lopez, C.; Maatz, H.; Reichart, D.; Sampaziotis, F.; et al. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 2020, 26, 681–687. [Google Scholar] [CrossRef]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef] [PubMed]
- Gordon, D.E.; Jang, G.M.; Bouhaddou, M.; Xu, J.; Obernier, K.; White, K.M.; O’Meara, M.J.; Rezelj, V.V.; Guo, J.Z.; Swaney, D.L.; et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 2020, 583, 459–468. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.J.; Okuda, K.; Edwards, C.E.; Martinez, D.R.; Asakura, T.; Dinnon, K.H., 3rd; Kato, T.; Lee, R.E.; Yount, B.L.; Mascenik, T.M.; et al. SARS-CoV-2 reverse genetics reveals a variable infection gradient in the respiratory tract. Cell 2020, 182, 429–446.e14. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Kleine-Weber, H.; Pohlmann, S. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol. Cell 2020, 78, 779–784.e5. [Google Scholar] [CrossRef] [PubMed]
- Teesalu, T.; Sugahara, K.N.; Kotamraju, V.R.; Ruoslahti, E. C-end rule peptides mediate neuropilin-1-dependent cell, vascular, and tissue penetration. Proc. Natl. Acad. Sci. USA 2009, 106, 16157–16162. [Google Scholar] [CrossRef]
- Daly, J.L.; Simonetti, B.; Klein, K.; Chen, K.E.; Williamson, M.K.; Anton-Plagaro, C.; Shoemark, D.K.; Simon-Gracia, L.; Bauer, M.; Hollandi, R.; et al. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science 2020, 370, 861–865. [Google Scholar] [CrossRef]
- Cantuti-Castelvetri, L.; Ojha, R.; Pedro, L.D.; Djannatian, M.; Franz, J.; Kuivanen, S.; van der Meer, F.; Kallio, K.; Kaya, T.; Anastasina, M.; et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science 2020, 370, 856–860. [Google Scholar] [CrossRef]
- Ackermann, M.; Verleden, S.E.; Kuehnel, M.; Haverich, A.; Welte, T.; Laenger, F.; Vanstapel, A.; Werlein, C.; Stark, H.; Tzankov, A.; et al. Pulmonary vascular endothelialitis, thrombosis, and angiogenesis in Covid-19. N. Engl. J. Med. 2020, 383, 120–128. [Google Scholar] [CrossRef]
- Amraie, R.; Napoleon, M.A.; Yin, W.; Berrigan, J.; Suder, E.; Zhao, G.; Olejnik, J.; Gummuluru, S.; Muhlberger, E.; Chitalia, V.; et al. CD209L/L-SIGN and CD209/DC-SIGN act as receptors for SARS-CoV-2 and are differentially expressed in lung and kidney epithelial and endothelial cells. bioRxiv 2020. [Google Scholar] [CrossRef]
- Clausen, T.M.; Sandoval, D.R.; Spliid, C.B.; Pihl, J.; Perrett, H.R.; Painter, C.D.; Narayanan, A.; Majowicz, S.A.; Kwong, E.M.; McVicar, R.N.; et al. SARS-CoV-2 infection depends on cellular heparan sulfate and ACE2. Cell 2020, 183, 1043–1057.e15. [Google Scholar] [CrossRef]
- Wang, K.; Chen, W.; Zhang, Z.; Deng, Y.; Lian, J.Q.; Du, P.; Wei, D.; Zhang, Y.; Sun, X.X.; Gong, L.; et al. CD147-spike protein is a novel route for SARS-CoV-2 infection to host cells. Signal Transduct. Target Ther. 2020, 5, 283. [Google Scholar] [CrossRef] [PubMed]
- Rabaan, A.A.; Al-Ahmed, S.H.; Haque, S.; Sah, R.; Tiwari, R.; Malik, Y.S.; Dhama, K.; Yatoo, M.I.; Bonilla-Aldana, D.K.; Rodriguez-Morales, A.J. SARS-CoV-2, SARS-CoV, and MERS-COV: A comparative overview. Infez. Med. 2020, 28, 174–184. [Google Scholar] [PubMed]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef]
- Zheng, J. SARS-CoV-2: An emerging coronavirus that causes a global threat. Int. J. Biol. Sci. 2020, 16, 1678–1685. [Google Scholar] [CrossRef]
- Zhou, F.; Yu, T.; Du, R.; Fan, G.; Liu, Y.; Liu, Z.; Xiang, J.; Wang, Y.; Song, B.; Gu, X.; et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: A retrospective cohort study. Lancet 2020, 395, 1054–1062. [Google Scholar] [CrossRef]
- Giannis, D.; Ziogas, I.A.; Gianni, P. Coagulation disorders in coronavirus infected patients: COVID-19, SARS-CoV-1, MERS-CoV and lessons from the past. J. Clin. Virol. 2020, 127, 104362. [Google Scholar] [CrossRef]
- Tang, N.; Li, D.; Wang, X.; Sun, Z. Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia. J. Thromb. Haemost. 2020, 18, 844–847. [Google Scholar] [CrossRef]
- Subramaniam, S.; Scharrer, I. Procoagulant activity during viral infections. Front. Biosci. 2018, 23, 1060–1081. [Google Scholar]
- Buja, L.M.; Wolf, D.A.; Zhao, B.; Akkanti, B.; McDonald, M.; Lelenwa, L.; Reilly, N.; Ottaviani, G.; Elghetany, M.T.; Trujillo, D.O.; et al. The emerging spectrum of cardiopulmonary pathology of the coronavirus disease 2019 (COVID-19): Report of 3 autopsies from Houston, Texas, and review of autopsy findings from other United States cities. Cardiovasc. Pathol. 2020, 48, 107233. [Google Scholar] [CrossRef]
- Guan, W.J.; Ni, Z.Y.; Hu, Y.; Liang, W.H.; Ou, C.Q.; He, J.X.; Liu, L.; Shan, H.; Lei, C.L.; Hui, D.S.C.; et al. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020, 382, 1708–1720. [Google Scholar] [CrossRef]
- Yan, C.H.; Faraji, F.; Prajapati, D.; Boone, C.E.; DeConde, A.S. Association of chemosensory dysfunction and COVID-19 in patients presenting with influenza-like symptoms. Int. Forum Allergy Rhinol. 2020, 10, 806–813. [Google Scholar] [CrossRef] [PubMed]
- Dell’Era, V.; Farri, F.; Garzaro, G.; Gatto, M.; Valletti, P.A.; Garzaro, M. Smell and taste disorders during COVID -19 outbreak: Cross-sectional study on 355 patients. Head Neck 2020, 42, 1591–1596. [Google Scholar] [CrossRef] [PubMed]
- Printza, A.; Constantinidis, J. The role of self-reported smell and taste disorders in suspected COVID-19. Eur. Arch. Oto-Rhino-Laryngol. 2020, 277, 2625–2630. [Google Scholar] [CrossRef] [PubMed]
- Afzelius, B.A. Ultrastructure of human nasal epithelium during an episode of coronavirus infection. Virchows. Archiv. 1994, 424, 295–300. [Google Scholar] [CrossRef]
- Dourmashkin, R.R.; Tyrrell, D.A.J. Attachment of two myxoviruses to ciliated epithelial cells. J. Gen. Virol. 1970, 9, 77–88. [Google Scholar] [CrossRef]
- Essaidi-Laziosi, M.; Brito, F.; Benaoudia, S.; Royston, L.; Cagno, V.; Fernandes-Rocha, M.; Piuz, I.; Zdobnov, E.; Huang, S.; Constant, S.; et al. Propagation of respiratory viruses in human airway epithelia reveals persistent virus-specific signatures. J. Allergy Clin. Immunol. 2018, 141, 2074–2084. [Google Scholar] [CrossRef]
- Nicholls, J.M.; Poon, L.L.M.; Lee, K.C.; Ng, W.F.; Lai, S.T.; Leung, C.Y.; Chu, C.M.; Hui, P.K.; Mak, K.L.; Lim, W.; et al. Lung pathology of fatal severe acute respiratory syndrome. Lancet 2003, 361, 1773–1778. [Google Scholar] [CrossRef]
- Zajkowska, J.; Hermanowska-Szpakowicz, T.; Pancewicz, S.; Kondrusik, M.; Grygorczuk, S. Severe acute respiratory syndrome (SARS)—New, unknown disease? Polski Merkur. Lek. 2004, 16, 183–187. [Google Scholar]
- Pei, F.; Zheng, J.; Gao, Z.-F.; Zhong, Y.-F.; Fang, W.-G.; Gong, E.-C.; Zou, W.-Z.; Wang, S.-L.; Gao, D.-X.; Xie, Z.; et al. Lung pathology and pathogenesis of severe acute respiratory syndrome: A report of six full autopsies. Zhonghua Bing Li Xue Za Zhi 2005, 34, 656–660. [Google Scholar]
- Chilvers, M.A.; Mckean, M.; Rutman, A.; Myint, B.; Silverman, M.; O’Callaghan, C. The effects of coronavirus on human nasal ciliated respiratory epithelium. Eur. Respir. J. 2001, 18, 965–970. [Google Scholar] [CrossRef]
- Haverkamp, A.-K.; Lehmbecker, A.; Spitzbarth, I.; Widagdo, W.; Haagmans, B.L.; Segalés, J.; Vergara-Alert, J.; Bensaid, A.; Brand, J.M.A.V.D.; Osterhaus, A.D.M.E.; et al. Experimental infection of dromedaries with Middle East respiratory syndrome-Coronavirus is accompanied by massive ciliary loss and depletion of the cell surface receptor dipeptidyl peptidase 4. Sci. Rep. 2018, 8, 9778. [Google Scholar] [CrossRef] [PubMed]
- Chilvers, M.A.; Rutman, A.; O’Callaghan, C.L. Functional analysis of cilia and ciliated epithelial ultrastructure in healthy children and young adults. Thorax 2003, 58, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Heidari, F.; Karimi, E.; Firouzifar, M.; Khamushian, P.; Ansari, R.; Ardehali, M.M. Anosmia as a Prominent Symptom of COVID-19 Infection. Rhinol. J. 2020, 58, 302–303. [Google Scholar] [CrossRef] [PubMed]
- Gane, S.; Kelly, C.; Hopkins, C. Isolated sudden onset anosmia in COVID-19 infection. A novel syndrome? Rhinology 2020, 58, 299–301. [Google Scholar] [CrossRef] [PubMed]
- Eliezer, M.; Hautefort, C.; Hamel, A.-L.; Verillaud, B.; Herman, P.; Houdart, E.; Eloit, C. Sudden and complete olfactory loss of function as a possible symptom of COVID-19. JAMA Otolaryngol. Head Neck Surg. 2020, 146, 674–675. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, P.M.; McEwen, D.P.; Martens, J.R. Olfactory cilia: Linking sensory cilia function and human disease. Chem. Senses 2009, 34, 451–464. [Google Scholar] [CrossRef] [PubMed]
- Williams, C.L.; Uytingco, C.R.; Green, W.W.; McIntyre, J.C.; Ukhanov, K.; Zimmerman, A.D.; Shively, D.T.; Zhang, L.; Nishimura, D.Y.; Sheffield, V.C.; et al. Gene therapeutic reversal of peripheral olfactory impairment in bardet-biedl syndrome. Mol. Ther. 2017, 25, 904–916. [Google Scholar] [CrossRef]
- McEwen, D.P.; Koenekoop, R.K.; Khanna, H.; Jenkins, P.M.; Lopez, I.; Swaroop, A.; Martens, J.R. Hypomorphic CEP290/NPHP6 mutations result in anosmia caused by the selective loss of G proteins in cilia of olfactory sensory neurons. Proc. Natl. Acad. Sci. USA 2007, 104, 15917–15922. [Google Scholar] [CrossRef]
- Li, W.; Li, M.; Ou, G. COVID-19, cilia, and smell. FEBS J. 2020, 287, 3672–3676. [Google Scholar] [CrossRef]
- Lin, D.I.; Aggarwal, P.; Diehl, J.A. Phosphorylation of MCM3 on Ser-112 regulates its incorporation into the MCM2-7 complex. Proc. Natl. Acad. Sci. USA 2008, 105, 8079–8084. [Google Scholar] [CrossRef]
- Enserink, J.M.; Kolodner, R.D. An overview of Cdk1-controlled targets and processes. Cell Div. 2010, 5, 11. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.-S.; Lu, L.X.; Ohi, M.D.; Creamer, K.M.; English, C.; Partridge, J.F.; Ohi, R.; Gould, K.L. Cdk1 phosphorylation of the kinetochore protein Nsk1 prevents error-prone chromosome segregation. J. Cell Biol. 2011, 195, 583–593. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, J.C.; Kiyomitsu, T.; Hori, T.; Backer, C.B.; Fukagawa, T.; Cheeseman, I.M. Aurora B kinase controls the targeting of the Astrin–SKAP complex to bioriented kinetochores. J. Cell Biol. 2010, 191, 269–280. [Google Scholar] [CrossRef] [PubMed]
- Dunsch, A.K.; Linnane, E.; Barr, F.A.; Gruneberg, U. The astrin–kinastrin/SKAP complex localizes to microtubule plus ends and facilitates chromosome alignment. J. Cell Biol. 2011, 192, 959–968. [Google Scholar] [CrossRef]
- Trulioff, A.; Ermakov, A.S.; Malashichev, Y. Primary cilia as a possible link between left-right asymmetry and neurodevelopmental diseases. Genes 2017, 8, 48. [Google Scholar] [CrossRef]
- Goggolidou, P.; Stevens, J.L.; Agueci, F.; Keynton, J.; Wheway, G.; Grimes, D.T.; Patel, S.H.; Hilton, H.; Morthorst, S.; Di Paolo, A.; et al. ATMIN is a transcriptional regulator of both lung morphogenesis and ciliogenesis. Development 2014, 141, 3966–3977. [Google Scholar] [CrossRef]
- Jurado, S.; Conlan, L.A.; Baker, E.K.; Ng, J.-L.; Tenis, N.; Hoch, N.C.; Gleeson, K.; Smeets, M.; Izon, D.; Heierhorst, J. ATM Substrate Chk2-interacting Zn2+ Finger (ASCIZ) is a bi-functional transcriptional activator and feedback sensor in the regulation of dynein light chain (DYNLL1) expression. J. Biol. Chem. 2012, 287, 3156–3164. [Google Scholar] [CrossRef]
- Goetz, S.C.; Anderson, K.V. The primary cilium: A signalling centre during vertebrate development. Nat. Rev. Genet. 2010, 11, 331–344. [Google Scholar] [CrossRef]
- Litingtung, Y.; Lei, L.; Westphal, H.; Chiang, C. Sonic hedgehog is essential to foregut development. Nat. Genet. 1998, 20, 58–61. [Google Scholar] [CrossRef]
- Motoyama, J.; Liu, J.; Mo, R.; Ding, Q.; Post, M.; Hui, C.-C. Essential function of Gli2 and Gli3 in the formation of lung, trachea and oesophagus. Nat. Genet. 1998, 20, 54–57. [Google Scholar] [CrossRef]
- Huber, C.; Cormier-Daire, V. Ciliary disorder of the skeleton. Am. J. Med. Genet. Part. C Semin. Med. Genet. 2012, 160C, 165–174. [Google Scholar] [CrossRef] [PubMed]
- Schmid, A.; Meili, D.; Salathe, M. Soluble adenylyl cyclase in health and disease. Biochim. Biophys. Acta 2014, 1842, 2584–2592. [Google Scholar] [CrossRef] [PubMed]
- Schmid, A.; Sutto, Z.; Schmid, N.; Novak, L.; Ivonnet, P.; Horvath, G.; Conner, G.; Fregien, N.; Salathe, M. Decreased soluble adenylyl cyclase activity in cystic fibrosis is related to defective apical bicarbonate exchange and affects ciliary beat frequency regulation. J. Biol. Chem. 2010, 285, 29998–30007. [Google Scholar] [CrossRef] [PubMed]
- Stephens, R.E.; Prior, G. Dynein from serotonin-activated cilia and flagella: Extraction characteristics and distinct sites for cAMP-dependent protein phosphorylation. J. Cell Sci. 1992, 103, 999–1012. [Google Scholar]
- Wang, Y.; Lam, C.S.; Wu, F.; Wang, W.; Duan, Y.; Huang, P. Regulation of CFTR channels by HCO3−-sensitive soluble adenylyl cyclase in human airway epithelial cells. Am. J. Physiol. Cell Physiol. 2005, 289, C1145–C1151. [Google Scholar] [CrossRef]
- Dubin, P.J.; Kolls, J.K. IL-23 mediates inflammatory responses to mucoid Pseudomonas aeruginosa lung infection in mice. Am. J. Physiol. Lung Cell Mol. Physiol. 2007, 292, L519–L528. [Google Scholar] [CrossRef]
- Happel, K.I.; Zheng, M.; Young, E.; Quinton, L.J.; Lockhart, E.; Ramsay, A.J.; Shellito, J.E.; Schurr, J.R.; Bagby, G.J.; Nelson, S.; et al. Cutting edge: Roles of toll-like receptor 4 and IL-23 in IL-17 expression in response to klebsiella pneumoniae infection. J. Immunol. 2003, 170, 4432–4436. [Google Scholar] [CrossRef]
- Kreindler, J.L.; Bertrand, C.A.; Lee, R.J.; Karasic, T.; Aujla, S.; Pilewski, J.M.; Frizzell, R.A.; Kolls, J.K. Interleukin-17A induces bicarbonate secretion in normal human bronchial epithelial cells. Am. J. Physiol. Cell. Mol. Physiol. 2009, 296, L257–L266. [Google Scholar] [CrossRef]
- Jiang, J.Y.; Falcone, J.L.; Curci, S.; Hofer, A.M. Direct visualization of cAMP signaling in primary cilia reveals up-regulation of ciliary GPCR activity following Hedgehog activation. Proc. Natl. Acad. Sci. USA 2019, 116, 12066–12071. [Google Scholar] [CrossRef]
- Bergeron, C.; Boulet, L.P. Structural changes in airway diseases: Characteristics, mechanisms, consequences, and pharmacologic modulation. Chest 2006, 129, 1068–1087. [Google Scholar] [CrossRef]
- Hogg, J.C.; Chu, F.; Utokaparch, S.; Woods, R.; Elliott, W.M.; Buzatu, L.; Cherniack, R.M.; Rogers, R.M.; Sciurba, F.C.; Coxson, H.O.; et al. The nature of small-airway obstruction in chronic obstructive pulmonary disease. N. Engl. J. Med. 2004, 350, 2645–2653. [Google Scholar] [CrossRef] [PubMed]
- Kuyper, L.M.; Paré, P.D.; Hogg, J.C.; Lambert, R.K.; Ionescu, D.; Woods, R.; Bai, T.R. Characterization of airway plugging in fatal asthma. Am. J. Med. 2003, 115, 6–11. [Google Scholar] [CrossRef]
- Vestbo, J.; Søorensen, T.; Lange, P.; Brix, A.; Torre, P.; Viskum, K. Long-term effect of inhaled budesonide in mild and moderate chronic obstructive pulmonary disease: A randomised controlled trial. Lancet 1999, 353, 1819–1823. [Google Scholar] [CrossRef]
- Thornton, D.J.; Gray, T.; Nettesheim, P.; Howard, M.; Koo, J.S.; Sheehan, J.K. Characterization of mucins from cultured normal human tracheobronchial epithelial cells. Am. J. Physiol. Lung Cell. Mol. Physiol. 2000, 278, L1118–L1128. [Google Scholar] [CrossRef]
- Zuhdi Alimam, M.; Piazza, F.M.; Selby, D.M.; Letwin, N.; Huang, L.; Rose, M.C. Muc-5/5ac mucin messenger RNA and protein expression is a marker of goblet cell metaplasia in murine airways. Am. J. Respir. Cell Mol. Biol. 2000, 22, 253–260. [Google Scholar] [CrossRef]
- Hovenberg, H.W.; Davies, J.R.; Carlstedt, I. Different mucins are produced by the surface epithelium and the submucosa in human trachea: Identification of MUC5AC as a major mucin from the goblet cells. Biochem. J. 1996, 318, 319–324. [Google Scholar] [CrossRef]
- Rose, M.C.; Voynow, J.A. Respiratory tract mucin genes and mucin glycoproteins in health and disease. Physiol. Rev. 2006, 86, 245–278. [Google Scholar] [CrossRef]
- Gray, T.; Nettesheim, P.; Loftin, C.; Koo, J.S.; Bonner, J.; Peddada, S.; Langenbach, R. Interleukin-1beta-induced mucin production in human airway epithelium is mediated by cyclooxygenase-2, prostaglandin E2 receptors, and cyclic AMP-protein kinase A signaling. Mol. Pharmacol. 2004, 66, 337–346. [Google Scholar] [CrossRef]
- Song, K.S.; Lee, W.J.; Chung, K.C.; Koo, J.S.; Yang, E.J.; Choi, J.Y.; Yoon, J.H. Interleukin-1 beta and tumor necrosis factor-alpha induce MUC5AC overexpression through a mechanism involving ERK/p38 mitogen-activated protein kinases-MSK1-CREB activation in human airway epithelial cells. J. Biol. Chem. 2003, 278, 23243–23250. [Google Scholar] [CrossRef]
- Koo, J.S.; Kim, Y.D.; Jetten, A.M.; Belloni, P.; Nettesheim, P. Overexpression of mucin genes induced by interleukin-1 beta, tumor necrosis factor-alpha, lipopolysaccharide, and neutrophil elastase is inhibited by a retinoic acid receptor alpha antagonist. Exp. Lung Res. 2002, 28, 315–332. [Google Scholar] [CrossRef]
- Perrais, M.; Pigny, P.; Copin, M.-C.; Aubert, J.-P.; Van Seuningen, I. Induction of MUC2 and MUC5AC Mucins by Factors of the Epidermal Growth Factor (EGF) Family Is Mediated by EGF Receptor/Ras/Raf/Extracellular Signal-regulated Kinase Cascade and Sp1. J. Biol. Chem. 2002, 277, 32258–32267. [Google Scholar] [CrossRef] [PubMed]
- Borchers, M.T.; Carty, M.P.; Leikauf, G.D. Regulation of human airway mucins by acrolein and inflammatory mediators. Am. J. Physiol. Content 1999, 276, L549–L555. [Google Scholar] [CrossRef] [PubMed]
- Lora, J.M.; Zhang, D.M.; Liao, S.M.; Burwell, T.; King, A.M.; Barker, P.A.; Singh, L.; Keaveney, M.; Morgenstern, J.; Gutierrez-Ramos, J.C.; et al. Tumor necrosis factor-alpha triggers mucus production in airway epithelium through an IkappaB kinase beta-dependent mechanism. J. Biol. Chem. 2005, 280, 36510–36517. [Google Scholar] [CrossRef] [PubMed]
- Li, J.D.; Feng, W.; Gallup, M.; Kim, J.H.; Gum, J.; Kim, Y.; Basbaum, C. Activation of NF-kappaB via a Src-dependent Ras-MAPK-pp90rsk pathway is required for Pseudomonas aeruginosa-induced mucin overproduction in epithelial cells. Proc. Natl. Acad. Sci. USA 1998, 95, 5718–5723. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Thai, P.; Zhao, Y.-H.; Ho, Y.-S.; DeSouza, M.M.; Wu, R. Stimulation of airway mucin gene expression by interleukin (IL)-17 through IL-6 paracrine/autocrine loop. J. Biol. Chem. 2003, 278, 17036–17043. [Google Scholar] [CrossRef] [PubMed]
- Longphre, M.; Li, D.; Gallup, M.; Drori, E.; Ordoñez, C.; Redman, T.; Wenzel, S.; Bice, D.E.; Fahy, J.; Basbaum, C. Allergen-induced IL-9 directly stimulates mucin transcription in respiratory epithelial cells. J. Clin. Investig. 1999, 104, 1375–1382. [Google Scholar] [CrossRef] [PubMed]
- Kuba, K.; Imai, Y.; Rao, S.; Gao, H.; Guo, F.; Guan, B.; Huan, Y.; Yang, P.; Zhang, Y.; Deng, W.; et al. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus–induced lung injury. Nat. Med. 2005, 11, 875–879. [Google Scholar] [CrossRef]
- Eguchi, S.; Kawai, T.; Scalia, R.; Rizzo, V. Understanding angiotensin II type 1 receptor signaling in vascular pathophysiology. Hypertension 2018, 71, 804–810. [Google Scholar] [CrossRef]
- Murakami, M.; Kamimura, D.; Hirano, T. Pleiotropy and specificity: Insights from the interleukin 6 family of cytokines. Immunity 2019, 50, 812–831. [Google Scholar] [CrossRef]
- De Wit, E.; van Doremalen, N.; Falzarano, D.; Munster, V.J. SARS and MERS: Recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016, 14, 523–534. [Google Scholar] [CrossRef]
- Tanaka, Y.; Sato, Y.; Sasaki, T. Suppression of coronavirus replication by cyclophilin inhibitors. Viruses 2013, 5, 1250–1260. [Google Scholar] [CrossRef] [PubMed]
- Radzikowska, U.; Ding, M.; Tan, G.; Zhakparov, D.; Peng, Y.; Wawrzyniak, P.; Wang, M.; Li, S.; Morita, H.; Altunbulakli, C.; et al. Distribution of ACE2, CD147, CD26, and other SARS-CoV-2 associated molecules in tissues and immune cells in health and in asthma, COPD, obesity, hypertension, and COVID-19 risk factors. Allergy 2020. [Google Scholar] [CrossRef] [PubMed]
- Hahn, J.N.; Kaushik, D.K.; Yong, V.W. The role of EMMPRIN in T cell biology and immunological diseases. J. Leukoc. Biol. 2015, 98, 33–48. [Google Scholar] [CrossRef] [PubMed]
- Huai, Q.; Kim, H.-Y.; Liu, Y.; Zhao, Y.; Mondragon, A.; Liu, J.O.; Ke, H. Crystal structure of calcineurin-cyclophilin-cyclosporin shows common but distinct recognition of immunophilin-drug complexes. Proc. Natl. Acad. Sci. USA 2002, 99, 12037–12042. [Google Scholar] [CrossRef] [PubMed]
- Clipstone, N.A.; Crabtree, G.R. Identification of calcineurin as a key signalling enzyme in T-lymphocyte activation. Nature 1992, 357, 695–697. [Google Scholar] [CrossRef] [PubMed]
- Jain, J.; McCafffrey, P.G.; Miner, Z.; Kerppola, T.K.; Lambert, J.N.; Verdine, G.L.; Curran, T.; Rao, A. The T-cell transcription factor NFATp is a substrate for calcineurin and interacts with Fos and Jun. Nature 1993, 365, 352–355. [Google Scholar] [CrossRef]
- Pratt, D.; Chen, J.; Welker, D.; Rivas, R.A.; Pillich, R.; Rynkov, V.; Ono, K.; Miello, C.; Hicks, L.; Szalma, S.; et al. NDEx, the network data exchange. Cell Syst. 2015, 1, 302–305. [Google Scholar] [CrossRef]
- Pratt, D.; Chen, J.; Pillich, R.; Rynkov, V.; Gary, A.; Demchak, B.; Ideker, T. NDEx 2.0: A clearinghouse for research on cancer pathways. Cancer Res. 2017, 77, e58–e61. [Google Scholar] [CrossRef]
- Pillich, R.T.; Chen, J.; Rynkov, V.; Welker, D.; Pratt, D. NDEx: A community resource for sharing and publishing of biological networks. Methods Mol. Biol. 2017, 1558, 271–301. [Google Scholar] [CrossRef]
- Perfetto, L.; Pastrello, C.; Del-Toro, N.; Duesbury, M.; Iannuccelli, M.; Kotlyar, M.; Licata, L.; Meldal, B.; Panneerselvam, K.; Panni, S.; et al. The IMEx Coronavirus interactome: An evolving map of Coronaviridae-Host molecular interactions. Database (Oxford) 2020, 2020, baaa096. [Google Scholar] [CrossRef]
- Luo, C.; Luo, H.; Zheng, S.; Gui, C.; Yue, L.; Yu, C.; Sun, T.; He, P.; Chen, J.; Shen, J.; et al. Nucleocapsid protein of SARS coronavirus tightly binds to human cyclophilin A. Biochem. Biophys. Res. Commun. 2004, 321, 557–565. [Google Scholar] [CrossRef] [PubMed]
- Bouhaddou, M.; Memon, D.; Meyer, B.; White, K.M.; Rezelj, V.V.; Correa Marrero, M.; Polacco, B.J.; Melnyk, J.E.; Ulferts, S.; Kaake, R.M.; et al. The global phosphorylation landscape of SARS-CoV-2 infection. Cell 2020, 182, 685–712.e9. [Google Scholar] [CrossRef] [PubMed]
- Pittet, J.F.; Griffiths, M.J.; Geiser, T.; Kaminski, N.; Dalton, S.L.; Huang, X.; Brown, L.A.; Gotwals, P.J.; Koteliansky, V.E.; Matthay, M.A.; et al. TGF-beta is a critical mediator of acute lung injury. J. Clin. Investig. 2001, 107, 1537–1544. [Google Scholar] [CrossRef] [PubMed]
- Mo, X.; Jian, W.; Su, Z.; Chen, M.; Peng, H.; Peng, P.; Lei, C.; Chen, R.; Zhong, N.; Li, S. Abnormal pulmonary function in COVID-19 patients at time of hospital discharge. Eur. Respir. J. 2020, 55, 2001217. [Google Scholar] [CrossRef]
- Stukalov, A.; Girault, V.; Grass, V.; Bergant, V.; Karayel, O.; Urban, C.; Haas, D.A.; Huang, Y.; Oubraham, L.; Wang, A. Multi-level proteomics reveals host-perturbation strategies of SARS-CoV-2 and SARS-CoV. bioRxiv 2020. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Lieberman, N.A.P.; Peddu, V.; Xie, H.; Shrestha, L.; Huang, M.L.; Mears, M.C.; Cajimat, M.N.; Bente, D.A.; Shi, P.Y.; Bovier, F.; et al. In vivo antiviral host transcriptional response to SARS-CoV-2 by viral load, sex, and age. PLoS Biol. 2020, 18, e3000849. [Google Scholar] [CrossRef]
- Anderson, S.D.; Daviskas, E.; Brannan, J.D.; Chan, H.-K. Repurposing excipients as active inhalation agents: The mannitol story. Adv. Drug Deliv. Rev. 2018, 133, 45–56. [Google Scholar] [CrossRef]
- Teper, A.; Jaques, A.; Charlton, B. Inhaled mannitol in patients with cystic fibrosis: A randomised open-label dose response trial. J. Cyst. Fibros. 2011, 10, 1–8. [Google Scholar] [CrossRef][Green Version]
- Bennett, W.D. Effect of beta-adrenergic agonists on mucociliary clearance. J. Allergy Clin. Immunol. 2002, 110, S291–S297. [Google Scholar] [CrossRef]
- Davis, B.; Marin, M.G.; Yee, J.W.; Nadel, J.A. Effect of terbutaline on movement of Cl- and Na+ across the trachea of the dog in vitro. Am. Rev. Respir. Dis. 1979, 120, 547–552. [Google Scholar] [PubMed]
- Shak, S.; Capon, D.J.; Hellmiss, R.; Marsters, S.A.; Baker, C.L. Recombinant human DNase I reduces the viscosity of cystic fibrosis sputum. Proc. Natl. Acad. Sci. USA 1990, 87, 9188–9192. [Google Scholar] [CrossRef] [PubMed]
- Yoo, Y.; Koh, Y.Y. Current treatment for primary ciliary dyskinesia conditions. Expert Opin. Pharmacother. 2004, 5, 369–377. [Google Scholar] [CrossRef] [PubMed]
- Knowles, M.R.; Church, N.L.; Waltner, W.E.; Yankaskas, J.R.; Gilligan, P.; King, M.; Edwards, L.J.; Helms, R.W.; Boucher, R.C. A pilot study of aerosolized amiloride for the treatment of lung disease in cystic fibrosis. N. Engl. J. Med. 1990, 322, 1189–1194. [Google Scholar] [CrossRef]
- Tomkiewicz, R.P.; App, E.M.; Zayas, J.G.; Ramirez, O.; Church, N.; Boucher, R.C.; Knowles, M.R.; King, M. Amiloride inhalation therapy in cystic fibrosis: Influence on ion content, hydration, and rheology of sputum. Am. Rev. Respir. Dis. 1993, 148, 1002–1007. [Google Scholar] [CrossRef]
- Kellerman, D.; Mospan, A.R.; Engels, J.; Schaberg, A.; Gorden, J.; Smiley, L. Denufosol: A review of studies with inhaled P2Y2 agonists that led to Phase 3. Pulm. Pharmacol. Ther. 2008, 21, 600–607. [Google Scholar] [CrossRef]
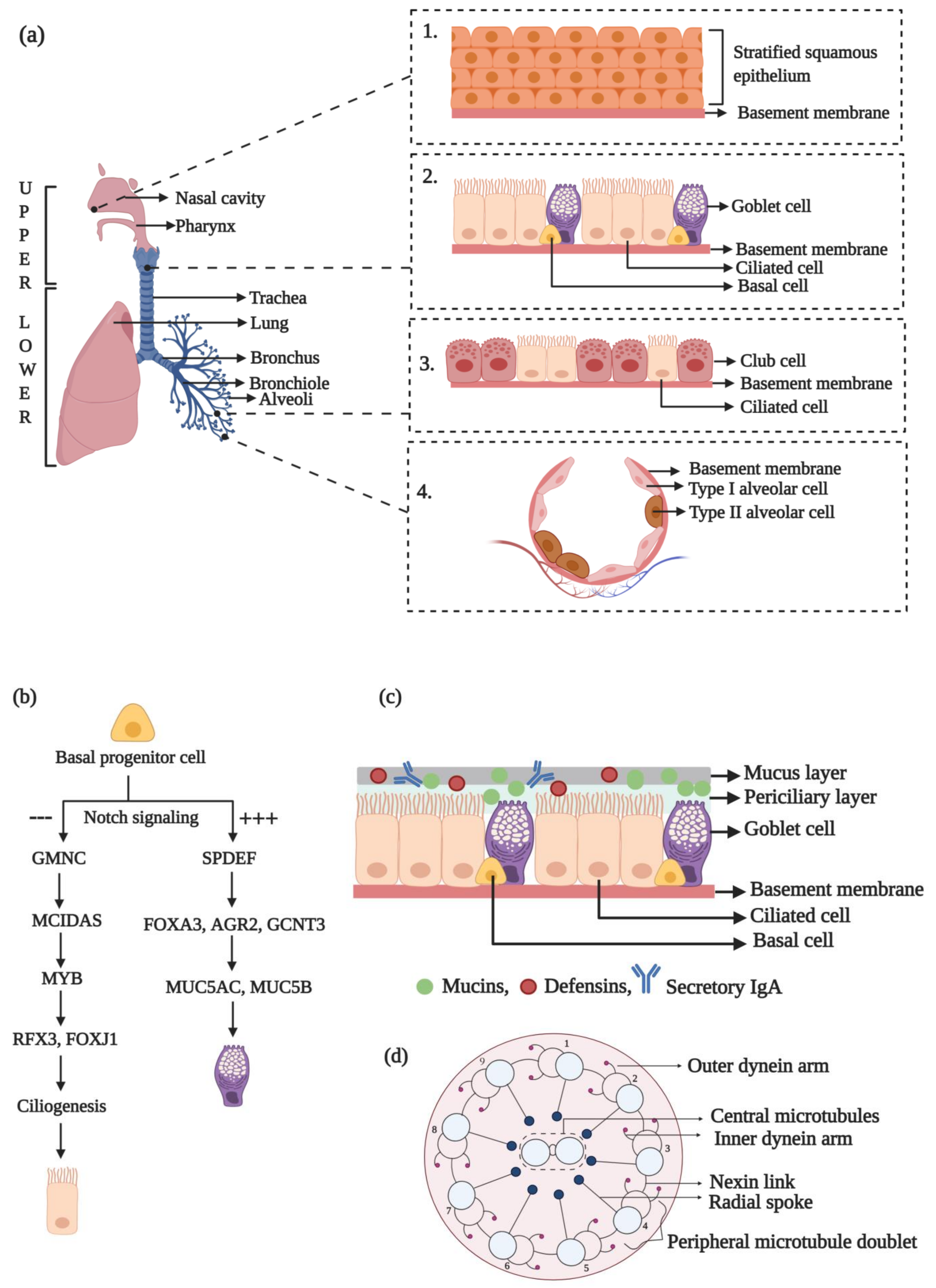

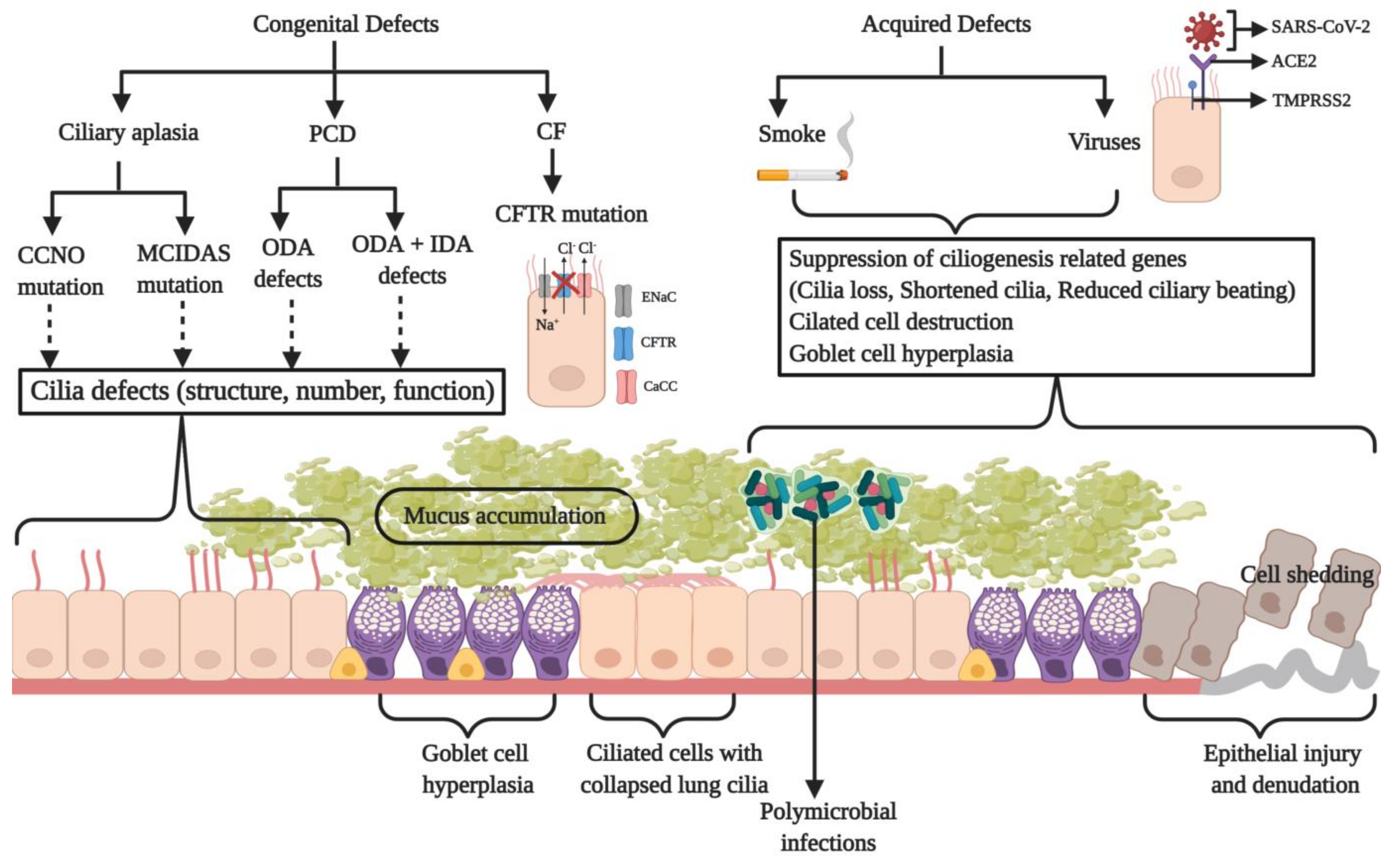
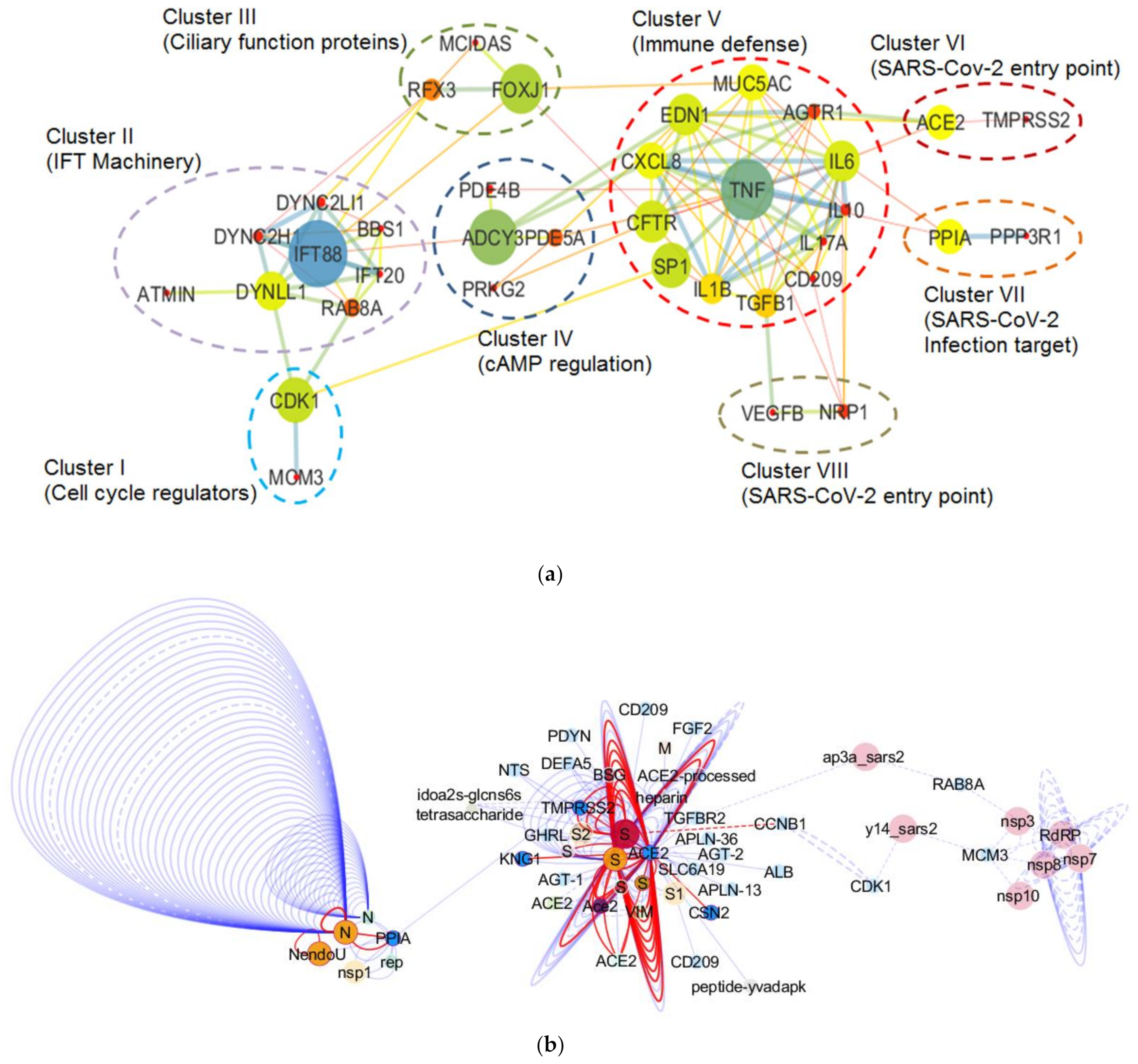
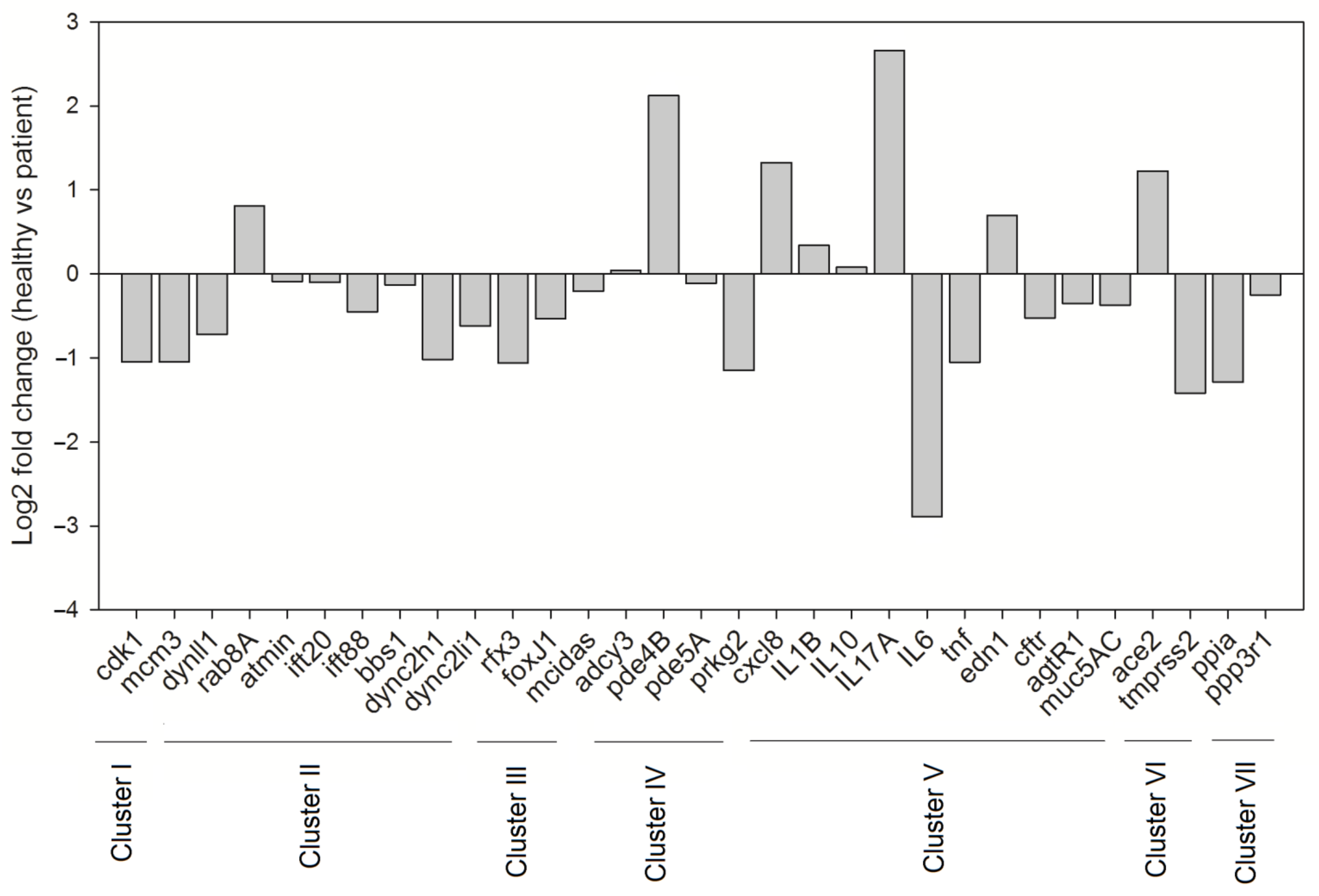
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adivitiya; Kaushik, M.S.; Chakraborty, S.; Veleri, S.; Kateriya, S. Mucociliary Respiratory Epithelium Integrity in Molecular Defense and Susceptibility to Pulmonary Viral Infections. Biology 2021, 10, 95. https://doi.org/10.3390/biology10020095
Adivitiya, Kaushik MS, Chakraborty S, Veleri S, Kateriya S. Mucociliary Respiratory Epithelium Integrity in Molecular Defense and Susceptibility to Pulmonary Viral Infections. Biology. 2021; 10(2):95. https://doi.org/10.3390/biology10020095
Chicago/Turabian StyleAdivitiya, Manish Singh Kaushik, Soura Chakraborty, Shobi Veleri, and Suneel Kateriya. 2021. "Mucociliary Respiratory Epithelium Integrity in Molecular Defense and Susceptibility to Pulmonary Viral Infections" Biology 10, no. 2: 95. https://doi.org/10.3390/biology10020095
APA StyleAdivitiya, Kaushik, M. S., Chakraborty, S., Veleri, S., & Kateriya, S. (2021). Mucociliary Respiratory Epithelium Integrity in Molecular Defense and Susceptibility to Pulmonary Viral Infections. Biology, 10(2), 95. https://doi.org/10.3390/biology10020095







