From Clonal Hematopoiesis to Therapy-Related Myeloid Neoplasms: The Silent Way of Cancer Progression
Abstract
:Simple Summary
Abstract
1. Introduction
2. CHIP (Clonal Hematopoiesis of Indeterminate Potential) and ARCH (Age-Related Clonal Hematopoiesis): Discovery, Biology, Definition and Risk Factors
3. When Clonal Hematopoiesis Becomes Clinically Evident: The Cases of ICUS (Idiopathic Cytopenia of Undetermined Significance) and CCUS (Clonal Cytopenia of Undetermined Significance)
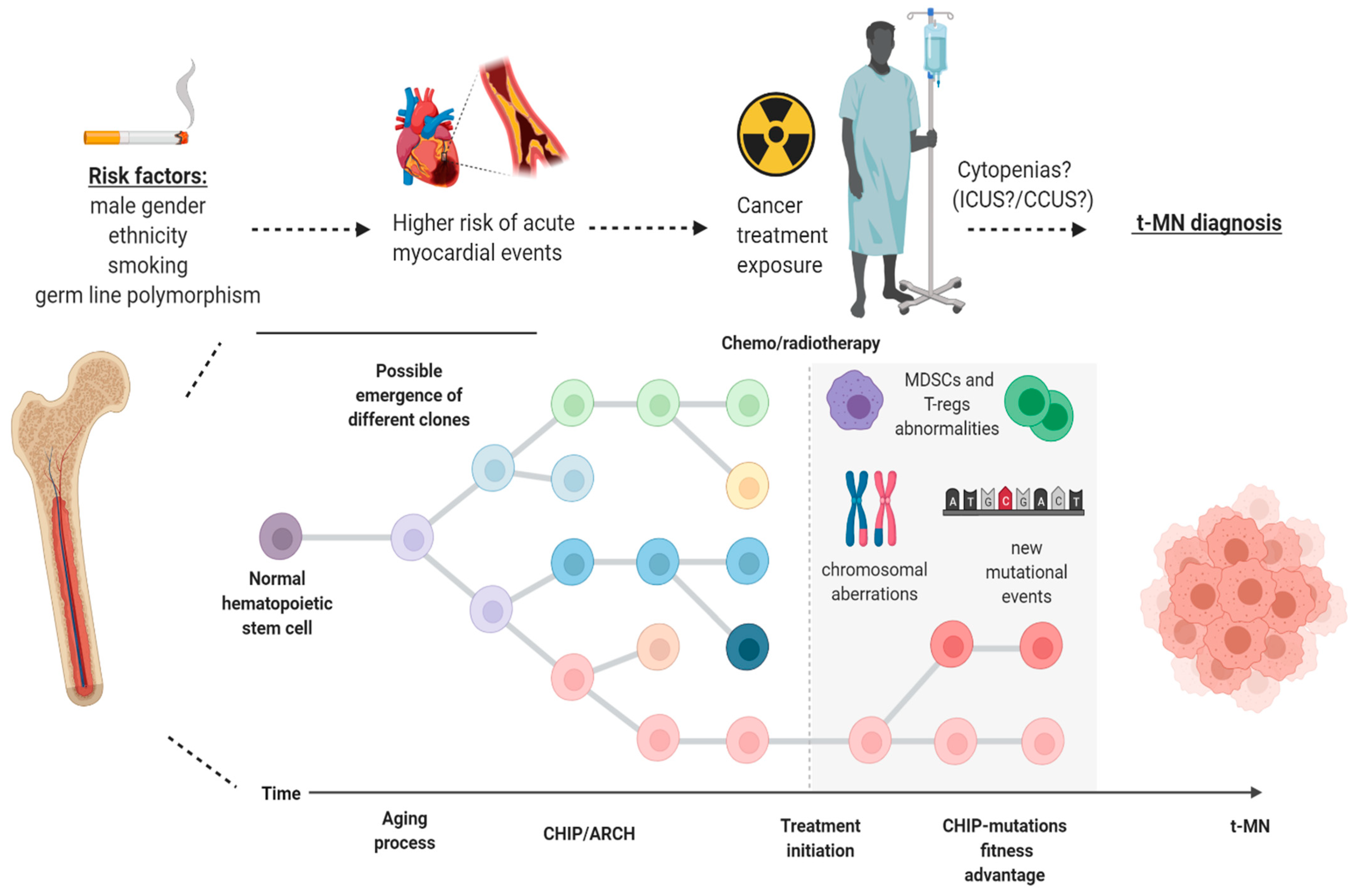
4. Therapy-Related Myeloid Neoplasm and Clonal Hematopoiesis: The Shift of a Paradigm
5. The Case of TP53 and PPM1D in Therapy-Related CHIP
6. Clinical Implications of CHIP: A Focus on Cardiovascular Risk
7. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
List of Gene Symbol Abbreviations
| Gene Symbol | Gene Name |
| ASXL1 | Additional sex combs-like 1 |
| CBL | Casitas B-lineage Lymphoma Proto-Oncogene |
| CBLB | Cbl Proto-Oncogene B |
| CHEK2 | Checkpoint kinase 2 |
| DNMT3A | DNA Methyltransferase 3 Alpha |
| EZH2 | Enhancer Of Zeste 2 Polycomb Repressive Complex 2 Subunit |
| FLT3 | Fms-like tyrosine kinase 3 |
| GNAS | Guanine nucleotide-binding protein G(s) subunit alpha |
| GNB1 | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
| IDH1/2 | Isocitrate dehydrogenase 1/2 |
| JAK2 | Janus kinase 2 |
| NPM1 | Nucleophosmin |
| PPM1D | Protein Phosphatase, Mg2+/Mn2+ Dependent 1D |
| PRPF8 | Pre-MRNA Processing Factor 8 |
| RAD21 | RAD21 Cohesin Complex Component |
| RAS | Rat Sarcoma Viral Oncogene Homolog |
| RUNX1 | Runt-related transcription factor 1 |
| SF3B1 | Splicing factor 3B subunit 1 |
| SRSF2 | Serine And Arginine Rich Splicing Factor 2 |
| TERT | Telomerase Reverse Transcriptase |
| TET2 | Tet methylcytosine dioxygenase 2 |
| TP53 | Tumor protein p53 |
| U2AF1 | U2 Small Nuclear RNA Auxiliary Factor 1 |
References
- Knudson, A.G. Mutation and Cancer: Statistical Study of Retinoblastoma. Proc. Natl. Acad. Sci. USA 1971, 68, 820–823. [Google Scholar] [CrossRef] [Green Version]
- Vogelstein, B.; Papadopoulos, N.; Velculescu, V.E.; Zhou, S.; Diaz, L.A.; Kinzler, K.W. Cancer Genome Landscapes. Science 2013, 339, 1546–1558. [Google Scholar] [CrossRef]
- Gibson, C.J.; Steensma, D.P. New Insights from Studies of Clonal Hematopoiesis. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2018, 24, 4633–4642. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee-Six, H.; Øbro, N.F.; Shepherd, M.S.; Grossmann, S.; Dawson, K.; Belmonte, M.; Osborne, R.J.; Huntly, B.J.P.; Martincorena, I.; Anderson, E.; et al. Population dynamics of normal human blood inferred from somatic mutations. Nature 2018, 561, 473–478. [Google Scholar] [CrossRef]
- Bowman, R.L.; Busque, L.; Levine, R.L. Clonal Hematopoiesis and Evolution to Hematopoietic Malignancies. Cell Stem Cell 2018, 22, 157–170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steensma, D.P.; Bejar, R.; Jaiswal, S.; Lindsley, R.C.; Sekeres, M.A.; Hasserjian, R.P.; Ebert, B.L. Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood 2015, 126, 9–16. [Google Scholar] [CrossRef] [Green Version]
- Steensma, D.P. Clinical Implications of Clonal Hematopoiesis. Mayo Clin. Proc. 2018, 93, 1122–1130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jaiswal, S.; Natarajan, P.; Silver, A.J.; Gibson, C.J.; Bick, A.G.; Shvartz, E.; McConkey, M.; Gupta, N.; Gabriel, S.; Ardissino, D.; et al. Clonal Hematopoiesis and Risk of Atherosclerotic Cardiovascular Disease. N. Engl. J. Med. 2017, 377, 111–121. [Google Scholar] [CrossRef]
- Murthy, G.S.G.; Hamadani, M.; Dhakal, B.; Hari, P.; Atallah, E. Incidence and survival of therapy related myeloid neoplasm in United States. Leuk. Res. 2018, 71, 95–99. [Google Scholar] [CrossRef]
- Desai, P.; Roboz, G.J. Clonal Hematopoiesis and therapy related MDS/AML. Best Pract. Res. Clin. Haematol. 2019, 32, 13–23. [Google Scholar] [CrossRef]
- Rowley, J.D. A New Consistent Chromosomal Abnormality in Chronic Myelogenous Leukaemia identified by Quinacrine Fluorescence and Giemsa Staining. Nature 1973, 243, 290–293. [Google Scholar] [CrossRef] [PubMed]
- Fey, M.F.; Liechti-Gallati, S.; von Rohr, A.; Borisch, B.; Theilkäs, L.; Schneider, V.; Oestreicher, M.; Nagel, S.; Ziemiecki, A.; Tobler, A. Clonality and X-inactivation patterns in hematopoietic cell populations detected by the highly informative M27 beta DNA probe. Blood 1994, 83, 931–938. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Busque, L.; Patel, J.P.; Figueroa, M.E.; Vasanthakumar, A.; Provost, S.; Hamilou, Z.; Mollica, L.; Li, J.; Viale, A.; Heguy, A.; et al. Recurrent somatic TET2 mutations in normal elderly individuals with clonal hematopoiesis. Nat. Genet. 2012, 44, 1179–1181. [Google Scholar] [CrossRef] [PubMed]
- Shlush, L.I.; Zandi, S.; Mitchell, A.; Chen, W.C.; Brandwein, J.M.; Gupta, V.; Kennedy, J.A.; Schimmer, A.D.; Schuh, A.C.; Yee, K.W.; et al. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature 2014, 506, 328–333. [Google Scholar] [CrossRef]
- Delhommeau, F.; Dupont, S.; Valle, V.D.; James, C.; Trannoy, S.; Massé, A.; Kosmider, O.; Le Couedic, J.-P.; Robert, F.; Alberdi, A.; et al. Mutation in TET2 in Myeloid Cancers. N. Engl. J. Med. 2009, 360, 2289–2301. [Google Scholar] [CrossRef] [PubMed]
- Xie, M.; Lu, C.; Wang, J.; McLellan, M.D.; Johnson, K.J.; Wendl, M.C.; McMichael, J.F.; Schmidt, H.K.; Yellapantula, V.; Miller, C.A.; et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat. Med. 2014, 20, 1472–1478. [Google Scholar] [CrossRef]
- Genovese, G.; Kähler, A.K.; Handsaker, R.E.; Lindberg, J.; Rose, S.A.; Bakhoum, S.F.; Chambert, K.; Mick, E.; Neale, B.M.; Fromer, M.; et al. Clonal Hematopoiesis and Blood-Cancer Risk Inferred from Blood DNA Sequence. N. Engl. J. Med. 2014, 371, 2477–2487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jaiswal, S.; Fontanillas, P.; Flannick, J.; Manning, A.; Grauman, P.V.; Mar, B.G.; Lindsley, R.C.; Mermel, C.H.; Burtt, N.; Chavez, A.; et al. Age-Related Clonal Hematopoiesis Associated with Adverse Outcomes. N. Engl. J. Med. 2014, 371, 2488–2498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kolata, G. Scientists discover a bone-deep risk for heart disease. New York Times, 30 January 2018. Available online: https://www.nytimes.com/2018/01/29/health/heart-disease-mutations-stem-cells.html(accessed on 29 December 2020).
- Jongen-Lavrencic, M.; Grob, T.; Hanekamp, D.; Kavelaars, F.G.; al Hinai, A.; Zeilemaker, A.; Erpelinck-Verschueren, C.A.J.; Gradowska, P.L.; Meijer, R.; Cloos, J.; et al. Molecular Minimal Residual Disease in Acute Myeloid Leukemia. N. Engl. J. Med. 2018, 378, 1189–1199. [Google Scholar] [CrossRef]
- Steensma, D.P.; Ebert, B.L. Clonal Hematopoiesis after Induction Chemotherapy for Acute Myeloid Leukemia. N. Engl. J. Med. 2018, 378, 1244–1245. [Google Scholar] [CrossRef]
- Ottone, T.; Alfonso, V.; Iaccarino, L.; Hasan, S.K.; Mancini, M.; Divona, M.; Lavorgna, S.; Cicconi, L.; Panetta, P.; Maurillo, L.; et al. Longitudinal detection of DNMT3A(R882H) transcripts in patients with acute myeloid leukemia. Am. J. Hematol. 2018, 93, E120–E123. [Google Scholar] [CrossRef] [PubMed]
- Steensma, D.P. Clinical consequences of clonal hematopoiesis of indeterminate potential. Blood Adv. 2018, 2, 3404–3410. [Google Scholar] [CrossRef]
- Steensma, D.P.; Ebert, B.L. Clonal hematopoiesis as a model for premalignant changes during aging. Exp. Hematol. 2020, 83, 48–56. [Google Scholar] [CrossRef] [Green Version]
- Kyle, R.A.; Larson, D.R.; Therneau, T.M.; Dispenzieri, A.; Kumar, S.; Cerhan, J.R.; Rajkumar, S.V. Long-Term Follow-up of Monoclonal Gammopathy of Undetermined Significance. N. Engl. J. Med. 2018, 378, 241–249. [Google Scholar] [CrossRef] [PubMed]
- Desai, P.; Mencia-Trinchant, N.; Savenkov, O.; Simon, M.S.; Cheang, G.; Lee, S.; Samuel, M.; Ritchie, E.K.; Guzman, M.L.; Ballman, K.V.; et al. Somatic mutations precede acute myeloid leukemia years before diagnosis. Nat. Med. 2018, 24, 1015–1023. [Google Scholar] [CrossRef]
- Abelson, S.; Collord, G.; Ng, S.W.K.; Weissbrod, O.; Cohen, N.M.; Niemeyer, E.; Barda, N.; Zuzarte, P.C.; Heisler, L.; Sundaravadanam, Y.; et al. Prediction of acute myeloid leukaemia risk in healthy individuals. Nature 2018, 559, 400–404. [Google Scholar] [CrossRef] [PubMed]
- Wong, T.N.; Ramsingh, G.; Young, A.L.; Miller, C.A.; Touma, W.; Welch, J.S.; Lamprecht, T.L.; Shen, D.; Hundal, J.; Fulton, R.S.; et al. Role of TP53 mutations in the origin and evolution of therapy-related acute myeloid leukaemia. Nature 2015, 518, 552–555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valent, P.; Bain, B.J.; Bennett, J.M.; Wimazal, F.; Sperr, W.R.; Mufti, G.; Horny, H.P. Idiopathic cytopenia of undetermined significance (ICUS) and idiopathic dysplasia of uncertain significance (IDUS), and their distinction from low risk MDS. Leuk. Res. 2012, 36, 1–5. [Google Scholar] [CrossRef]
- Valent, P.; Horny, H.P.; Bennett, J.M.; Fonatsch, C.; Germing, U.; Greenberg, P.; Haferlach, T.; Haase, D.; Kolb, H.J.; Krieger, O.; et al. Definitions and standards in the diagnosis and treatment of the myelodysplastic syndromes: Consensus statements and report from a working conference. Leuk. Res. 2007, 31, 727–736. [Google Scholar] [CrossRef] [PubMed]
- Steensma, D.P. The Clinical Challenge of Idiopathic Cytopenias of Undetermined Significance (ICUS) and Clonal Cytopenias of Undetermined Significance (CCUS). Curr. Hematol. Malig. Rep. 2019, 14, 536–542. [Google Scholar] [CrossRef]
- Malcovati, L.; Cazzola, M. The shadowlands of MDS: Idiopathic cytopenias of undetermined significance (ICUS) and clonal hematopoiesis of indeterminate potential (CHIP). Hematol. Am. Soc. Hematol. Educ. Program 2015, 2015, 299–307. [Google Scholar] [CrossRef] [PubMed]
- Mufti, G.J.; Bennett, J.M.; Goasguen, J.; Bain, B.J.; Baumann, I.; Brunning, R.; Cazzola, M.; Fenaux, P.; Germing, U.; Hellström-Lindberg, E.; et al. Diagnosis and classification of myelodysplastic syndrome: International Working Group on Morphology of myelodysplastic syndrome (IWGM-MDS) consensus proposals for the definition and enumeration of myeloblasts and ring sideroblasts. Haematologica 2008, 93, 1712–1717. [Google Scholar] [CrossRef]
- Arber, D.A.; Orazi, A.; Hasserjian, R.; Thiele, J.; Borowitz, M.J.; Le Beau, M.M.; Bloomfield, C.D.; Cazzola, M.; Vardiman, J.W. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 2016, 127, 2391–2405. [Google Scholar] [CrossRef]
- Valent, P.; Orazi, A.; Steensma, D.P.; Ebert, B.L.; Haase, D.; Malcovati, L.; van de Loosdrecht, A.A.; Haferlach, T.; Westers, T.M.; Wells, D.A.; et al. Proposed minimal diagnostic criteria for myelodysplastic syndromes (MDS) and potential pre-MDS conditions. Oncotarget 2017, 8, 73483–73500. [Google Scholar] [CrossRef] [Green Version]
- Malcovati, L.; Gallì, A.; Travaglino, E.; Ambaglio, I.; Rizzo, E.; Molteni, E.; Elena, C.; Ferretti, V.V.; Catricalà, S.; Bono, E.; et al. Clinical significance of somatic mutation in unexplained blood cytopenia. Blood 2017, 129, 3371–3378. [Google Scholar] [CrossRef]
- Kern, W.; Meggendorfer, M.; Haferlach, C.; Haferlach, T. Integrated Diagnostic Approach for Suspected Myelodysplastic Syndrome As a Basis for Advancement of Diagnostic Criteria. Blood 2016, 128, 299. [Google Scholar] [CrossRef]
- Fianchi, L.; Criscuolo, M.; Fabiani, E.; Falconi, G.; Maraglino, A.M.E.; Voso, M.T.; Pagano, L. Therapy-related myeloid neoplasms: Clinical perspectives. Onco Target 2018, 11, 5909–5915. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuendgen, A.; Nomdedeu, M.; Tuechler, H.; Garcia-Manero, G.; Komrokji, R.S.; Sekeres, M.A.; Porta, M.G.D.; Cazzola, M.; DeZern, A.E.; Roboz, G.J.; et al. Therapy-related myelodysplastic syndromes deserve specific diagnostic sub-classification and risk-stratification-an approach to classification of patients with t-MDS. Leukemia 2020, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Criscuolo, M.; Chiusolo, P.; Giammarco, S.; Giachelia, M.; Fianchi, L.; Fabiani, E.; Falconi, G.; Hohaus, S.; Sica, S.; Leone, G.; et al. Methylenetetrahydrofolate reductase polymorphisms in myelodysplastic syndromes and therapy-related myeloid neoplasms. Leuk. Lymphoma 2014, 55, 2942–2944. [Google Scholar] [CrossRef]
- Fabiani, E.; Fianchi, L.; Falconi, G.; Boncompagni, R.; Criscuolo, M.; Guidi, F.; La Brocca, A.; Hohaus, S.; Leone, G.; Voso, M.T. The BCL2L10 Leu21Arg variant and risk of therapy-related myeloid neoplasms and de novo myelodysplastic syndromes. Leuk. Lymphoma 2014, 55, 1538–1543. [Google Scholar] [CrossRef]
- Voso, M.T.; Fabiani, E.; Zang, Z.; Fianchi, L.; Falconi, G.; Padella, A.; Martini, M.; Li Zhang, S.; Santangelo, R.; Larocca, L.M.; et al. Fanconi anemia gene variants in therapy-related myeloid neoplasms. Blood Cancer J. 2015, 5, e323. [Google Scholar] [CrossRef] [Green Version]
- Zawit, M.; Durrani, J.; Shen, W.; Adema, V.; Kerr, C.M.; Awada, H.; Kongkiatkamon, S.; Gurnari, C.; Pagliuca, S.; Terkawi, L.; et al. Impact of Pathogenic Germ Line Variants in Adults with Acquired Bone Marrow Failure Syndromes Vs. Myeloid Neoplasia. Blood 2020, 136, 106. [Google Scholar] [CrossRef]
- Gurnari, C.; Pagliuca, S.; Patel, B.J.; Awada, H.; Kerr, C.M.; Shen, W.; Kongkiatkamon, S.; Terkawi, L.; Zawit, M.; Durrani, J.; et al. The Genomic Landscape of Myeloid Neoplasms Evolved from AA/PNH. Blood 2020, 136, 337–347. [Google Scholar] [CrossRef]
- Takahashi, K.; Wang, F.; Kantarjian, H.; Doss, D.; Khanna, K.; Thompson, E.; Zhao, L.; Patel, K.; Neelapu, S.; Gumbs, C.; et al. Preleukaemic clonal haemopoiesis and risk of therapy-related myeloid neoplasms: A case-control study. Lancet. Oncol. 2017, 18, 100–111. [Google Scholar] [CrossRef] [Green Version]
- Gillis, N.K.; Ball, M.; Zhang, Q.; Ma, Z.; Zhao, Y.; Yoder, S.J.; Balasis, M.E.; Mesa, T.E.; Sallman, D.A.; Lancet, J.E.; et al. Clonal haemopoiesis and therapy-related myeloid malignancies in elderly patients: A proof-of-concept, case-control study. Lancet. Oncol. 2017, 18, 112–121. [Google Scholar] [CrossRef]
- Arends, C.M.; Galan-Sousa, J.; Hoyer, K.; Chan, W.; Jäger, M.; Yoshida, K.; Seemann, R.; Noerenberg, D.; Waldhueter, N.; Fleischer-Notter, H.; et al. Hematopoietic lineage distribution and evolutionary dynamics of clonal hematopoiesis. Leukemia 2018, 32, 1908–1919. [Google Scholar] [CrossRef] [PubMed]
- Voso, M.T.; Pandzic, T.; Iskas, M.; Denčić-Fekete, M.; De Bellis, E.; Scarfo, L.; Ljungström, V.; Del Poeta, G.; Ranghetti, P.; Laidou, S.; et al. Clonal Hematopoiesis Is Associated with Increased Risk for Therapy-Related Myeloid Neoplasms in Chronic Lymphocytic Leukemia Patients Treated with Chemo(immuno)Therapy. Blood 2020, 136, 19–20. [Google Scholar] [CrossRef]
- Abkowitz, J.L. Clone Wars—The Emergence of Neoplastic Blood-Cell Clones with Aging. N. Engl. J. Med. 2014, 371, 2523–2525. [Google Scholar] [CrossRef]
- Gibson, C.J.; Lindsley, R.C.; Tchekmedyian, V.; Mar, B.G.; Shi, J.; Jaiswal, S.; Bosworth, A.; Francisco, L.; He, J.; Bansal, A.; et al. Clonal Hematopoiesis Associated With Adverse Outcomes After Autologous Stem-Cell Transplantation for Lymphoma. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2017, 35, 1598–1605. [Google Scholar] [CrossRef]
- Coombs, C.C.; Zehir, A.; Devlin, S.M.; Kishtagari, A.; Syed, A.; Jonsson, P.; Hyman, D.M.; Solit, D.B.; Robson, M.E.; Baselga, J.; et al. Therapy-Related Clonal Hematopoiesis in Patients with Non-hematologic Cancers Is Common and Associated with Adverse Clinical Outcomes. Cell Stem Cell 2017, 21, 374–382. [Google Scholar] [CrossRef] [Green Version]
- Alexandrov, L.B.; Nik-Zainal, S.; Wedge, D.C.; Aparicio, S.A.; Behjati, S.; Biankin, A.V.; Bignell, G.R.; Bolli, N.; Borg, A.; Børresen-Dale, A.L.; et al. Signatures of mutational processes in human cancer. Nature 2013, 500, 415–421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshida, K.; French, B.; Yoshida, N.; Hida, A.; Ohishi, W.; Kusunoki, Y. Radiation exposure and longitudinal changes in peripheral monocytes over 50 years: The Adult Health Study of atomic-bomb survivors. Br. J. Haematol. 2019, 185, 107–115. [Google Scholar] [CrossRef] [PubMed]
- Midic, D.; Rinke, J.; Perner, F.; Müller, V.; Hinze, A.; Pester, F.; Landschulze, J.; Ernst, J.; Gruhn, B.; Matziolis, G.; et al. Prevalence and dynamics of clonal hematopoiesis caused by leukemia-associated mutations in elderly individuals without hematologic disorders. Leukemia 2020, 34, 2198–2205. [Google Scholar] [CrossRef] [PubMed]
- Bolton, K.L.; Ptashkin, R.N.; Gao, T.; Braunstein, L.; Devlin, S.M.; Kelly, D.; Patel, M.; Berthon, A.; Syed, A.; Yabe, M.; et al. Cancer therapy shapes the fitness landscape of clonal hematopoiesis. Nat. Genet. 2020, 52, 1219–1226. [Google Scholar] [CrossRef] [PubMed]
- McNerney, M.E.; Godley, L.A.; Le Beau, M.M. Therapy-related myeloid neoplasms: When genetics and environment collide. Nat. Rev. Cancer 2017, 17, 513–527. [Google Scholar] [CrossRef]
- Wong, T.N.; Miller, C.A.; Jotte, M.R.M.; Bagegni, N.; Baty, J.D.; Schmidt, A.P.; Cashen, A.F.; Duncavage, E.J.; Helton, N.M.; Fiala, M.; et al. Cellular stressors contribute to the expansion of hematopoietic clones of varying leukemic potential. Nat. Commun. 2018, 9, 455. [Google Scholar] [CrossRef]
- Chen, S.; Gao, R.; Yao, C.; Kobayashi, M.; Liu, S.Z.; Yoder, M.C.; Broxmeyer, H.; Kapur, R.; Boswell, H.S.; Mayo, L.D.; et al. Genotoxic stresses promote clonal expansion of hematopoietic stem cells expressing mutant p53. Leukemia 2018, 32, 850–854. [Google Scholar] [CrossRef] [PubMed]
- Fabiani, E.; Falconi, G.; Fianchi, L.; Criscuolo, M.; Ottone, T.; Cicconi, L.; Hohaus, S.; Sica, S.; Postorino, M.; Neri, A.; et al. Clonal evolution in therapy-related neoplasms. Oncotarget 2017, 8, 12031–12040. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Welch, J.S. Patterns of mutations in TP53 mutated AML. Best Pract. Res. Clin. Haematol. 2018, 31, 379–383. [Google Scholar] [CrossRef] [PubMed]
- Bernard, E.; Nannya, Y.; Hasserjian, R.P.; Devlin, S.M.; Tuechler, H.; Medina-Martinez, J.S.; Yoshizato, T.; Shiozawa, Y.; Saiki, R.; Malcovati, L.; et al. Implications of TP53 allelic state for genome stability, clinical presentation and outcomes in myelodysplastic syndromes. Nat. Med. 2020, 26, 1549–1556. [Google Scholar] [CrossRef]
- Sallman, D.A.; McLemore, A.F.; Aldrich, A.L.; Komrokji, R.S.; McGraw, K.L.; Dhawan, A.; Geyer, S.; Hou, H.-A.; Eksioglu, E.A.; Sullivan, A.; et al. TP53 mutations in myelodysplastic syndromes and secondary AML confer an immunosuppressive phenotype. Blood 2020, 136, 2812–2823. [Google Scholar] [CrossRef] [PubMed]
- Hsu, J.I.; Dayaram, T.; Tovy, A.; De Braekeleer, E.; Jeong, M.; Wang, F.; Zhang, J.; Heffernan, T.P.; Gera, S.; Kovacs, J.J.; et al. PPM1D Mutations Drive Clonal Hematopoiesis in Response to Cytotoxic Chemotherapy. Cell Stem Cell 2018, 23, 700–713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kahn, J.D.; Miller, P.G.; Silver, A.J.; Sellar, R.S.; Bhatt, S.; Gibson, C.; McConkey, M.; Adams, D.; Mar, B.; Mertins, P.; et al. PPM1D-truncating mutations confer resistance to chemotherapy and sensitivity to PPM1D inhibition in hematopoietic cells. Blood 2018, 132, 1095–1105. [Google Scholar] [CrossRef] [PubMed]
- Kleiblova, P.; Shaltiel, I.A.; Benada, J.; Ševčík, J.; Pecháčková, S.; Pohlreich, P.; Voest, E.E.; Dundr, P.; Bartek, J.; Kleibl, Z.; et al. Gain-of-function mutations of PPM1D/Wip1 impair the p53-dependent G1 checkpoint. J. Cell Biol. 2013, 201, 511–521. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Chen, L.H.; Wan, H.; Yang, R.; Wang, Z.; Feng, J.; Yang, S.; Jones, S.; Wang, S.; Zhou, W.; et al. Exome sequencing identifies somatic gain-of-function PPM1D mutations in brainstem gliomas. Nat. Genet. 2014, 46, 726–730. [Google Scholar] [CrossRef] [Green Version]
- Basiorka, A.A.; McGraw, K.L.; Eksioglu, E.A.; Chen, X.; Johnson, J.; Zhang, L.; Zhang, Q.; Irvine, B.A.; Cluzeau, T.; Sallman, D.A.; et al. The NLRP3 inflammasome functions as a driver of the myelodysplastic syndrome phenotype. Blood 2016, 128, 2960–2975. [Google Scholar] [CrossRef]
- Baldwin, A.G.; Brough, D.; Freeman, S. Inhibiting the Inflammasome: A Chemical Perspective. J. Med. Chem. 2016, 59, 1691–1710. [Google Scholar] [CrossRef]
- El-Sharkawy, L.Y.; Brough, D.; Freeman, S. Inhibiting the NLRP3 Inflammasome. Molecules 2020, 25, 5533. [Google Scholar] [CrossRef]
- Cimmino, L.; Dolgalev, I.; Wang, Y.; Yoshimi, A.; Martin, G.H.; Wang, J.; Ng, V.; Xia, B.; Witkowski, M.T.; Mitchell-Flack, M.; et al. Restoration of TET2 Function Blocks Aberrant Self-Renewal and Leukemia Progression. Cell 2017, 170, 1079–1095. [Google Scholar] [CrossRef] [Green Version]
- Guan, Y.; Greenberg, E.F.; Hasipek, M.; Chen, S.; Liu, X.; Kerr, C.M.; Gackowski, D.; Zarakowska, E.; Radivoyevitch, T.; Gu, X.; et al. Context dependent effects of ascorbic acid treatment in TET2 mutant myeloid neoplasia. Commun. Biol. 2020, 3, 493. [Google Scholar] [CrossRef]
- Cook, E.K.; Luo, M.; Rauh, M.J. Clonal hematopoiesis and inflammation: Partners in leukemogenesis and comorbidity. Exp. Hematol. 2020, 83, 85–94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Franceschi, C.; Bonafè, M.; Valensin, S.; Olivieri, F.; De Luca, M.; Ottaviani, E.; De Benedictis, G. Inflamm-aging. An evolutionary perspective on immunosenescence. Ann. N. Y. Acad. Sci. 2000, 908, 244–254. [Google Scholar] [CrossRef] [PubMed]
- Tsagaratou, A.; González-Avalos, E.; Rautio, S.; Scott-Browne, J.P.; Togher, S.; Pastor, W.A.; Rothenberg, E.V.; Chavez, L.; Lähdesmäki, H.; Rao, A. TET proteins regulate the lineage specification and TCR-mediated expansion of iNKT cells. Nat. Immunol. 2017, 18, 45–53. [Google Scholar] [CrossRef] [PubMed]
- GeneCards®: The Human Gene Database Version 5.0. Available online: https://www.genecards.org (accessed on 30 January 2021).
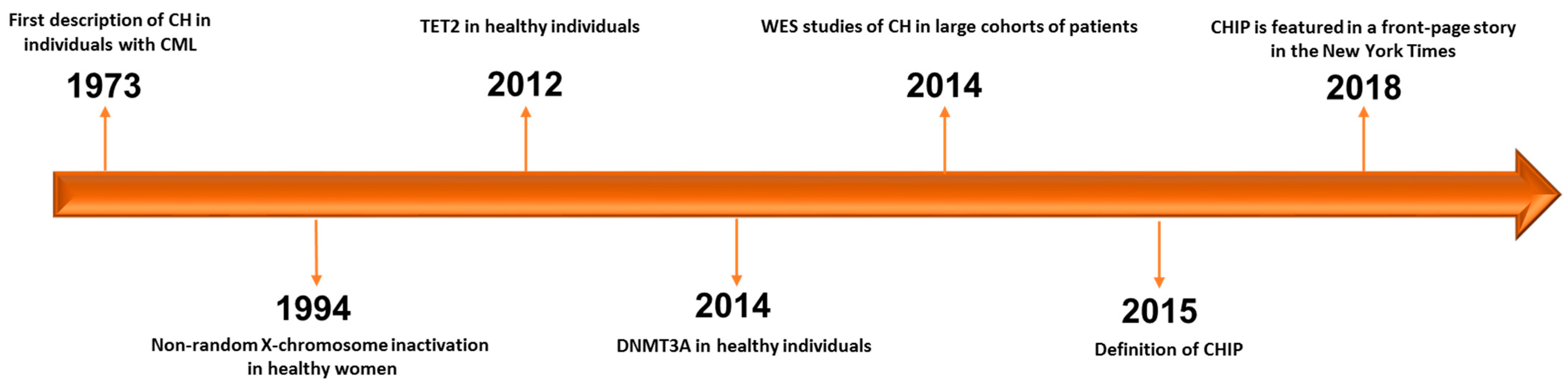
| Acronym | Condition | Description/Definition |
|---|---|---|
| ARCH | Age-related clonal hematopoiesis | Defined by detectable clonal hematopoiesis (marked by the presence of somatic mutations in the blood or bone marrow) occurring in elderly individuals.A specific VAF cut-off has not been defined and the clinical significance is undefined. |
| CHIP | Clonal hematopoiesis of indeterminate potential | Defined by somatic mutations of driver myeloid genes in the blood or bone marrow, present at ≥2% VAF in individuals without a WHO-defined hematologic disorder. |
| IDUS | Idiopathic dysplasia of undetermined significance | The finding of unexplained morphologic dysplasia of blood cells in individuals who are not cytopenic (also within clonal hematopoiesis). |
| ICUS | Idiopathic cytopenia of undetermined significance | Unexplained cytopenia(s) in patients who do not meet the diagnostic criteria for a myelodysplastic syndrome or other WHO-defined hematologic disorders (also within clonal hematopoiesis). |
| CCUS | Clonal cytopenia of undetermined significance | Unexplained cytopenia(s) in patients who do not meet the diagnostic criteria for a myelodysplastic syndrome or other WHO-defined hematologic disorders, but have somatic mutations of driver myeloid genes in the blood or bone marrow, present at ≥2% VAF. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gurnari, C.; Fabiani, E.; Falconi, G.; Travaglini, S.; Ottone, T.; Cristiano, A.; Voso, M.T. From Clonal Hematopoiesis to Therapy-Related Myeloid Neoplasms: The Silent Way of Cancer Progression. Biology 2021, 10, 128. https://doi.org/10.3390/biology10020128
Gurnari C, Fabiani E, Falconi G, Travaglini S, Ottone T, Cristiano A, Voso MT. From Clonal Hematopoiesis to Therapy-Related Myeloid Neoplasms: The Silent Way of Cancer Progression. Biology. 2021; 10(2):128. https://doi.org/10.3390/biology10020128
Chicago/Turabian StyleGurnari, Carmelo, Emiliano Fabiani, Giulia Falconi, Serena Travaglini, Tiziana Ottone, Antonio Cristiano, and Maria Teresa Voso. 2021. "From Clonal Hematopoiesis to Therapy-Related Myeloid Neoplasms: The Silent Way of Cancer Progression" Biology 10, no. 2: 128. https://doi.org/10.3390/biology10020128
APA StyleGurnari, C., Fabiani, E., Falconi, G., Travaglini, S., Ottone, T., Cristiano, A., & Voso, M. T. (2021). From Clonal Hematopoiesis to Therapy-Related Myeloid Neoplasms: The Silent Way of Cancer Progression. Biology, 10(2), 128. https://doi.org/10.3390/biology10020128






