A Multifocal Pediatric Papillary Thyroid Carcinoma (PTC) Harboring the AGK-BRAF and RET/PTC3 Fusion in a Mutually Exclusive Pattern Reveals Distinct Levels of Genomic Instability and Nuclear Organization
Abstract
Simple Summary
Abstract
1. Introduction
2. Results
2.1. Patient Description
2.2. 3D Analysis of Nuclear Telomere Organization Indicates Increased Telomere-Related Genomic Instability in the AGK-BRAF Positive Focus
2.3. Three-Dimensional Structured Illumination Microscopy (3D-SIM) Measurements Show Significant Changes in DNA Structure in the AGK-BRAF-Positive Focus
3. Discussion
4. Materials and Methods
4.1. Patient Samples
4.2. 3D Telomere Q-FISH, Image Acquisition and Analysis using TeloView® Software Platform
4.3. 3D-SIM Slide Preparation, Image Acquisition and Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 3D-SIM | 3D super-resolution structured Illumination Microscopy |
| Q-FISH | Quantitative fluorescet in situ hybridization |
| PTC | Papillary thyroid carcinoma |
| SIM | Structured illumination microscopy |
| TA | Telomeric aggregates |
References
- Paulson, V.A.; Rudzinski, E.R.; Hawkins, D.S. Thyroid cancer in the pediatric population. Genes 2019, 10, 723. [Google Scholar] [CrossRef]
- Howlader, N.; Noone, A.; Krapcho, M.; Miller, D.; Brest, A.; Yu, M.; Ruhl, J.; Tatalovich, Z.; Mariotto, A.; Lewis, D.; et al. Cancer Statistics Review, 1975–2017—SEER Statistics. Available online: https://seer.cancer.gov/csr/1975_2017/ (accessed on 13 July 2020).
- Cordioli, M.I.; Moraes, L.; Cury, A.N.; Cerutti, J.M. Are we really at the dawn of understanding sporadic pediatric thyroid carcinoma? Endocr. Relat. Cancer 2015, 22, R311–R324. [Google Scholar] [CrossRef] [PubMed]
- Prescott, J.D.; Zeiger, M.A. The RET oncogene in papillary thyroid carcinoma. Cancer 2015. [Google Scholar] [CrossRef]
- Ricarte-Filho, J.C.; Li, S.; Garcia-Rendueles, M.E.R.; Montero-Conde, C.; Voza, F.; Knauf, J.A.; Heguy, A.; Viale, A.; Bogdanova, T.; Thomas, G.A.; et al. Identification of kinase fusion oncogenes in post-Chernobyl radiation-induced thyroid cancers. J. Clin. Investig. 2013, 123, 4935–4944. [Google Scholar] [CrossRef]
- Sisdelli, L.; Cordioli, M.I.C.V.; Vaisman, F.; Moraes, L.; Colozza-Gama, G.A.; Alves, P.A.G.; Araújo, M.L.; Alves, M.T.S.; Monte, O.; Longui, C.A.; et al. AGK-BRAF is associated with distant metastasis and younger age in pediatric papillary thyroid carcinoma. Pediatric Blood Cancer 2019, 66, 1–7. [Google Scholar] [CrossRef]
- Cordioli, M.I.; Moraes, L.; Carvalheira, G.; Sisdelli, L.; Alves, M.T.S.; Delcelo, R.; Monte, O.; Longui, C.A.; Cury, A.N.; Cerutti, J.M. AGK-BRAF gene fusion is a recurrent event in sporadic pediatric thyroid carcinoma. Cancer Med. 2016, 5, 1535–1541. [Google Scholar] [CrossRef]
- Mai, S. The three-dimensional cancer nucleus. Genes Chromosom. Cancer 2019, 58, 462–473. [Google Scholar] [CrossRef] [PubMed]
- Schermelleh, L.; Ferrand, A.; Huser, T.; Eggeling, C.; Sauer, M.; Biehlmaier, O.; Drummen, G.P.C. Super-resolution microscopy demystified. Nat. Cell Biol. 2019, 21, 72–84. [Google Scholar] [CrossRef]
- Cordioli, M.I.; Moraes, L.; Bastos, A.U.; Besson, P.; de Alves, M.T.S.; Delcelo, R.; Monte, O.; Longui, C.; Cury, A.N.; Cerutti, J.M. Fusion Oncogenes Are the Main Genetic Events Found in Sporadic Papillary Thyroid Carcinomas from Children. Thyroid 2017, 27, 182–188. [Google Scholar] [CrossRef] [PubMed]
- Galati, A.; Micheli, E.; Cacchione, S. Chromatin structure in telomere dynamics. Front. Oncol. 2013, 3, 46. [Google Scholar] [CrossRef]
- Righolt, C.H.; Guffei, A.; Knecht, H.; Young, I.T.; Stallinga, S.; Van Vliet, L.J.; Mai, S. Differences in nuclear DNA organization between lymphocytes, hodgkin and reed-sternberg cells revealed by structured illumination microscopy. J. Cell. Biochem. 2014, 115, 1441–1448. [Google Scholar] [CrossRef]
- Chow, K.H.; Factor, R.E.; Ullman, K.S. The nuclear envelope environment and its cancer connections. Nat. Rev. Cancer 2012, 12, 196–209. [Google Scholar] [CrossRef]
- Cui, Y.; Borysova, M.K.; Johnson, J.O.; Guadagno, T.M. Oncogenic B-RafV600E induces spindle abnormalities, supernumerary centrosomes, and aneuploidy in human melanocytic cells. Cancer Res. 2010, 70, 675–684. [Google Scholar] [CrossRef]
- Fagin, J.A.; Wells, S.A. Biologic and clinical perspectives on thyroid cancer. N. Engl. J. Med. 2016, 375, 1054–1067. [Google Scholar] [CrossRef] [PubMed]
- Rizzotto, A.; Tollis, S.; Pham, N.; Wildenhain, J.; Zuleger, N.; Keys, J.; Batrakou, D.; Culley, J.; Zheng, S.; Lammerding, J.; et al. Chemical-Genetic Interrogation of Nuclear Size Control Reveals Cancer-Specific Effects on Cell Migration and Invasion. bioRxiv 2020. [Google Scholar] [CrossRef]
- Zink, D.; Fischer, A.H.; Nickerson, J.A. Nuclear structure in cancer cells. Nat. Rev. Cancer 2004, 4, 677–687. [Google Scholar] [CrossRef]
- Vermolen, B.J.; Garini, Y.; Mai, S.; Mougey, V.; Fest, T.; Chuang, T.C.-Y.; Chuang, A.Y.-C.; Wark, L.; Young, I.T. Characterizing the three-dimensional organization of telomeres. Cytom. Part A 2005, 67A, 144–150. [Google Scholar] [CrossRef]
- Gadji, M.; Fortin, D.; Tsanaclis, A.M.; Garini, Y.; Katzir, N.; Wienburg, Y.; Yan, J.; Klewes, L.; Klonisch, T.; Drouin, R.; et al. Three-dimensional nuclear telomere architecture is associated with differential time to progression and overall survival in glioblastoma patients. Neoplasia 2010, 12, 183–191. [Google Scholar] [CrossRef]
- Righolt, C.H.; Knecht, H.; Mai, S. DNA Superresolution Structure of Reed-Sternberg Cells Differs between Long-Lasting Remission Versus Relapsing Hodgkin’s Lymphoma Patients. J. Cell. Biochem. 2016, 117, 1633–1637. [Google Scholar] [CrossRef] [PubMed]
- Sathitruangsak, C.; Righolt, C.H.; Klewes, L.; Tammur, P.; Ilus, T.; Tamm, A.; Punab, M.; Olujohungbe, A.; Mai, S. Quantitative superresolution microscopy reveals differences in nuclear dna organization of multiple myeloma and monoclonal gammopathy of undetermined significance. J. Cell. Biochem. 2015, 116, 704–710. [Google Scholar] [CrossRef] [PubMed]
- Rangel-Pozzo, A.; Kuzyk, A.; Gartner, J.; Mai, S. MYCN overexpression is linked to significant differences in nuclear DNA organization in neuroblastoma. SPG BioMed 2019. [Google Scholar] [CrossRef]
- Dolde, C.E.; Mukherjee, M.; Cho, C.; Resar, L.M.S. HMG-I/Y in human breast cancer cell lines. Breast Cancer Res. Treat. 2002, 71, 181–191. [Google Scholar] [CrossRef] [PubMed]
- Chin, S.; Daigo, Y.; Huang, H.; Iyer, N.G.; Callagy, G.; Kranjac, T.; Gonzalez, M.; Sangan, T.; Earl, H.; Caldas, C. A simple and reliable pretreatment protocol facilitates fluorescent in situ hybridisation on tissue microarrays of paraffin wax embedded tumour samples. J. Clin. Pathol. Mol. Pathol. 2003, 56, 275–279. [Google Scholar] [CrossRef] [PubMed]
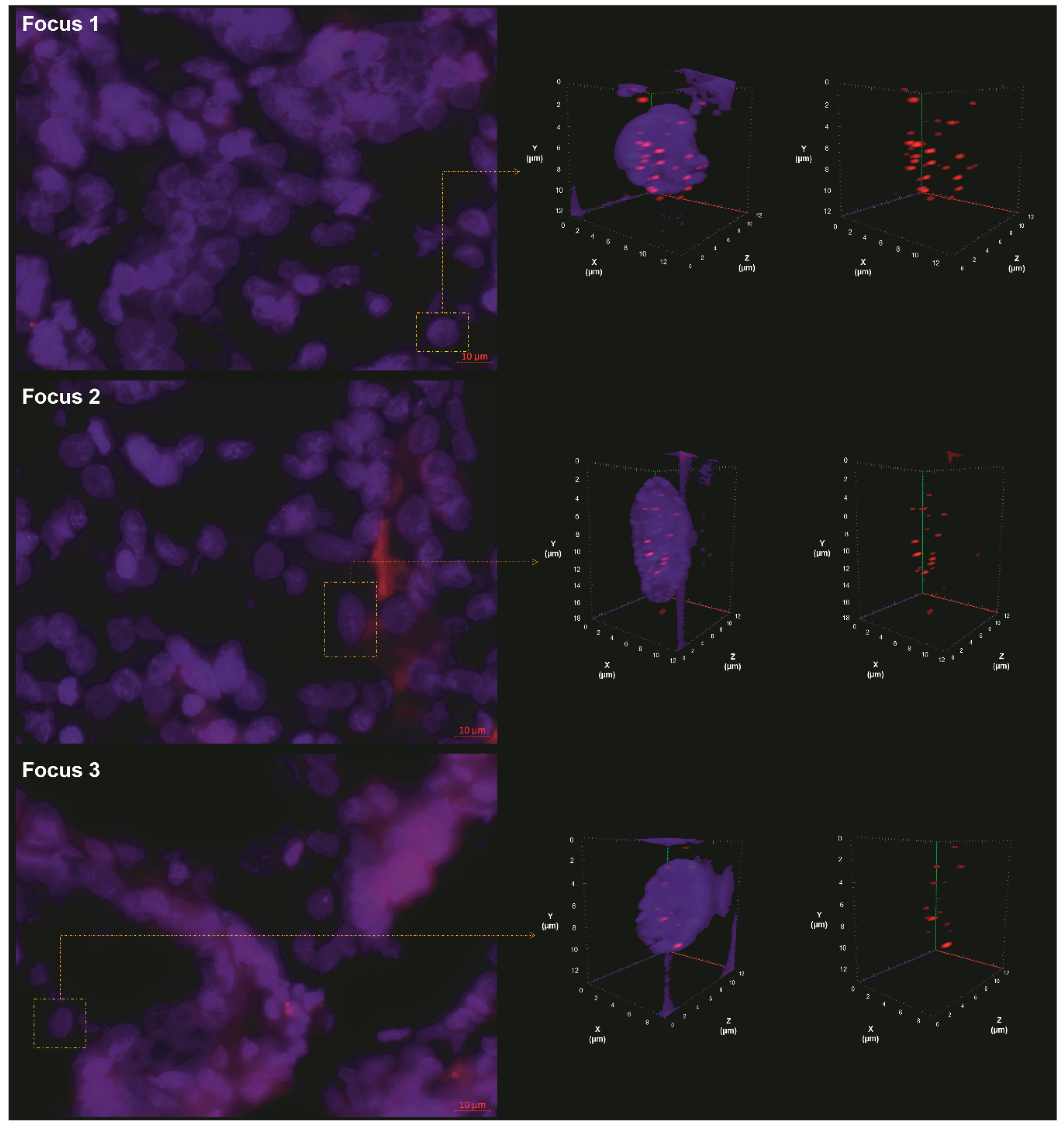
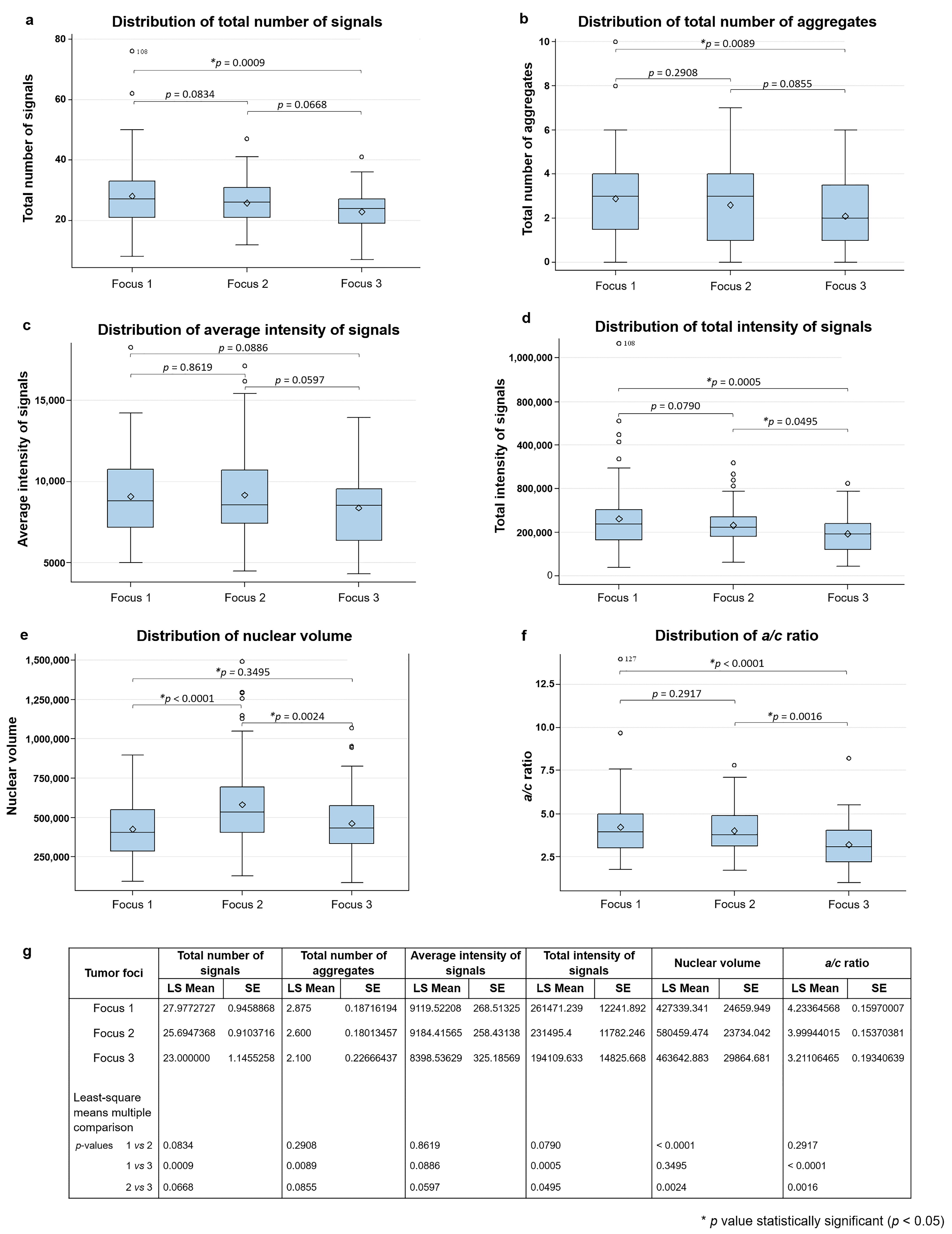
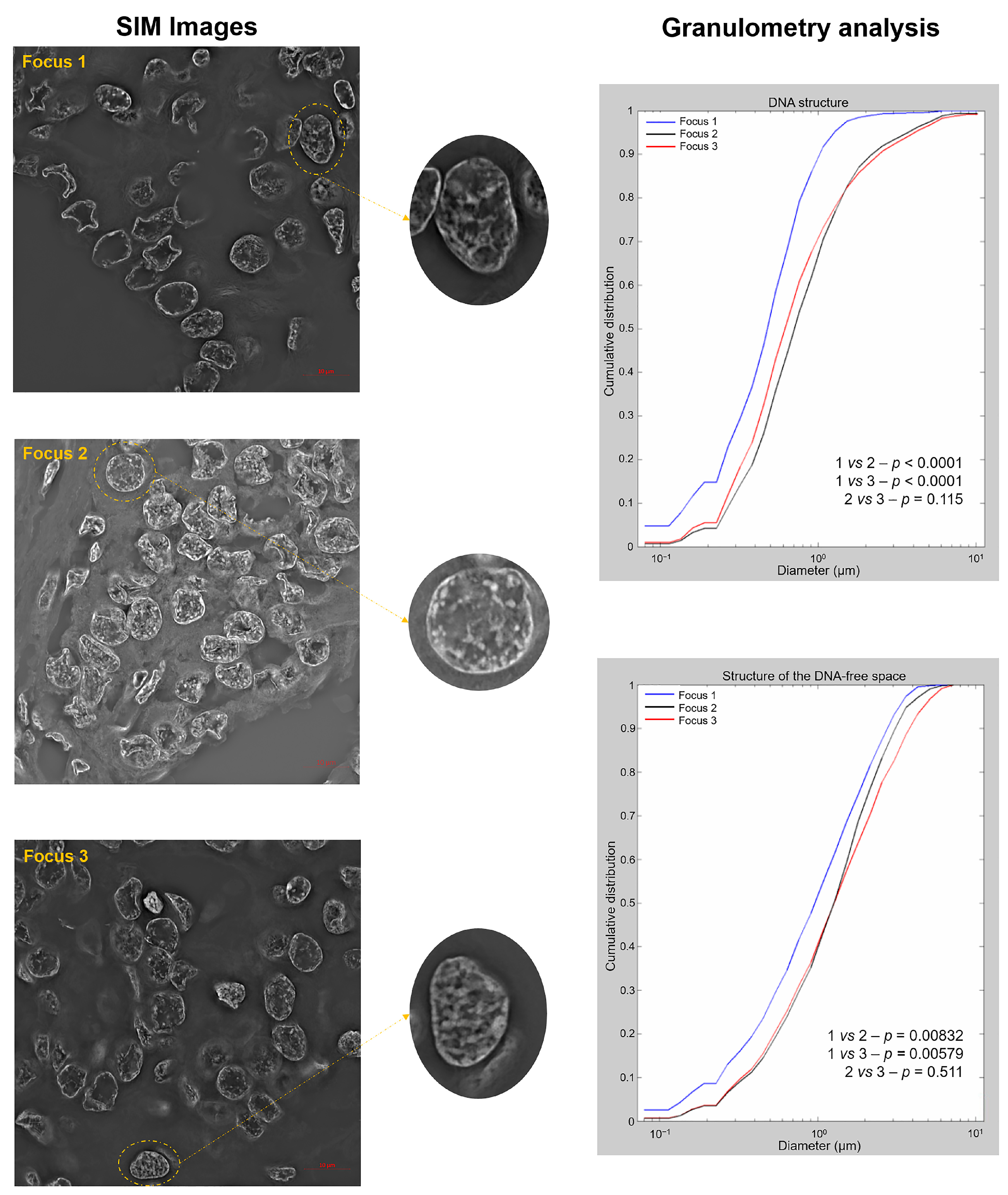
| Genetic Alteration | |
|---|---|
| Focus 1 | AGK-BRAF |
| Focus 2 | No alterations ** |
| Focus 3 | RET/PTC3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sisdelli, L.; Cordioli, M.I.V.; Vaisman, F.; Monte, O.; Longui, C.A.; Cury, A.N.; Freitas, M.O.; Rangel-Pozzo, A.; Mai, S.; Cerutti, J.M. A Multifocal Pediatric Papillary Thyroid Carcinoma (PTC) Harboring the AGK-BRAF and RET/PTC3 Fusion in a Mutually Exclusive Pattern Reveals Distinct Levels of Genomic Instability and Nuclear Organization. Biology 2021, 10, 125. https://doi.org/10.3390/biology10020125
Sisdelli L, Cordioli MIV, Vaisman F, Monte O, Longui CA, Cury AN, Freitas MO, Rangel-Pozzo A, Mai S, Cerutti JM. A Multifocal Pediatric Papillary Thyroid Carcinoma (PTC) Harboring the AGK-BRAF and RET/PTC3 Fusion in a Mutually Exclusive Pattern Reveals Distinct Levels of Genomic Instability and Nuclear Organization. Biology. 2021; 10(2):125. https://doi.org/10.3390/biology10020125
Chicago/Turabian StyleSisdelli, Luiza, Maria Isabel V. Cordioli, Fernanda Vaisman, Osmar Monte, Carlos A. Longui, Adriano N. Cury, Monique O. Freitas, Aline Rangel-Pozzo, Sabine Mai, and Janete M. Cerutti. 2021. "A Multifocal Pediatric Papillary Thyroid Carcinoma (PTC) Harboring the AGK-BRAF and RET/PTC3 Fusion in a Mutually Exclusive Pattern Reveals Distinct Levels of Genomic Instability and Nuclear Organization" Biology 10, no. 2: 125. https://doi.org/10.3390/biology10020125
APA StyleSisdelli, L., Cordioli, M. I. V., Vaisman, F., Monte, O., Longui, C. A., Cury, A. N., Freitas, M. O., Rangel-Pozzo, A., Mai, S., & Cerutti, J. M. (2021). A Multifocal Pediatric Papillary Thyroid Carcinoma (PTC) Harboring the AGK-BRAF and RET/PTC3 Fusion in a Mutually Exclusive Pattern Reveals Distinct Levels of Genomic Instability and Nuclear Organization. Biology, 10(2), 125. https://doi.org/10.3390/biology10020125






