The Central Role of the F-Actin Surface in Myosin Force Generation
Simple Summary
Abstract
1. The Role of Actin in the Cell
The Topology of the Actin Filament
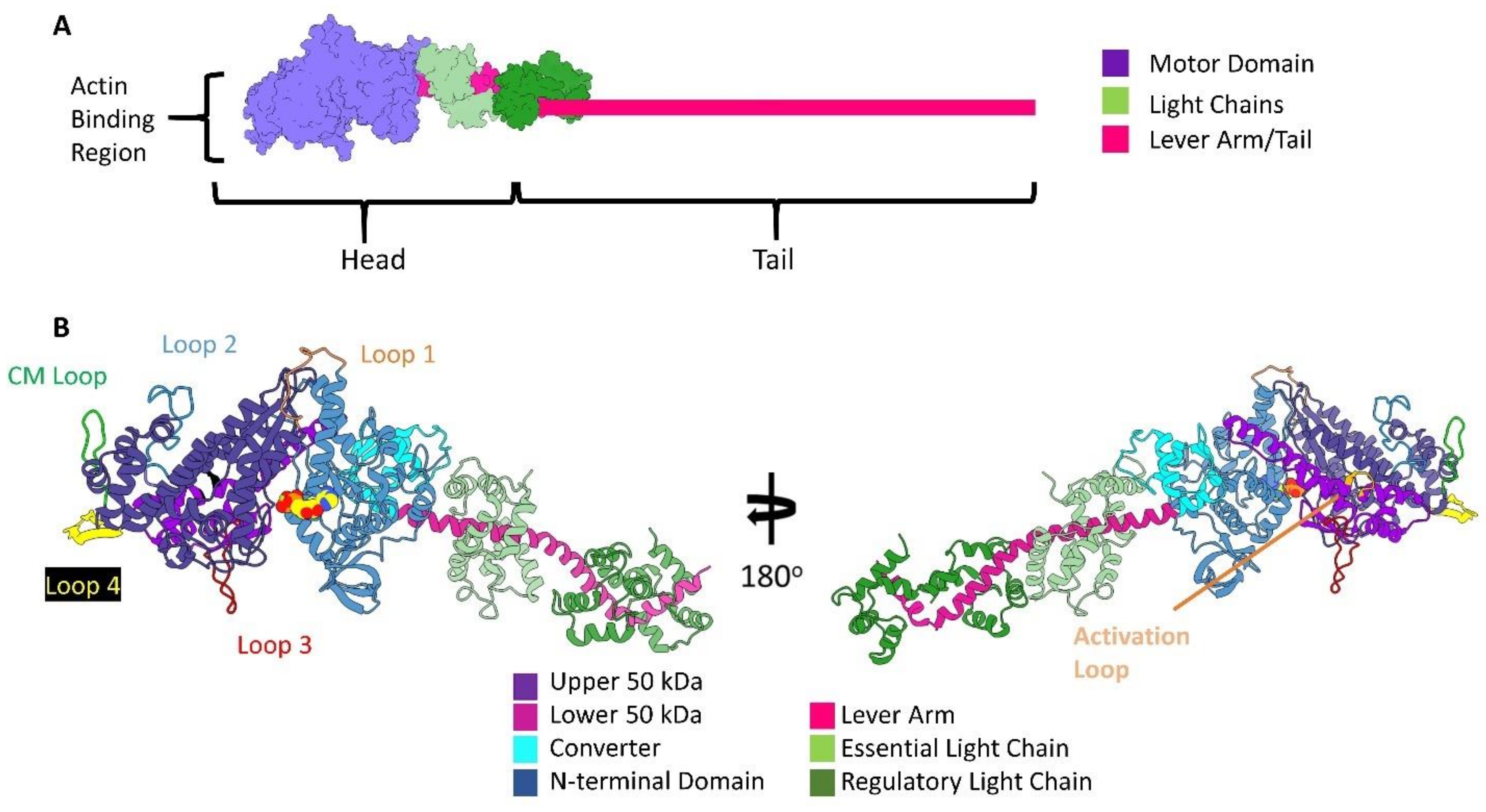
2. Introduction to Myosin Structure
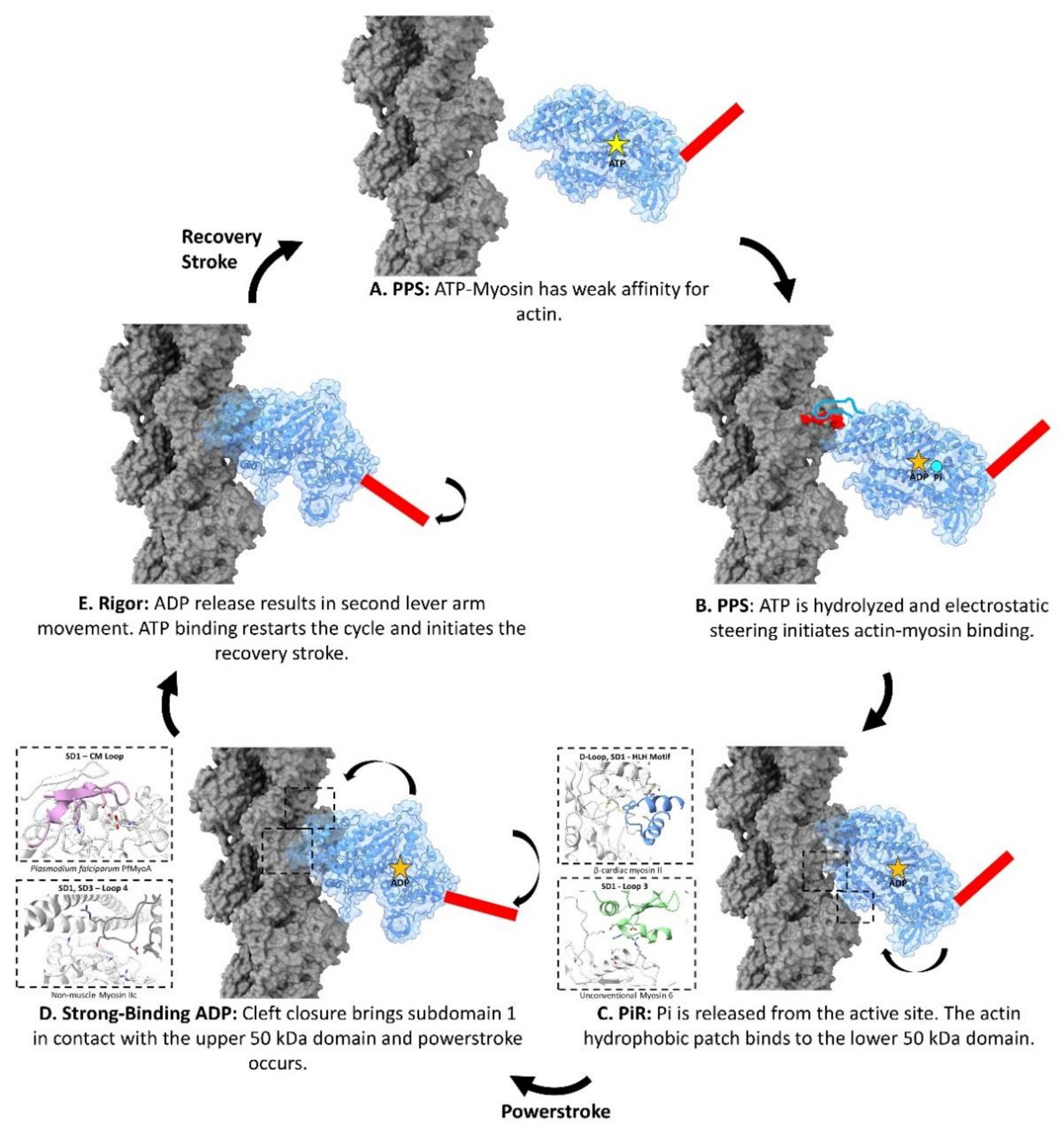
3. The Sequential Binding of Myosin to Actin
4. Electrostatic Steering Initiates Actin-Myosin Binding
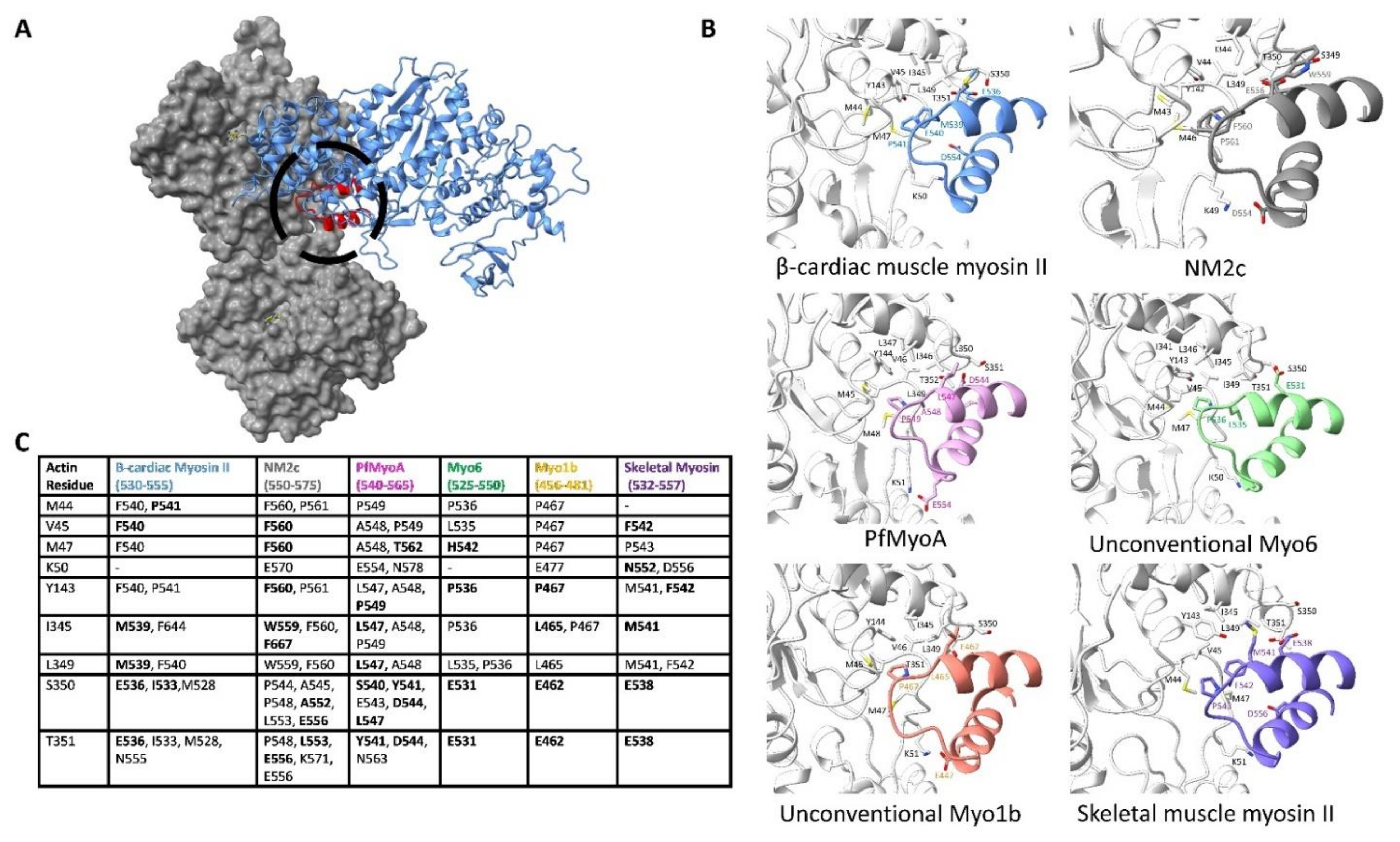
4.1. Conserved Actin-Loop 2 Interactions
4.2. Isoform-Specific Actin-Loop 2 Interactions
5. Actin D-Loop Hydrophobic Patch Binds to the PiR State Lower 50 kDa Domain
5.1. The D-Loop Hydrophobic Patch Makes Conserved Contacts with the Hydrophobic HLH Motif
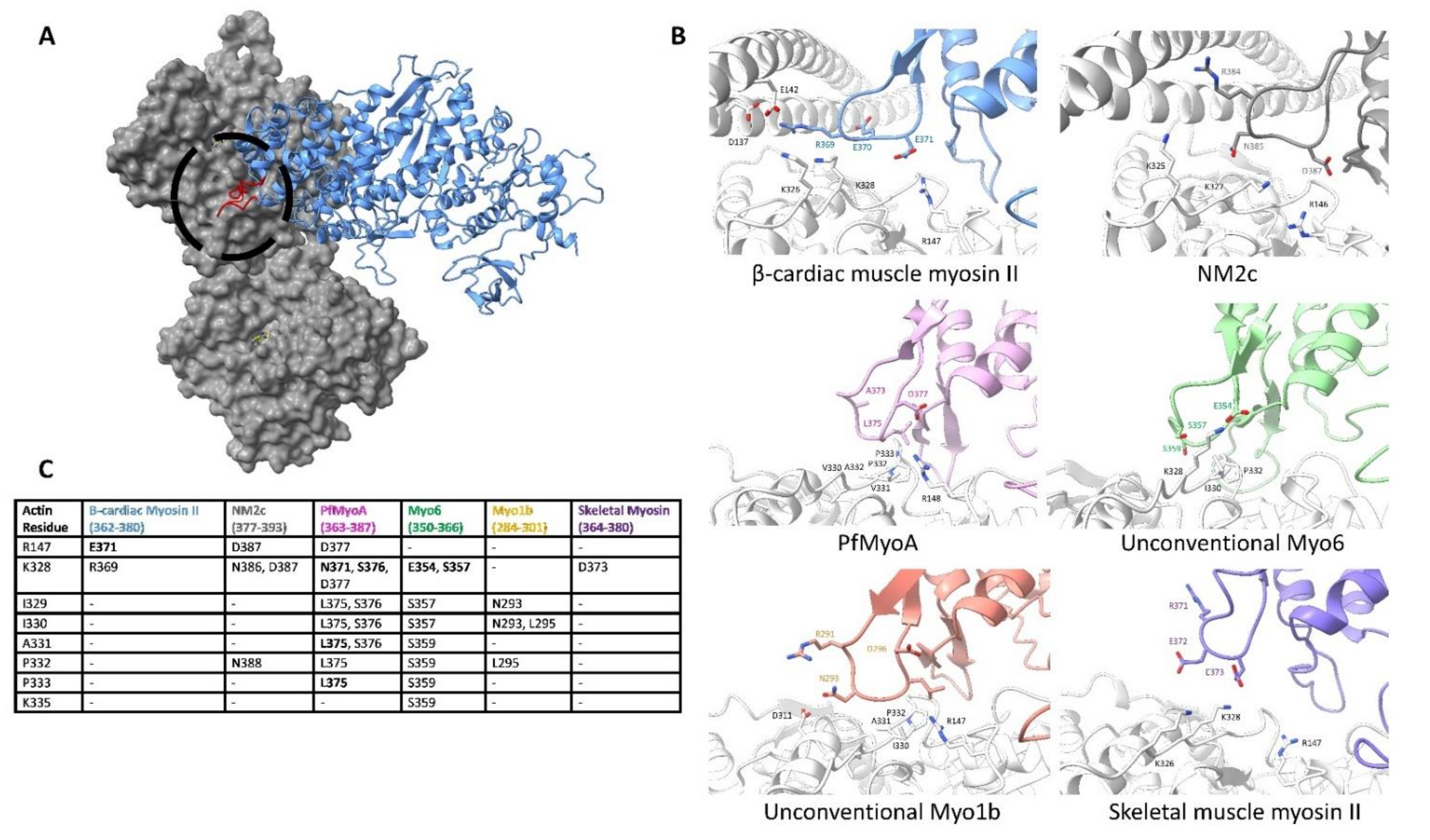
5.2. Subdomain 1 Makes Isoform-Specific Contacts with Loop 3 upon Lower 50 kDa Binding
5.3. Subdomain 1 Binds to the Activation Loop
6. The Actin Surface Prompts Myosin Cleft Closure
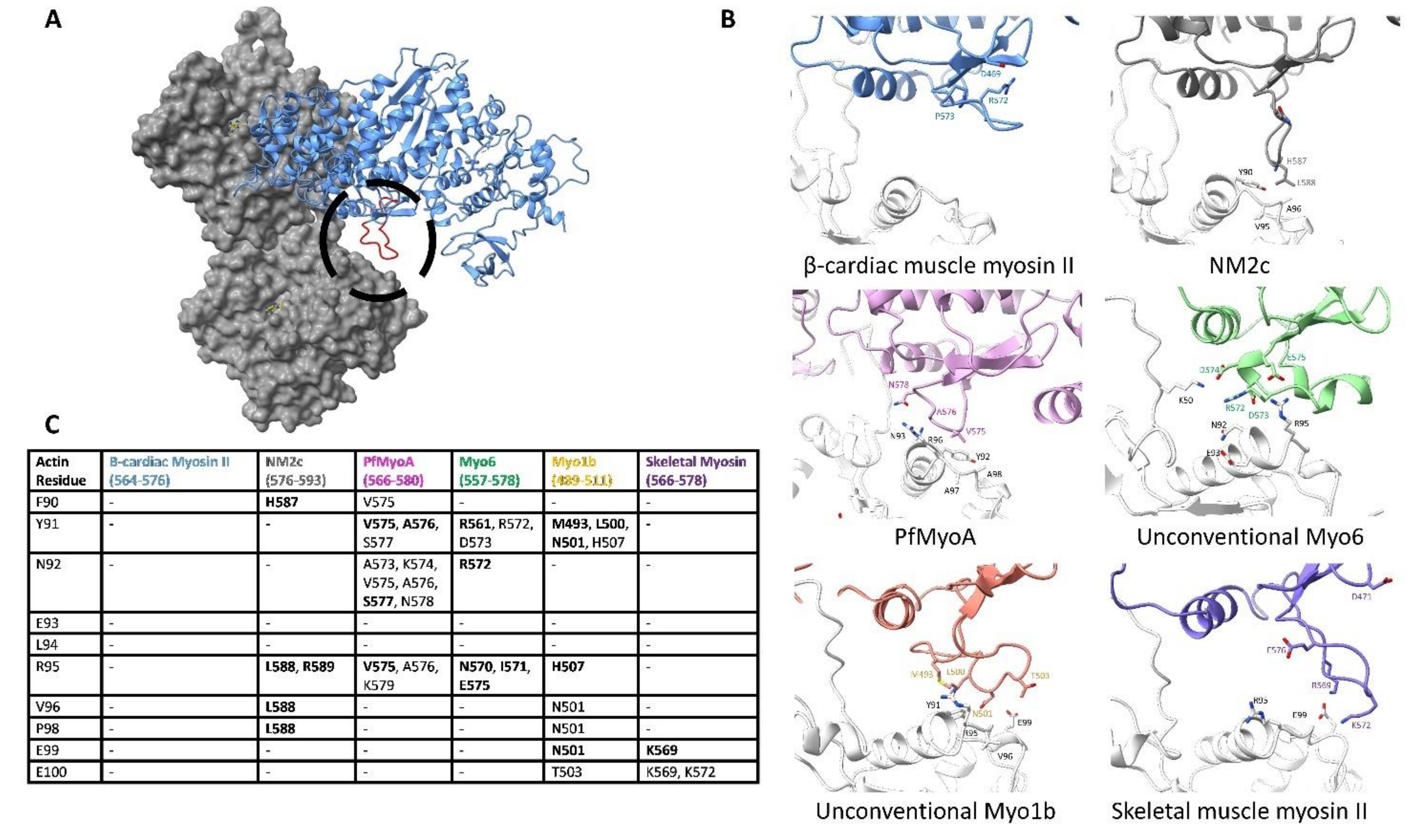
6.1. The N-Terminus of Actin and the C-Terminal Base of Loop 2 Initiate Cleft Closure
6.2. Cleft Closure Results in F-Actin Subdomain 1 Interacting with the CM Loop
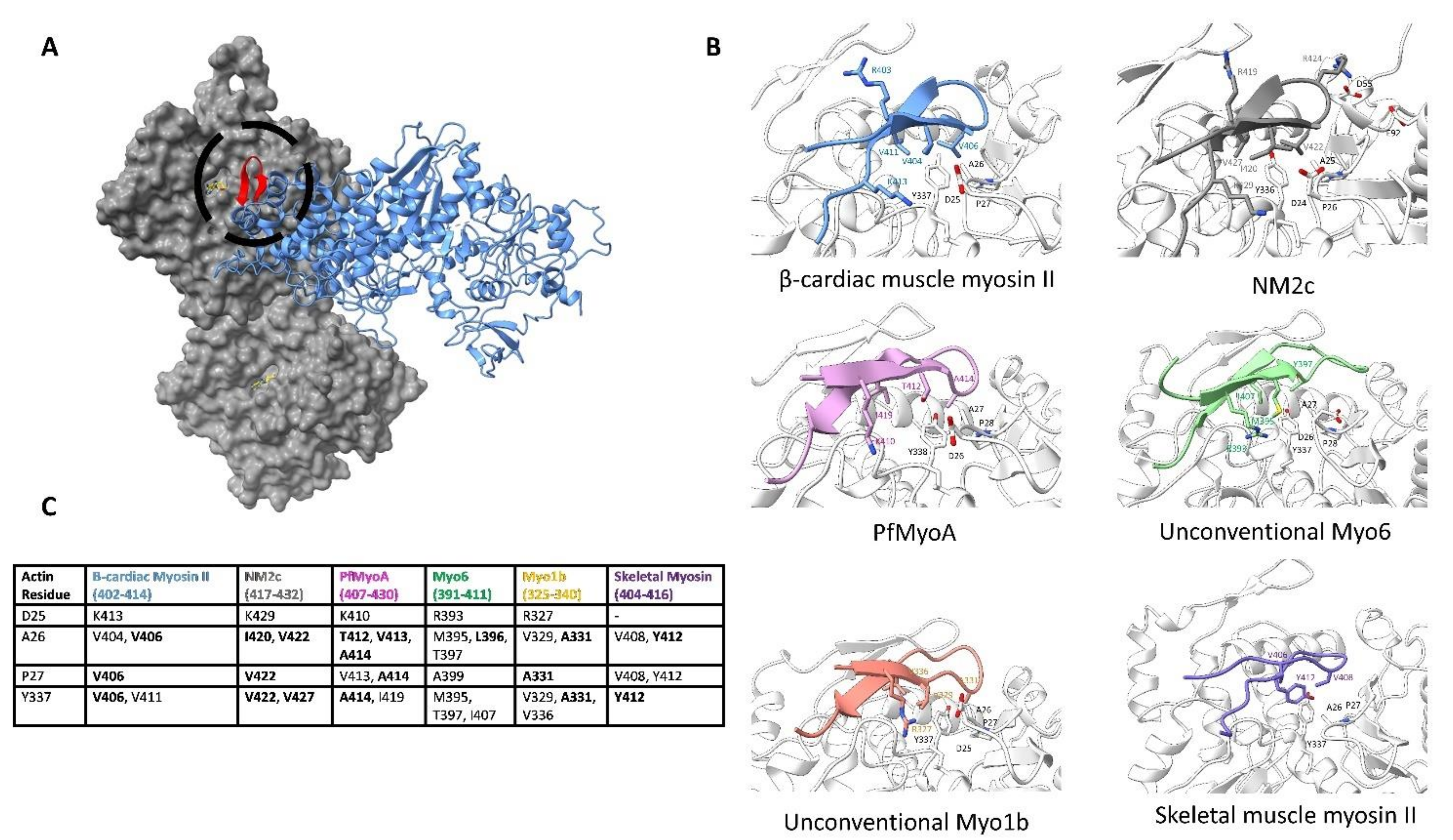
6.3. Actin Subdomain 3 Binds to Loop 4 to Form the Edge of the Actin-Myosin Interface
7. Conclusions and Perspectives
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gunning, P.W.; Ghoshdastider, U.; Whitaker, S.; Popp, D.; Robinson, R.C. The evolution of compositionally and functionally distinct actin filaments. J. Cell Sci. 2015, 128, 2009–2019. [Google Scholar] [CrossRef] [PubMed]
- Dominguez, R.; Holmes, K.C. Actin Structure and Function. Annu. Rev. Biophys. 2011, 40, 169–186. [Google Scholar] [CrossRef]
- Geeves, M.A.; Holmes, K.C. Structural mechanism of muscle contraction. Annu. Rev. Biochem. 1999, 68, 687–728. [Google Scholar] [CrossRef]
- Miralles, F.; Visa, N. Actin in transcription and transcription regulation. Curr. Opin. Cell Biol. 2006, 18, 261–266. [Google Scholar] [CrossRef]
- Pollard, T.D.; Cooper, J.A. Actin, a Central Player in Cell Shape and Movement. Science 2009, 326, 1208–1212. [Google Scholar] [CrossRef] [PubMed]
- Titus, M.A. Myosin-Driven Intracellular Transport. Cold Spring Harb. Perspect. Biol. 2018, 10, a021972. [Google Scholar] [CrossRef]
- Pollard, T.D.; Cooper, J.A. Actin and actin-binding proteins. A critical evaluation of mechanisms and functions. Annu. Rev. Biochem. 1986, 55, 987–1035. [Google Scholar] [CrossRef] [PubMed]
- Pollard, T.D. Actin and Actin-Binding Proteins. Cold Spring Harb. Perspect. Biol. 2016, 8, a018226. [Google Scholar] [CrossRef]
- Dominguez, R. Actin-binding proteins— A unifying hypothesis. Trends Biochem. Sci. 2004, 29, 572–578. [Google Scholar] [CrossRef]
- Bravo-Cordero, J.J.; Magalhaes, M.A.O.; Eddy, R.J.; Hodgson, L.; Condeelis, J. Functions of cofilin in cell locomotion and invasion. Nat. Rev. Mol. Cell Biol. 2013, 14, 405–415. [Google Scholar] [CrossRef]
- Unsain, N.; Stefani, F.D.; Cáceres, A. The Actin/Spectrin Membrane-Associated Periodic Skeleton in Neurons. Front. Synaptic Neurosci. 2018, 10, 10. [Google Scholar] [CrossRef]
- Burgess, D.R. Cytokinesis: New roles for myosin. Curr. Biol. CB 2005, 15, R310–R311. [Google Scholar] [CrossRef]
- DePina, A.S.; Langford, G.M. Vesicle transport: The role of actin filaments and myosin motors. Microsc. Res. Tech. 1999, 47, 93–106. [Google Scholar] [CrossRef]
- Sweeney, H.L.; Holzbaur, E.L.F. Motor Proteins. Cold Spring Harb. Perspect. Biol. 2018, 10, a021931. [Google Scholar] [CrossRef]
- Hartman, M.A.; Spudich, J.A. The myosin superfamily at a glance. J. Cell Sci. 2012, 125, 1627–1632. [Google Scholar] [CrossRef]
- Hartman, M.A.; Finan, D.; Sivaramakrishnan, S.; Spudich, J.A. Principles of unconventional myosin function and targeting. Annu. Rev. Cell Dev. Biol. 2011, 27, 133–155. [Google Scholar] [CrossRef] [PubMed]
- Bond, L.M.; Brandstaetter, H.; Sellers, J.R.; Kendrick-Jones, J.; Buss, F. Myosin motor proteins are involved in the final stages of the secretory pathways. Biochem. Soc. Trans. 2011, 39, 1115–1119. [Google Scholar] [CrossRef] [PubMed]
- Ross, J.L.; Ali, M.Y.; Warshaw, D.M. Cargo transport: Molecular motors navigate a complex cytoskeleton. Curr. Opin. Cell Biol. 2008, 20, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Oda, T.; Maéda, Y. Multiple Conformations of F-actin. Structure 2010, 18, 761–767. [Google Scholar] [CrossRef][Green Version]
- Oda, T.; Iwasa, M.; Aihara, T.; Maéda, Y.; Narita, A. The nature of the globular- to fibrous-actin transition. Nature 2009, 457, 441–445. [Google Scholar] [CrossRef]
- Moore, P.B.; Huxley, H.E.; DeRosier, D.J. Three-dimensional reconstruction of F-actin, thin filaments and decorated thin filaments. J. Mol. Biol. 1970, 50, 279–295. [Google Scholar] [CrossRef]
- von der Ecken, J.; Müller, M.; Lehman, W.; Manstein, D.J.; Penczek, P.A.; Raunser, S. Structure of the F-actin-tropomyosin complex. Nature 2015, 519, 114–117. [Google Scholar] [CrossRef]
- Lehman, W.; Rynkiewicz, M.; Moore, J.R. A new twist on tropomyosin binding to actin filaments: Perspectives on thin filament function, assembly and biomechanics. J. Muscle Res. Cell Motil. 2019, 41, 23–38. [Google Scholar] [CrossRef]
- von der Ecken, J.; Heissler, S.M.; Pathan-Chhatbar, S.; Manstein, D.J.; Raunser, S. Cryo-EM structure of a human cytoplasmic actomyosin complex at near-atomic resolution. Nature 2016, 534, 724–728. [Google Scholar] [CrossRef]
- Kabsch, W.; Mannherz, H.G.; Suck, D.; Pai, E.F.; Holmes, K.C. Atomic structure of the actin: DNase I complex. Nature 1990, 347, 37–44. [Google Scholar] [CrossRef]
- Holmes, K.C.; Popp, D.; Gebhard, W.; Kabsch, W. Atomic model of the actin filament. Nature 1990, 347, 44–49. [Google Scholar] [CrossRef]
- Merino, F.; Pospich, S.; Funk, J.; Wagner, T.; Küllmer, F.; Arndt, H.-D.; Bieling, P.; Raunser, S. Structural transitions of F-actin upon ATP hydrolysis at near-atomic resolution revealed by cryo-EM. Nat. Struct. Mol. Biol. 2018, 25, 528–537. [Google Scholar] [CrossRef] [PubMed]
- Chou, S.Z.; Pollard, T.D. Mechanism of actin polymerization revealed by cryo-EM structures of actin filaments with three different bound nucleotides. Proc. Natl. Acad. Sci. USA 2019, 116, 4265–4274. [Google Scholar] [CrossRef] [PubMed]
- Belyy, A.; Merino, F.; Sitsel, O.; Raunser, S. Structure of the Lifeact-F-actin complex. PLoS Biol. 2020, 18, e3000925. [Google Scholar] [CrossRef]
- Bañuelos, S.; Saraste, M.; Carugo, K.D. Structural comparisons of calponin homology domains: Implications for actin binding. Structure 1998, 6, 1419–1431. [Google Scholar] [CrossRef]
- Yin, L.-M.; Schnoor, M.; Jun, C.-D. Structural Characteristics, Binding Partners and Related Diseases of the Calponin Homology (CH) Domain. Front. Cell Dev. Biol. 2020, 8, 342. [Google Scholar] [CrossRef]
- Stradal, T.; Kranewitter, W.; Winder, S.J.; Gimona, M. CH domains revisited. FEBS Lett. 1998, 431, 134–137. [Google Scholar] [CrossRef]
- Kumari, A.; Kesarwani, S.; Javoor, M.G.; Vinothkumar, K.R.; Sirajuddin, M. Structural insights into actin filament recognition by commonly used cellular actin markers. EMBO J. 2020, 39, e104006. [Google Scholar] [CrossRef] [PubMed]
- Avery, A.W.; Fealey, M.E.; Wang, F.; Orlova, A.; Thompson, A.R.; Thomas, D.D.; Hays, T.S.; Egelman, E.H. Structural basis for high-affinity actin binding revealed by a β-III-spectrin SCA5 missense mutation. Nat. Commun. 2017, 8, 1350. [Google Scholar] [CrossRef]
- Iwamoto, D.V.; Huehn, A.; Simon, B.; Huet-Calderwood, C.; Baldassarre, M.; Sindelar, C.V.; Calderwood, D.A. Structural basis of the filamin A actin-binding domain interaction with F-actin. Nat. Struct. Mol. Biol. 2018, 25, 918–927. [Google Scholar] [CrossRef]
- Schwebach, C.L.; Kudryashova, E.; Zheng, W.; Orchard, M.; Smith, H.; Runyan, L.A.; Egelman, E.H.; Kudryashov, D.S. Osteogenesis imperfecta mutations in plastin 3 lead to impaired calcium regulation of actin bundling. Bone Res. 2020, 8, 21. [Google Scholar] [CrossRef]
- Hampton, C.M.; Taylor, D.W.; Taylor, K.A. Novel structures for α-actinin: F-actin interactions and their implications for actin-membrane attachment and tension sensing in the cytoskeleton. J. Mol. Biol. 2007, 368, 92–104. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.M.; Mallela, K.M.G. The N-Terminal Actin-Binding Tandem Calponin-Homology (CH) Domain of Dystrophin Is in a Closed Conformation in Solution and When Bound to F-actin. Biophys. J. 2012, 103, 1970–1978. [Google Scholar] [CrossRef]
- Boczkowska, M.; Rebowski, G.; Kremneva, E.; Lappalainen, P.; Dominguez, R. How Leiomodin and Tropomodulin use a common fold for different actin assembly functions. Nat. Commun. 2015, 6, 8314. [Google Scholar] [CrossRef]
- Mierke, C.T. The role of vinculin in the regulation of the mechanical properties of cells. Cell Biochem. Biophys. 2009, 53, 115–126. [Google Scholar] [CrossRef]
- Peng, X.; Nelson, E.S.; Maiers, J.L.; DeMali, K.A. New Insights into Vinculin Function and Regulation. Int. Rev. Cell Mol. Biol. 2011, 287, 191–231. [Google Scholar] [CrossRef]
- Golji, J.; Mofrad, M.R.K. The Interaction of Vinculin with Actin. PLoS Comput. Biol. 2013, 9, e1002995. [Google Scholar] [CrossRef] [PubMed]
- Mei, L.; Espinosa de los Reyes, S.; Reynolds, M.J.; Leicher, R.; Liu, S.; Alushin, G.M. Molecular mechanism for direct actin force-sensing by α-catenin. eLife 2020, 9, e62514. [Google Scholar] [CrossRef] [PubMed]
- Risi, C.; Belknap, B.; Forgacs-Lonart, E.; Harris, S.P.; Schröder, G.F.; White, H.D.; Galkin, V.E. N-Terminal Domains of Cardiac Myosin Binding Protein C Cooperatively Activate the Thin Filament. Structure 2018, 26, 1604–1611.e4. [Google Scholar] [CrossRef] [PubMed]
- Risi, C.M.; Patra, M.; Belknap, B.; Harris, S.P.; White, H.D.; Galkin, V.E. Interaction of the C2 Ig-like Domain of Cardiac Myosin Binding Protein-C with F-actin. J. Mol. Biol. 2021, 433, 167178. [Google Scholar] [CrossRef] [PubMed]
- Hatch, V.; Zhi, G.; Smith, L.; Stull, J.T.; Craig, R.; Lehman, W. Myosin light chain kinase binding to a unique site on F-actin revealed by three-dimensional image reconstruction. J. Cell Biol. 2001, 154, 611–618. [Google Scholar] [CrossRef]
- Lehman, W.; Li, X.; Kiani, F.A.; Moore, J.R.; Campbell, S.G.; Fischer, S.; Rynkiewicz, M.J. Precise Binding of Tropomyosin on Actin Involves Sequence-Dependent Variance in Coiled-Coil Twisting. Biophys. J. 2018, 115, 1082–1092. [Google Scholar] [CrossRef]
- Lehman, W.; Li, X.; Orzechowski, M.; Fischer, S. The structural dynamics of α-tropomyosin on F-actin shape the overlap complex between adjacent tropomyosin molecules. Arch. Biochem. Biophys. 2014, 552–553, 68–73. [Google Scholar] [CrossRef][Green Version]
- Li, X.E.; Tobacman, L.S.; Mun, J.Y.; Craig, R.; Fischer, S.; Lehman, W. Tropomyosin position on F-actin revealed by EM reconstruction and computational chemistry. Biophys. J. 2011, 100, 1005–1013. [Google Scholar] [CrossRef]
- Gooding, C.; Smith, C.W.J. Tropomyosin exons as models for alternative splicing. Adv. Exp. Med. Biol. 2008, 644, 27–42. [Google Scholar] [CrossRef]
- Gunning, P.; O’Neill, G.; Hardeman, E. Tropomyosin-based regulation of the actin cytoskeleton in time and space. Physiol. Rev. 2008, 88, 1–35. [Google Scholar] [CrossRef]
- Hanson, J.; Lowy, J. The structure of F-actin and of actin filaments isolated from muscle. J. Mol. Biol. 1963, 6, 46-IN5. [Google Scholar] [CrossRef]
- Huxley, H.E. The Mechanism of Muscular Contraction. Science 1969, 164, 1356–1366. [Google Scholar] [CrossRef] [PubMed]
- Lehman, W.; Craig, R.; Vibert, P. Ca(2+)-induced tropomyosin movement in Limulus thin filaments revealed by three-dimensional reconstruction. Nature 1994, 368, 65–67. [Google Scholar] [CrossRef]
- Varland, S.; Vandekerckhove, J.; Drazic, A. Actin Post-translational Modifications: The Cinderella of Cytoskeletal Control. Trends Biochem. Sci. 2019, 44, 502–516. [Google Scholar] [CrossRef]
- Grintsevich, E.E.; Yesilyurt, H.G.; Rich, S.K.; Hung, R.-J.; Terman, J.R.; Reisler, E. F-actin dismantling through a redox-driven synergy between Mical and cofilin. Nat. Cell Biol. 2016, 18, 876–885. [Google Scholar] [CrossRef]
- Grintsevich, E.E.; Ge, P.; Sawaya, M.R.; Yesilyurt, H.G.; Terman, J.R.; Zhou, Z.H.; Reisler, E. Catastrophic disassembly of actin filaments via Mical-mediated oxidation. Nat. Commun. 2017, 8, 2183. [Google Scholar] [CrossRef] [PubMed]
- Houdusse, A.; Sweeney, H.L. How myosin generates force on actin filaments. Trends Biochem. Sci. 2016, 41, 989–997. [Google Scholar] [CrossRef]
- Heissler, S.M.; Sellers, J.R. Kinetic Adaptations of Myosins for Their Diverse Cellular Functions. Traffic 2016, 17, 839–859. [Google Scholar] [CrossRef]
- Masters, T.A.; Kendrick-Jones, J.; Buss, F. Myosins: Domain Organisation, Motor Properties, Physiological Roles and Cellular Functions. Handb. Exp. Pharmacol. 2017, 235, 77–122. [Google Scholar] [CrossRef]
- Uyeda, T.Q.; Abramson, P.D.; Spudich, J.A. The neck region of the myosin motor domain acts as a lever arm to generate movement. Proc. Natl. Acad. Sci. USA 1996, 93, 4459–4464. [Google Scholar] [CrossRef] [PubMed]
- Sata, M.; Stafford, W.F.; Mabuchi, K.; Ikebe, M. The motor domain and the regulatory domain of myosin solely dictate enzymatic activity and phosphorylation-dependent regulation, respectively. Proc. Natl. Acad. Sci. USA 1997, 94, 91–96. [Google Scholar] [CrossRef]
- Rayment, I.; Rypniewski, W.R.; Smith, R.; Tomchick, D.R.; Benning, M.M.; Winkelmann, D.A.; Wesenberg, G.; Holden, H.M. Three-Dimensional Structure of Myosin Subfragment-1: A Molecular Motor. Science 1993, 261, 10. [Google Scholar] [CrossRef]
- Hokanson, D.E.; Laakso, J.M.; Lin, T.; Sept, D.; Ostap, E.M. Myo1c binds phosphoinositides through a putative pleckstrin homology domain. Mol. Biol. Cell 2006, 17, 4856–4865. [Google Scholar] [CrossRef]
- de Jonge, J.J.; Batters, C.; O’Loughlin, T.; Arden, S.D.; Buss, F. The MYO6 interactome: Selective motor-cargo complexes for diverse cellular processes. FEBS Lett. 2019, 593, 1494–1507. [Google Scholar] [CrossRef] [PubMed]
- Ojima, K. Myosin: Formation and maintenance of thick filaments. Anim. Sci. J. 2019, 90, 801–807. [Google Scholar] [CrossRef]
- Heissler, S.M.; Sellers, J.R. Myosin light chains: Teaching old dogs new tricks. Bioarchitecture 2015, 4, 169–188. [Google Scholar] [CrossRef]
- Dausse, E.; Komajda, M.; Fetler, L.; Dubourg, O.; Dufour, C.; Carrier, L.; Wisnewsky, C.; Bercovici, J.; Hengstenberg, C.; Al-Mahdawi, S. Familial hypertrophic cardiomyopathy. Microsatellite haplotyping and identification of a hot spot for mutations in the beta-myosin heavy chain gene. J. Clin. Investig. 1993, 92, 2807–2813. [Google Scholar] [CrossRef] [PubMed]
- Greber-Platzer, S.; Marx, M.; Fleischmann, C.; Suppan, C.; Dobner, M.; Wimmer, M. Beta-myosin heavy chain gene mutations and hypertrophic cardiomyopathy in Austrian children. J. Mol. Cell. Cardiol. 2001, 33, 141–148. [Google Scholar] [CrossRef]
- Kamisago, M.; Sharma, S.D.; DePalma, S.R.; Solomon, S.; Sharma, P.; McDonough, B.; Smoot, L.; Mullen, M.P.; Woolf, P.K.; Wigle, E.D.; et al. Mutations in sarcomere protein genes as a cause of dilated cardiomyopathy. N. Engl. J. Med. 2000, 343, 1688–1696. [Google Scholar] [CrossRef]
- Richard, P.; Charron, P.; Carrier, L.; Ledeuil, C.; Cheav, T.; Pichereau, C.; Benaiche, A.; Isnard, R.; Dubourg, O.; Burban, M.; et al. Hypertrophic Cardiomyopathy. Circulation 2003, 107, 2227–2232. [Google Scholar] [CrossRef]
- Toydemir, R.M.; Rutherford, A.; Whitby, F.G.; Jorde, L.B.; Carey, J.C.; Bamshad, M.J. Mutations in embryonic myosin heavy chain (MYH3) cause Freeman-Sheldon syndrome and Sheldon-Hall syndrome. Nat. Genet. 2006, 38, 561–565. [Google Scholar] [CrossRef]
- Geisterfer-Lowrance, A.A.; Kass, S.; Tanigawa, G.; Vosberg, H.P.; McKenna, W.; Seidman, C.E.; Seidman, J.G. A molecular basis for familial hypertrophic cardiomyopathy: A beta cardiac myosin heavy chain gene missense mutation. Cell 1990, 62, 999–1006. [Google Scholar] [CrossRef]
- Fujii, T.; Namba, K. Structure of actomyosin rigour complex at 5.2 Å resolution and insights into the ATPase cycle mechanism. Nat. Commun. 2017, 8, 1–11. [Google Scholar] [CrossRef]
- Wulf, S.F.; Ropars, V.; Fujita-Becker, S.; Oster, M.; Hofhaus, G.; Trabuco, L.G.; Pylypenko, O.; Sweeney, H.L.; Houdusse, A.M.; Schröder, R.R. Force-producing ADP state of myosin bound to actin. Proc. Natl. Acad. Sci. USA 2016, 113, E1844–E1852. [Google Scholar] [CrossRef]
- Trivedi, D.V.; Muretta, J.M.; Swenson, A.M.; Davis, J.P.; Thomas, D.D.; Yengo, C.M. Direct measurements of the coordination of lever arm swing and the catalytic cycle in myosin V. Proc. Natl. Acad. Sci. USA 2015, 112, 14593–14598. [Google Scholar] [CrossRef] [PubMed]
- Llinas, P.; Isabet, T.; Song, L.; Ropars, V.; Zong, B.; Benisty, H.; Sirigu, S.; Morris, C.; Kikuti, C.; Safer, D.; et al. How Actin Initiates the Motor Activity of Myosin. Dev. Cell 2015, 33, 401–412. [Google Scholar] [CrossRef] [PubMed]
- Geeves, M.A. Review: The ATPase mechanism of myosin and actomyosin. Biopolymers 2016, 105, 483–491. [Google Scholar] [CrossRef] [PubMed]
- Muretta, J.M.; Petersen, K.J.; Thomas, D.D. Direct real-time detection of the actin-activated power stroke within the myosin catalytic domain. Proc. Natl. Acad. Sci. USA 2013, 110, 7211–7216. [Google Scholar] [CrossRef]
- Mentes, A.; Huehn, A.; Liu, X.; Zwolak, A.; Dominguez, R.; Shuman, H.; Ostap, E.M.; Sindelar, C.V. High-resolution cryo-EM structures of actin-bound myosin states reveal the mechanism of myosin force sensing. Proc. Natl. Acad. Sci. USA 2018, 115, 1292–1297. [Google Scholar] [CrossRef]
- Fischer, S.; Windshügel, B.; Horak, D.; Holmes, K.C.; Smith, J.C. Structural mechanism of the recovery stroke in the myosin molecular motor. Proc. Natl. Acad. Sci. USA 2005, 102, 6873–6878. [Google Scholar] [CrossRef]
- Planelles-Herrero, V.J.; Hartman, J.J.; Robert-Paganin, J.; Malik, F.I.; Houdusse, A. Mechanistic and structural basis for activation of cardiac myosin force production by omecamtiv mecarbil. Nat. Commun. 2017, 8, 190. [Google Scholar] [CrossRef] [PubMed]
- Doran, M.H.; Pavadai, E.; Rynkiewicz, M.J.; Walklate, J.; Bullitt, E.; Moore, J.R.; Regnier, M.; Geeves, M.A.; Lehman, W. Cryo-EM and Molecular Docking Shows Myosin Loop 4 Contacts Actin and Tropomyosin on Thin Filaments. Biophys. J. 2020, 119, 821–830. [Google Scholar] [CrossRef] [PubMed]
- Risi, C.; Schäfer, L.U.; Belknap, B.; Pepper, I.; White, H.D.; Schröder, G.F.; Galkin, V.E. High-Resolution Cryo-EM Structure of the Cardiac Actomyosin Complex. Structure 2021, 29, 50–60.e4. [Google Scholar] [CrossRef]
- Gurel, P.S.; Kim, L.Y.; Ruijgrok, P.V.; Omabegho, T.; Bryant, Z.; Alushin, G.M. Cryo-EM structures reveal specialization at the myosin VI-actin interface and a mechanism of force sensitivity. eLife 2017, 6, e31125. [Google Scholar] [CrossRef]
- Banerjee, C.; Hu, Z.; Huang, Z.; Warrington, J.A.; Taylor, D.W.; Trybus, K.M.; Lowey, S.; Taylor, K.A. The structure of the actin-smooth muscle myosin motor domain complex in the rigor state. J. Struct. Biol. 2017, 200, 325–333. [Google Scholar] [CrossRef] [PubMed]
- Behrmann, E.; Müller, M.; Penczek, P.A.; Mannherz, H.G.; Manstein, D.J.; Raunser, S. Structure of the Rigor Actin-Tropomyosin-Myosin Complex. Cell 2012, 150, 327–338. [Google Scholar] [CrossRef]
- Robert-Paganin, J.; Xu, X.-P.; Swift, M.F.; Auguin, D.; Robblee, J.P.; Lu, H.; Fagnant, P.M.; Krementsova, E.B.; Trybus, K.M.; Houdusse, A.; et al. The actomyosin interface contains an evolutionary conserved core and an ancillary interface involved in specificity. Nat. Commun. 2021, 12, 1892. [Google Scholar] [CrossRef]
- Furch, M.; Remmel, B.; Geeves, M.A.; Manstein, D.J. Stabilization of the Actomyosin Complex by Negative Charges on Myosin. Biochemistry 2000, 39, 11602–11608. [Google Scholar] [CrossRef]
- Joel, P.B.; Trybus, K.M.; Sweeney, H.L. Two conserved lysines at the 50/20-kDa junction of myosin are necessary for triggering actin activation. J. Biol. Chem. 2001, 276, 2998–3003. [Google Scholar] [CrossRef]
- Onishi, H.; Mikhailenko, S.V.; Morales, M.F. Toward understanding actin activation of myosin ATPase: The role of myosin surface loops. Proc. Natl. Acad. Sci. USA 2006, 103, 6136–6141. [Google Scholar] [CrossRef]
- Murphy, C.T.; Spudich, J.A. The sequence of the myosin 50–20 K loop affects Myosin’s affinity for actin throughout the actin-myosin ATPase cycle and its maximum ATPase activity. Biochemistry 1999, 38, 3785–3792. [Google Scholar] [CrossRef]
- van den Boom, F.; Düssmann, H.; Uhlenbrock, K.; Abouhamed, M.; Bähler, M. The Myosin IXb Motor Activity Targets the Myosin IXb RhoGAP Domain as Cargo to Sites of Actin Polymerization. Mol. Biol. Cell 2007, 18, 1507–1518. [Google Scholar] [CrossRef]
- Struchholz, S.; Elfrink, K.; Pieper, U.; Kalhammer, G.; Honnert, U.; Grützner, A.; Linke, W.A.; Liao, W.; Bähler, M. Functional Role of the Extended Loop 2 in the Myosin 9b Head for Binding F-actin *. J. Biol. Chem. 2009, 284, 3663–3671. [Google Scholar] [CrossRef]
- Elfrink, K.; Liao, W.; Pieper, U.; Oeding, S.J.; Bähler, M. The Loop2 Insertion of Type IX Myosin Acts as an Electrostatic Actin Tether that Permits Processive Movement. PLoS ONE 2014, 9, e84874. [Google Scholar] [CrossRef]
- Yengo, C.M.; Sweeney, H.L. Functional role of loop 2 in myosin V. Biochemistry 2004, 43, 2605–2612. [Google Scholar] [CrossRef] [PubMed]
- Robert-Paganin, J.; Pylypenko, O.; Kikuti, C.; Sweeney, H.L.; Houdusse, A. Force Generation by Myosin Motors: A Structural Perspective. Chem. Rev. 2020, 120, 5–35. [Google Scholar] [CrossRef]
- Rayment, I.; Holden, H.M.; Whittaker, M.; Yohn, C.B.; Lorenz, M.; Holmes, K.C.; Milligan, R.A. Structure of the actin-myosin complex and its implications for muscle contraction. Science 1993, 261, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, N.; Ohkura, R.; Sutoh, K. Dictyostelium myosin II as a model to study the actin-myosin interactions during force generation. J. Muscle Res. Cell Motil. 2002, 23, 697–702. [Google Scholar] [CrossRef] [PubMed]
- Spudich, J.A.; Aksel, T.; Bartholomew, S.R.; Nag, S.; Kawana, M.; Yu, E.C.; Sarkar, S.S.; Sung, J.; Sommese, R.F.; Sutton, S.; et al. Effects of hypertrophic and dilated cardiomyopathy mutations on power output by human β-cardiac myosin. J. Exp. Biol. 2016, 219, 161–167. [Google Scholar] [CrossRef]
- Milligan, R.A.; Whittaker, M.; Safer, D. Molecular structure of F-actin and location of surface binding sites. Nature 1990, 348, 217–221. [Google Scholar] [CrossRef]
- Milligan, R.A. Protein-protein interactions in the rigor actomyosin complex. Proc. Natl. Acad. Sci. USA 1996, 93, 21–26. [Google Scholar] [CrossRef]
- Van Dijk, J.; Furch, M.; Lafont, C.; Manstein, D.J.; Chaussepied, P. Functional characterization of the secondary actin binding site of myosin II. Biochemistry 1999, 38, 15078–15085. [Google Scholar] [CrossRef]
- Várkuti, B.H.; Yang, Z.; Malnasi-Csizmadia, A. Structural model of weak binding actomyosin in the prepowerstroke state. J. Biol. Chem. 2015, 290, 1679–1688. [Google Scholar] [CrossRef] [PubMed]
- Várkuti, B.H.; Yang, Z.; Kintses, B.; Erdélyi, P.; Bárdos-Nagy, I.; Kovács, A.L.; Hári, P.; Kellermayer, M.; Vellai, T.; Málnási-Csizmadia, A. A novel actin binding site of myosin required for effective muscle contraction. Nat. Struct. Mol. Biol. 2012, 19, 299–306. [Google Scholar] [CrossRef]
- Coureux, P.-D.; Wells, A.L.; Ménétrey, J.; Yengo, C.M.; Morris, C.A.; Sweeney, H.L.; Houdusse, A. A structural state of the myosin V motor without bound nucleotide. Nature 2003, 425, 419–423. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, N.; Ohkura, R.; Sutoh, K. Insertion or Deletion of a Single Residue in the Strut Sequence of Dictyostelium Myosin II Abolishes Strong Binding to Actin *. J. Biol. Chem. 2000, 275, 38705–38709. [Google Scholar] [CrossRef] [PubMed]
- Woo, A.; Rakowski, H.; Liew, J.C.; Zhao, M.-S.; Liew, C.-C.; Parker, T.G.; Zeller, M.; Wigle, E.D.; Sole, M.J. Mutations of the β myosin heavy chain gene in hypertrophic cardiomyopathy: Critical functional sites determine prognosis. Heart 2003, 89, 1179–1185. [Google Scholar] [CrossRef]
- Walsh, R.; Thomson, K.L.; Ware, J.S.; Funke, B.H.; Woodley, J.; McGuire, K.J.; Mazzarotto, F.; Blair, E.; Seller, A.; Taylor, J.C.; et al. Reassessment of Mendelian gene pathogenicity using 7,855 cardiomyopathy cases and 60,706 reference samples. Genet. Med. 2017, 19, 192–203. [Google Scholar] [CrossRef]
- Gyimesi, M.; Tsaturyan, A.K.; Kellermayer, M.S.Z.; Málnási-Csizmadia, A. Kinetic characterization of the function of myosin loop 4 in the actin-myosin interaction. Biochemistry 2008, 47, 283–291. [Google Scholar] [CrossRef]
- Lieto-Trivedi, A.; Dash, S.; Coluccio, L.M. Myosin surface loop 4 modulates inhibition of actomyosin 1b ATPase activity by tropomyosin. Biochemistry 2007, 46, 2779–2786. [Google Scholar] [CrossRef]
- Gunning, P.W.; Hardeman, E.C.; Lappalainen, P.; Mulvihill, D.P. Tropomyosin—Master regulator of actin filament function in the cytoskeleton. J. Cell Sci. 2015, 128, 2965–2974. [Google Scholar] [CrossRef]
- Gunning, P.W.; Hardeman, E.C. Tropomyosins. Curr. Biol. 2017, 27, R8–R13. [Google Scholar] [CrossRef] [PubMed]
- Cope, M.J.; Whisstock, J.; Rayment, I.; Kendrick-Jones, J. Conservation within the myosin motor domain: Implications for structure and function. Structure 1996, 4, 969–987. [Google Scholar] [CrossRef]
- Kollmar, M.; Mühlhausen, S. Myosin repertoire expansion coincides with eukaryotic diversification in the Mesoproterozoic era. BMC Evol. Biol. 2017, 17, 211. [Google Scholar] [CrossRef] [PubMed]
- Sebé-Pedrós, A.; Grau-Bové, X.; Richards, T.A.; Ruiz-Trillo, I. Evolution and classification of myosins, a paneukaryotic whole-genome approach. Genome Biol. Evol. 2014, 6, 290–305. [Google Scholar] [CrossRef]
- Müller, M.; Diensthuber, R.P.; Chizhov, I.; Claus, P.; Heissler, S.M.; Preller, M.; Taft, M.H.; Manstein, D.J. Distinct functional interactions between actin isoforms and nonsarcomeric myosins. PLoS ONE 2013, 8, e70636. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Doran, M.H.; Lehman, W. The Central Role of the F-Actin Surface in Myosin Force Generation. Biology 2021, 10, 1221. https://doi.org/10.3390/biology10121221
Doran MH, Lehman W. The Central Role of the F-Actin Surface in Myosin Force Generation. Biology. 2021; 10(12):1221. https://doi.org/10.3390/biology10121221
Chicago/Turabian StyleDoran, Matthew H., and William Lehman. 2021. "The Central Role of the F-Actin Surface in Myosin Force Generation" Biology 10, no. 12: 1221. https://doi.org/10.3390/biology10121221
APA StyleDoran, M. H., & Lehman, W. (2021). The Central Role of the F-Actin Surface in Myosin Force Generation. Biology, 10(12), 1221. https://doi.org/10.3390/biology10121221






