Effects of Salinity on Physiological, Biochemical and Gene Expression Parameters of Black Tiger Shrimp (Penaeus monodon): Potential for Farming in Low-Salinity Environments
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Shrimp Collection
2.2. Experimental Tank Setting and Acclimation
2.3. Salinity Stress Experiment
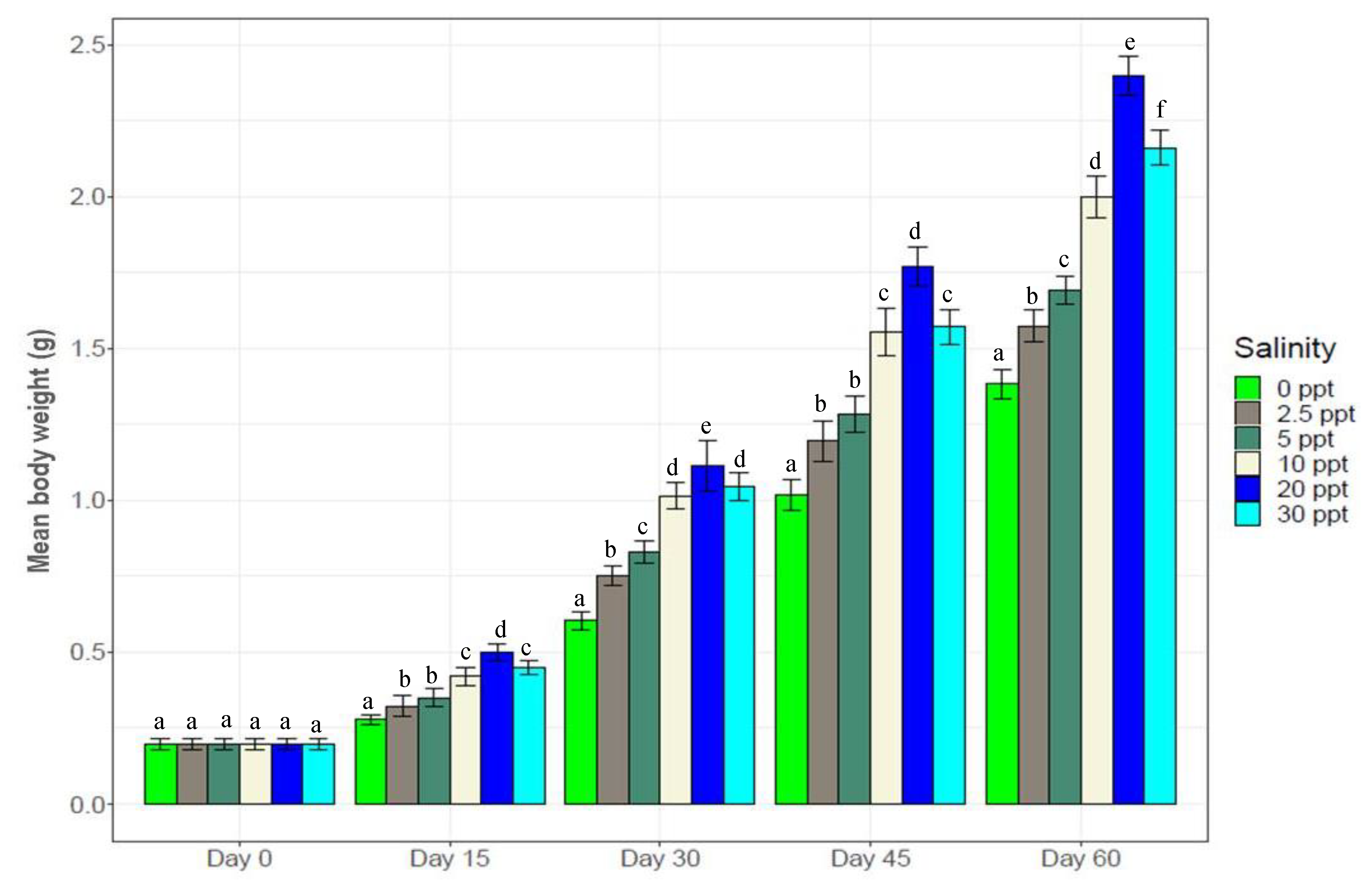
2.4. Growth and Survival Performance
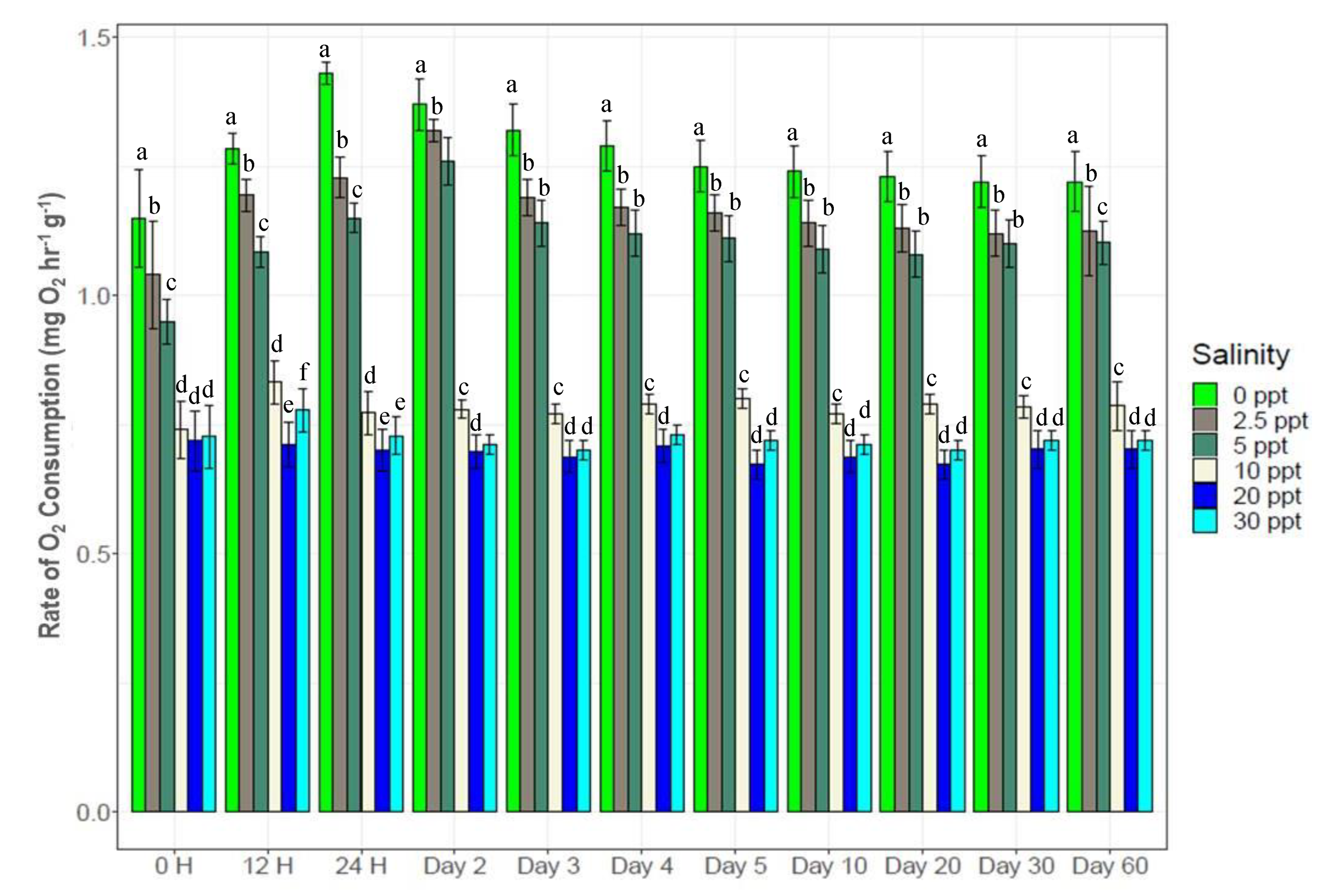
2.5. Rates of Oxygen Consumption
2.6. Assaying Hemolymph Osmolality
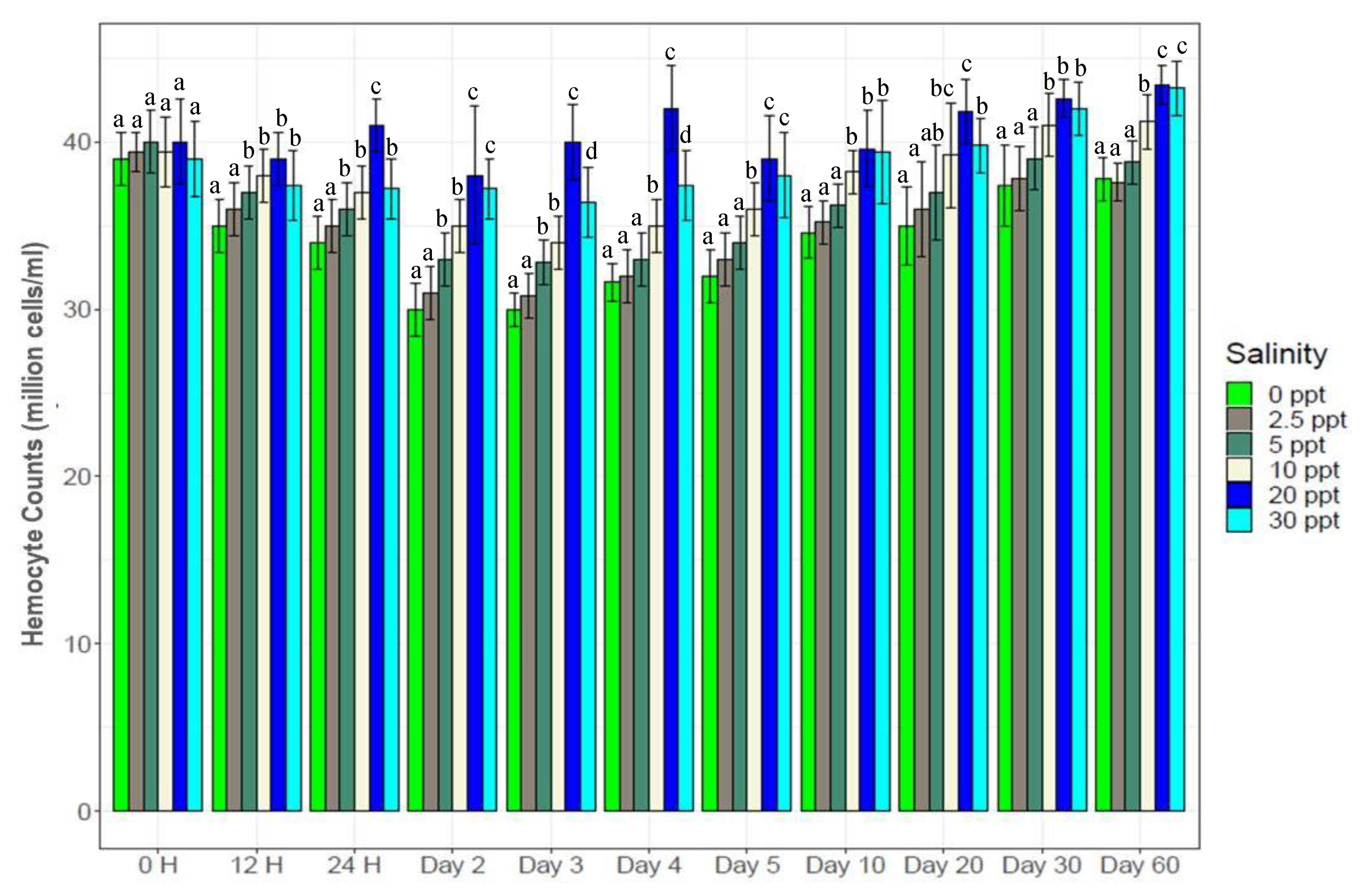
2.7. Total Hemocyte Counts
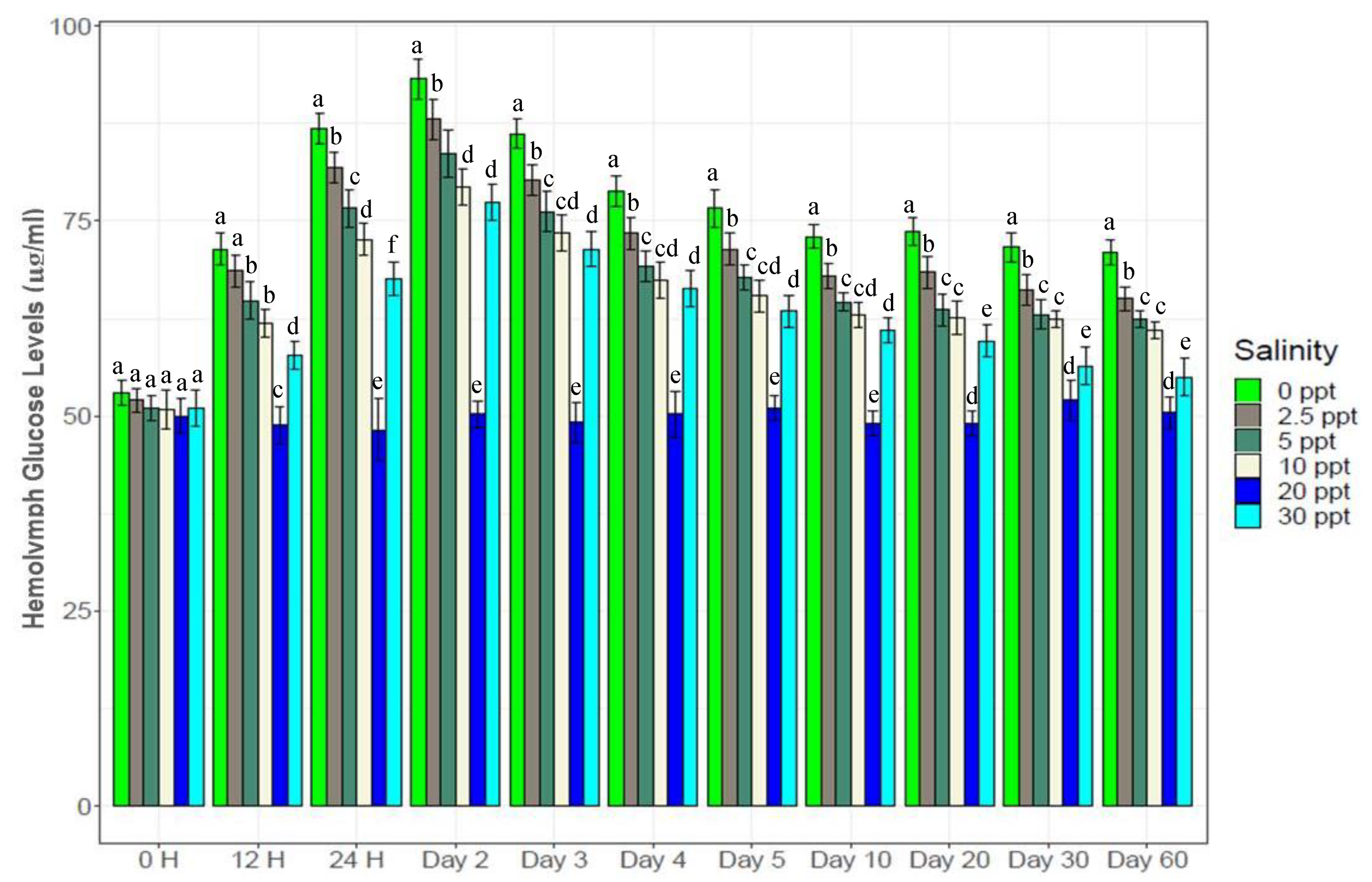
2.8. Assaying Glucose and Serotonin Levels in the Hemolymph
2.9. Free Amino Acid (FAA) Levels in Hemolymph
2.10. Free Fatty Acid (FFA) Levels
2.11. Gene Expression Study
2.12. Data Analysis
3. Results
3.1. Growth and Survival Performance of Black Tiger Shrimp at Different Salinity Levels
3.2. Changes in O2 Consumption Rates
3.3. Hemolymph Osmolality Changes
3.4. Total Hemocytes in the Hemolymph
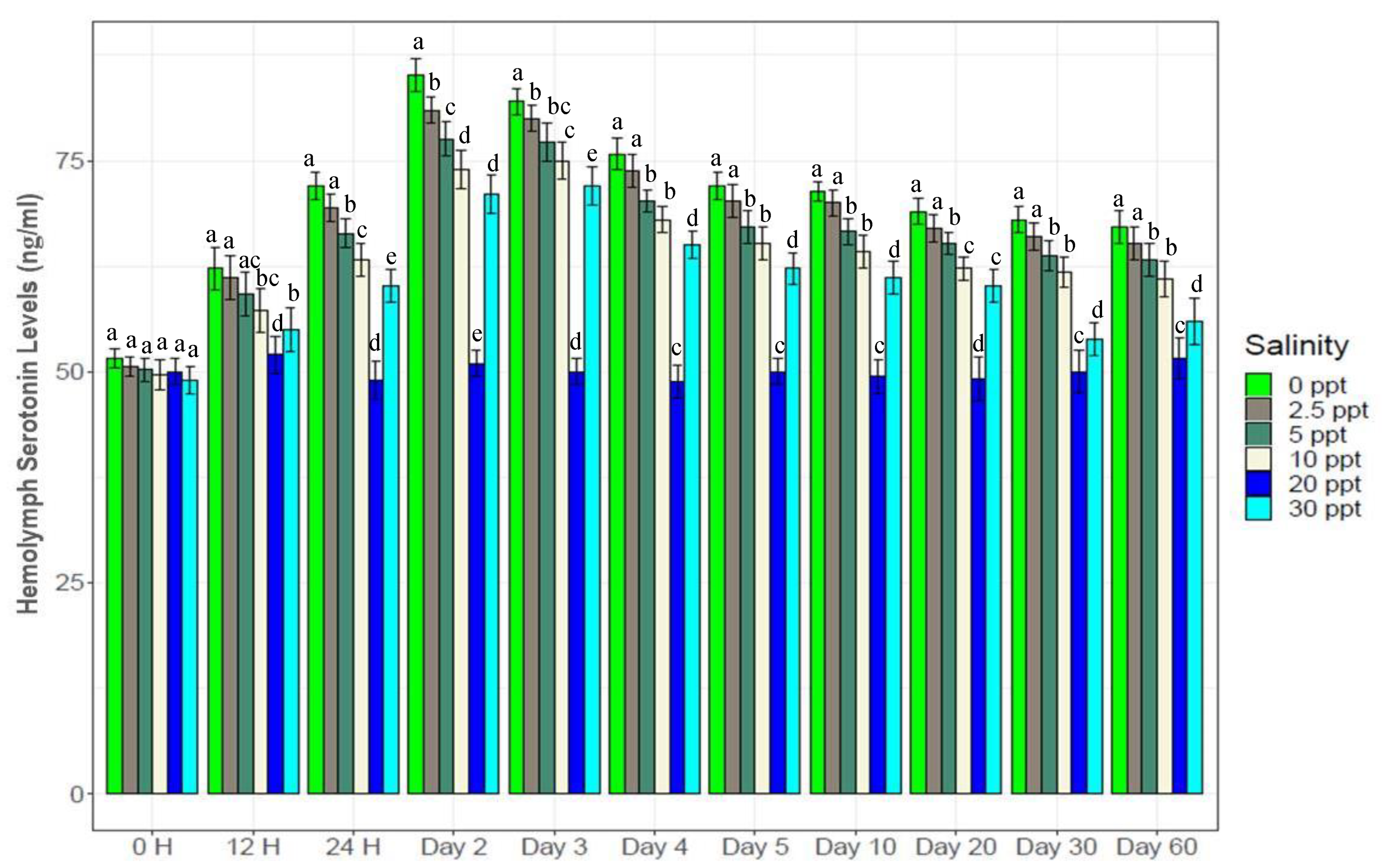
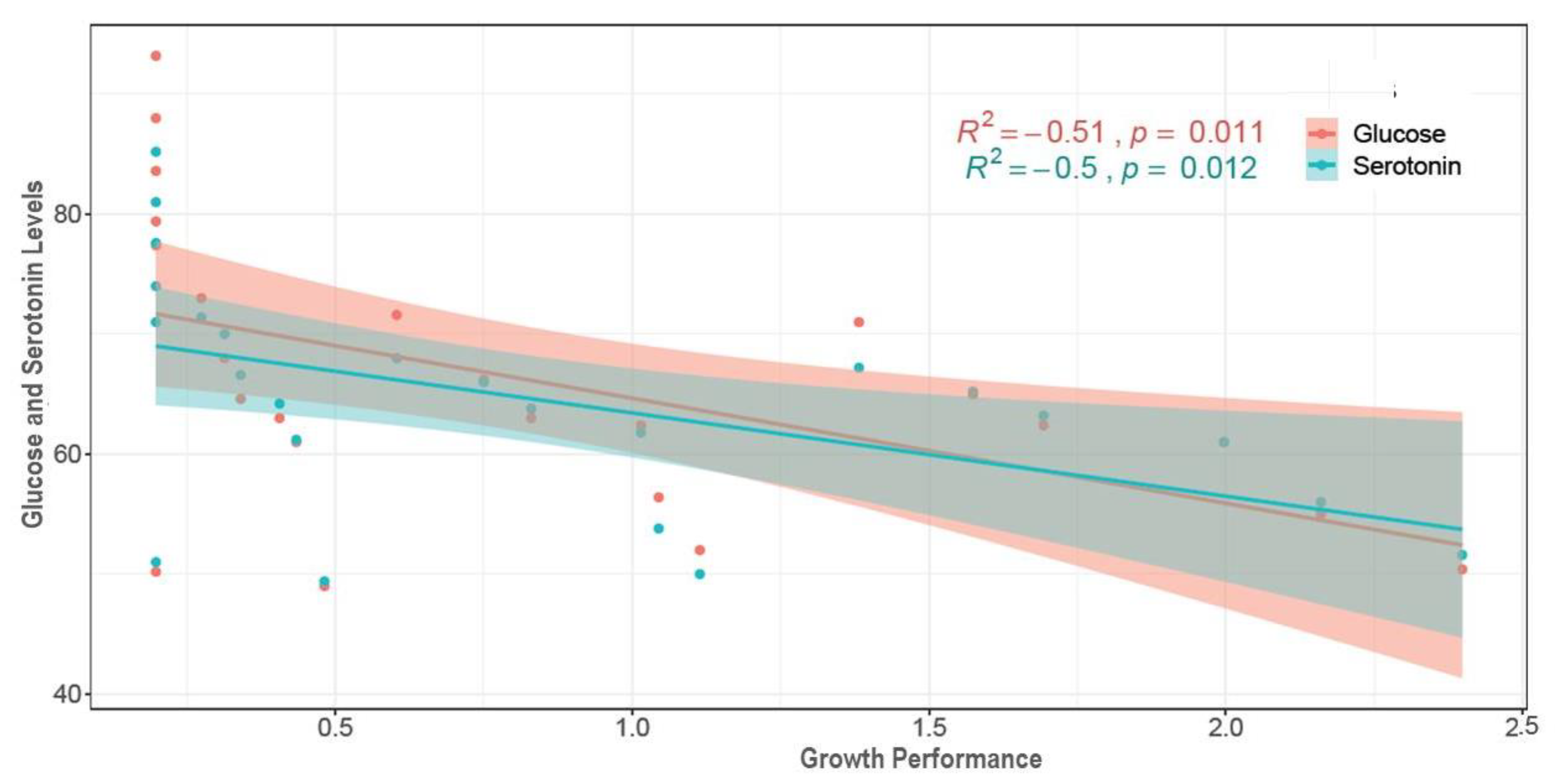
3.5. Hemolymph Glucose and Serotonin Levels
3.6. Salinity Induced Changes in Free Amino Acids (FAAs) and Free Fatty Acids (FFAs)
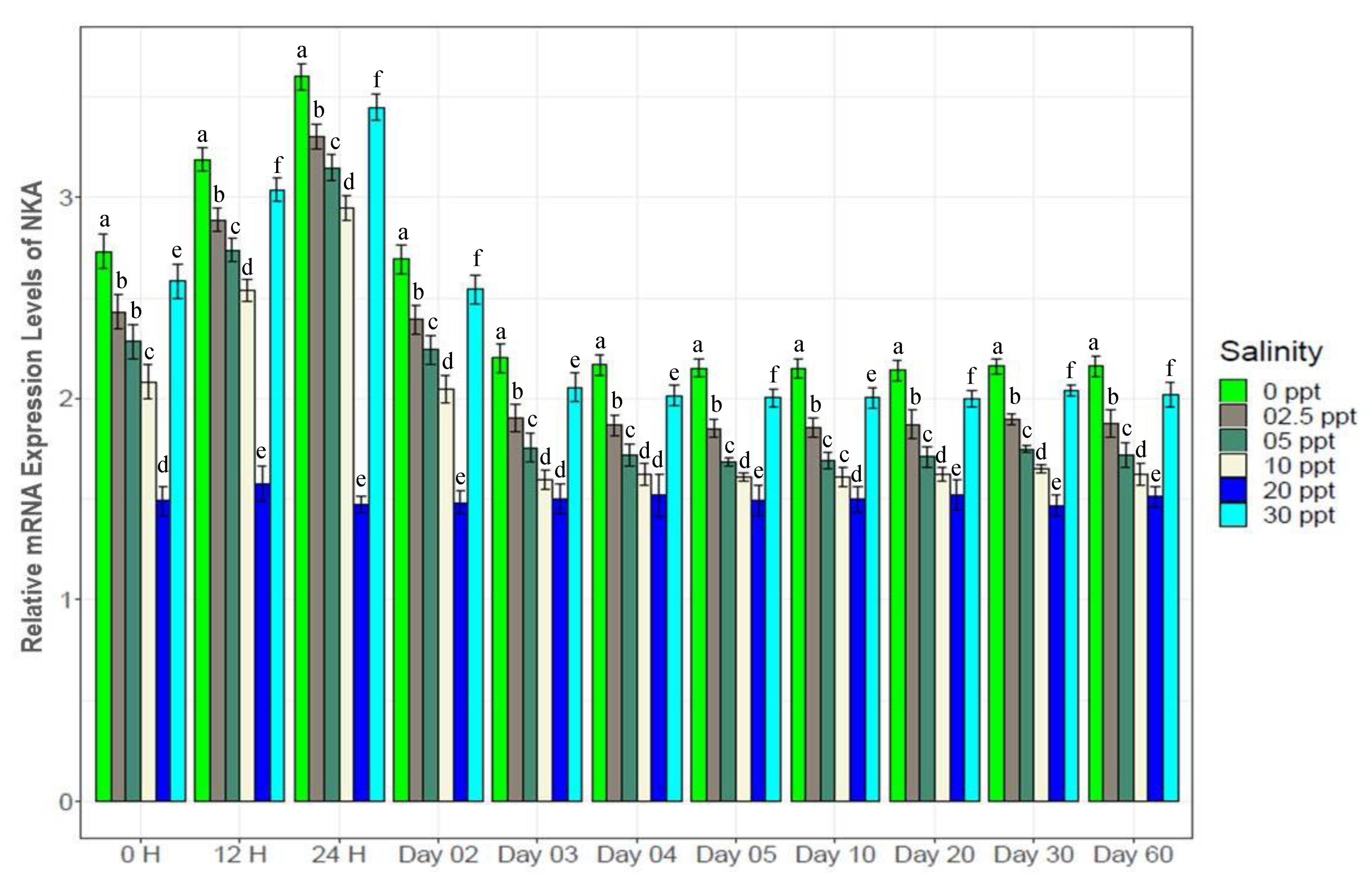
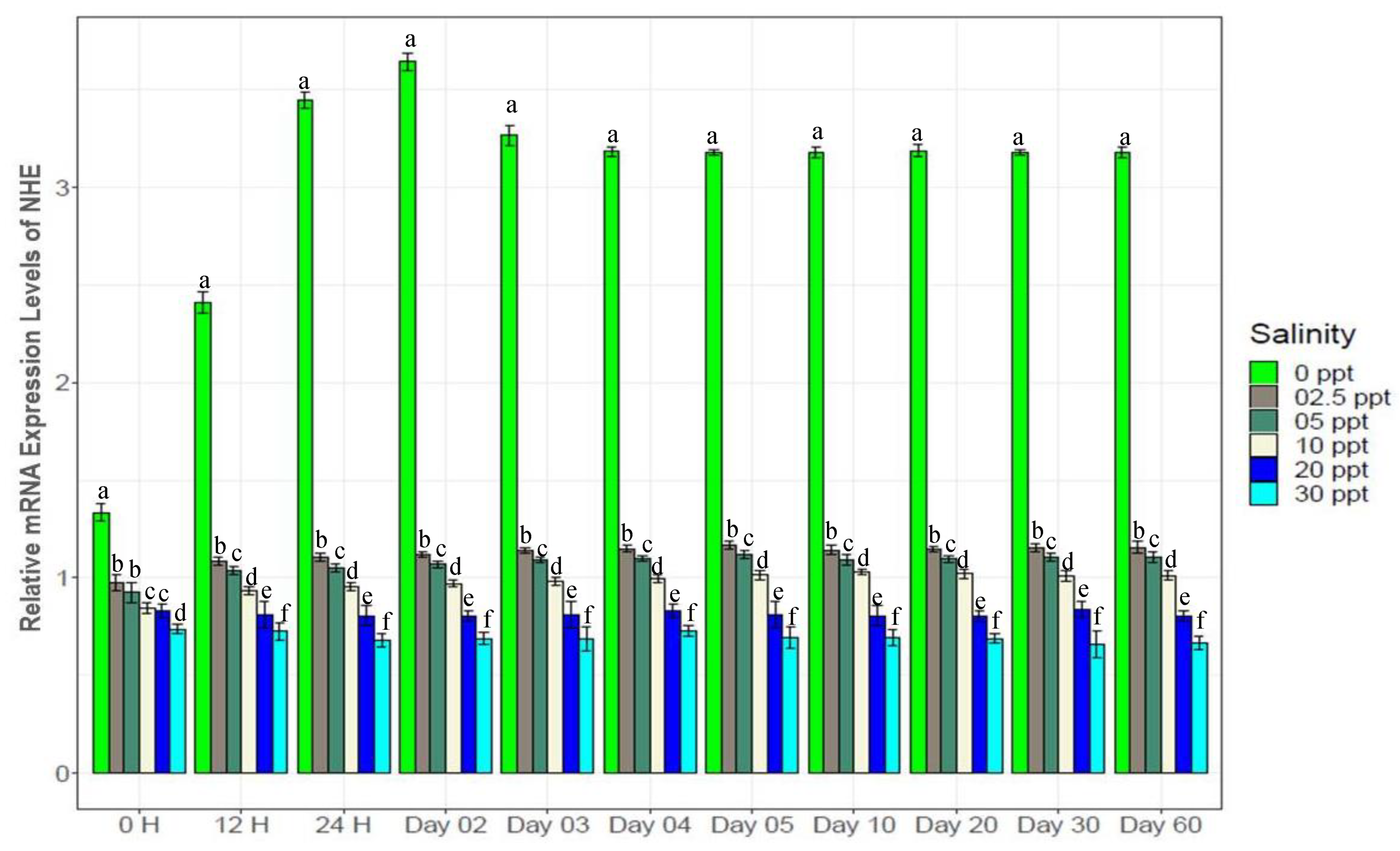
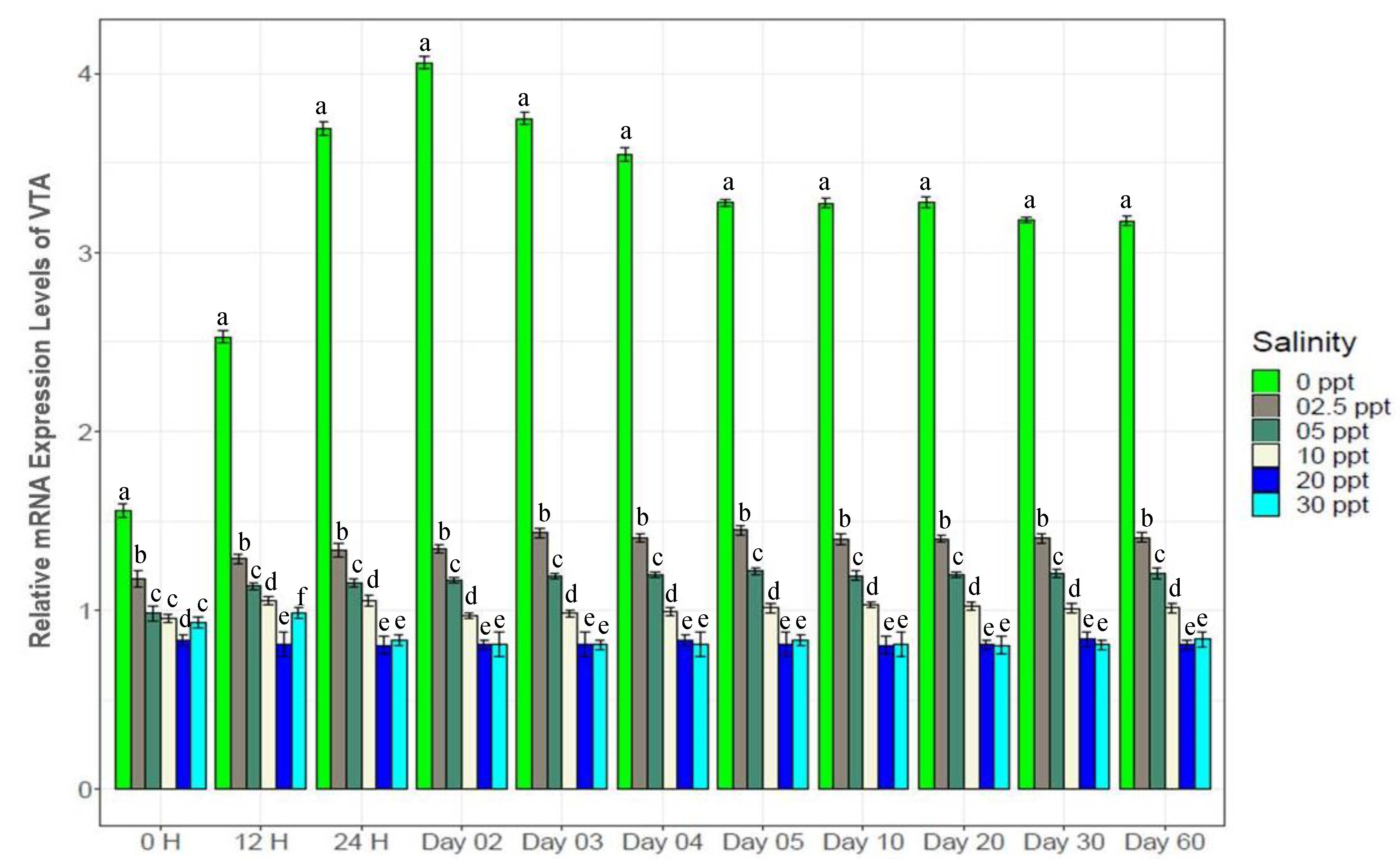
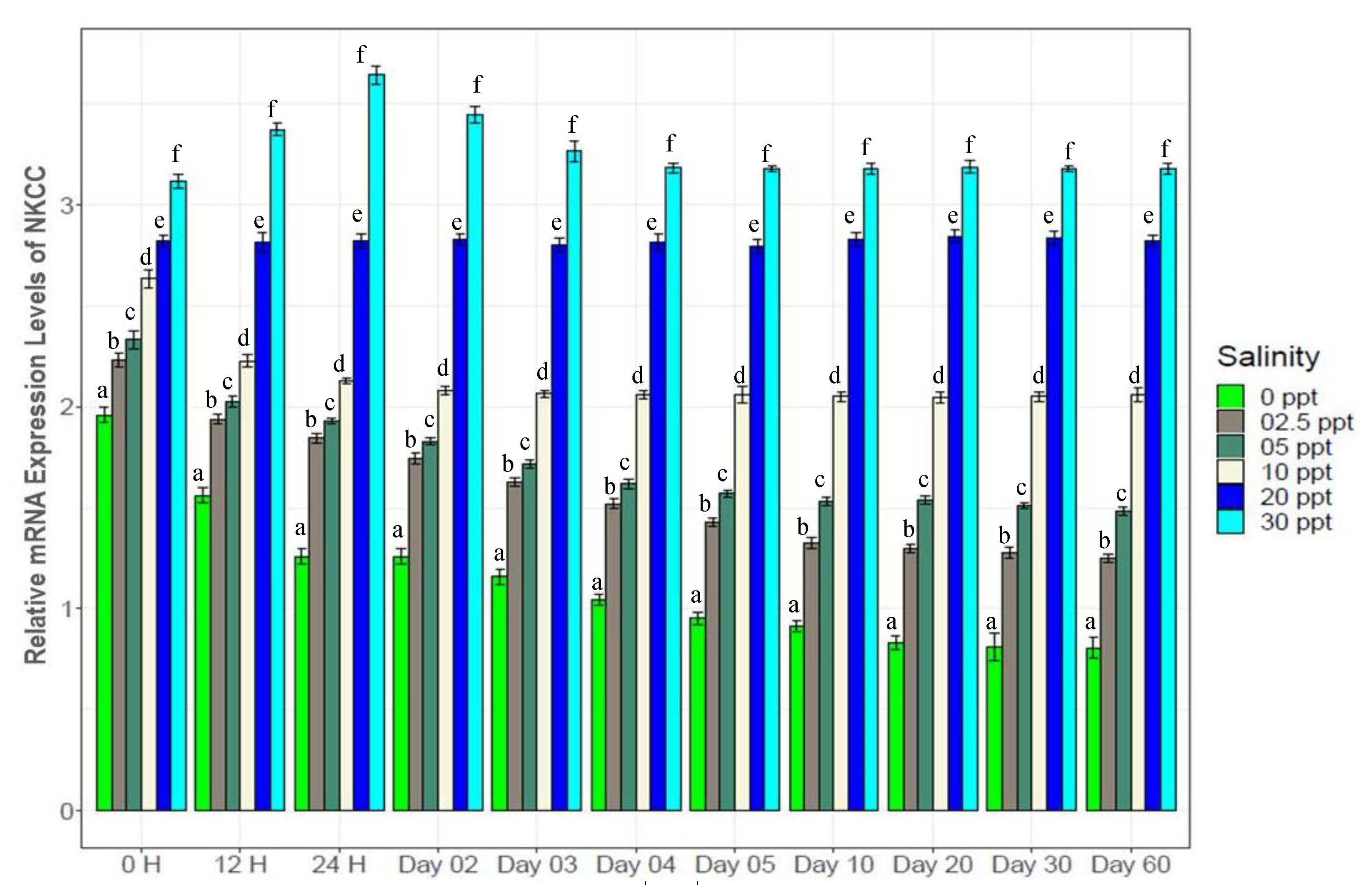
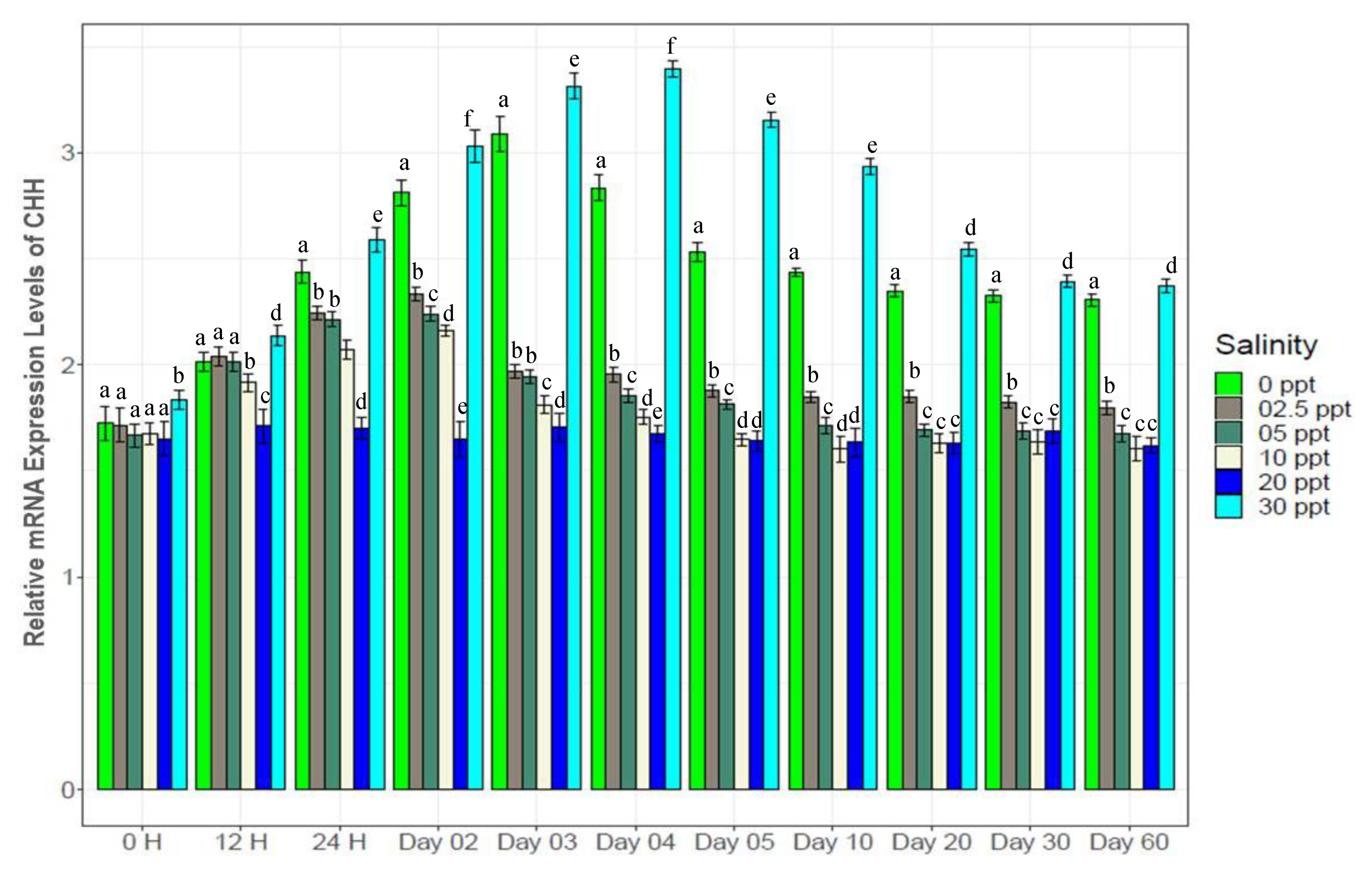
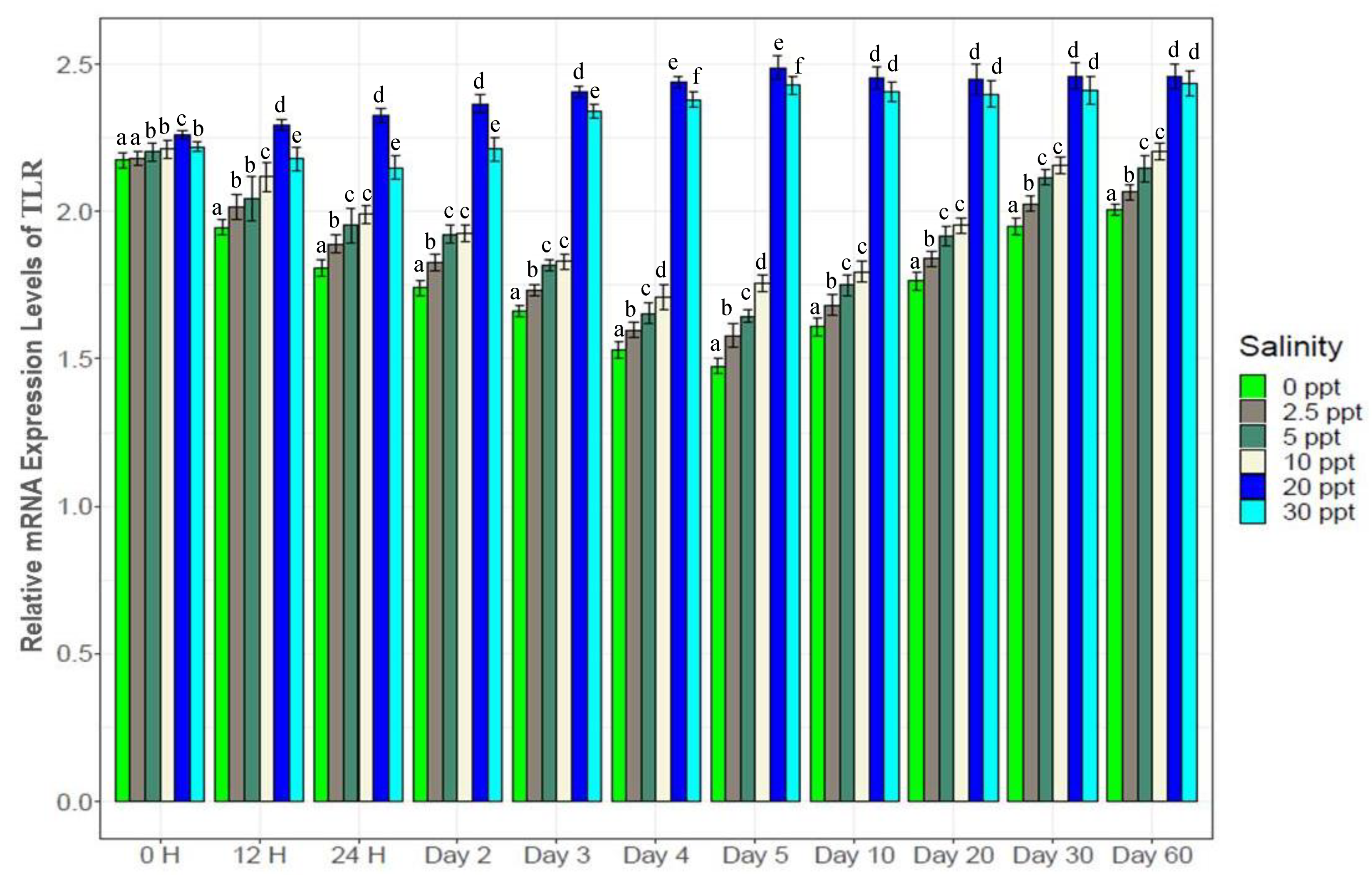
3.7. Changes in Gene Expression
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ye, L.; Jiang, S.; Zhu, X.; Yang, Q.; Wen, W.; Wu, K. Effects of salinity on growth and energy budget of juvenile Penaeus monodon. Aquaculture 2009, 290, 140–144. [Google Scholar] [CrossRef]
- Jaffer, Y.D.; Saraswathy, R.; Ishfaq, M.; Antony, J.; Bundela, D.S.; Sharma, P.C. Effect of low salinity on the growth and survival of juvenile Pacific white shrimp, Penaeus vannamei: A revival. Aquaculture 2020, 515, 734561. [Google Scholar] [CrossRef]
- Rahi, M.L.; Sabbir, W.; Salin, K.R.; Aziz, D.; Hurwood, D.A. Physiological, biochemical and genetic responses of black tiger shrimp (Penaeus monodon) to differential exposure to white spot syndrome virus and Vibrio parahemolyticus. Aquaculture 2021, 546, 737337. [Google Scholar] [CrossRef]
- Phuoc, L.H.; Cortell, M.; Thanh, N.C.; Nauwynck, H.; Pensaert, N.; Alday-Sanz, V.; Broeck, W.V.D.; Sorgellos, P.; Bossier, P. Effect of dose and challenge routes of Vibrio spp. on co-infection with white spot syndrome virus in Penaeus vannamei. Aquaculture 2009, 290, 61–68. [Google Scholar] [CrossRef]
- Tantulo, U.; Fotedar, R. Comparison of growth, osmoregulatory capacity, ionic regulation and organosomatic indices of black tiger prawn (Penaeus monodon Fabricius, 1798) juveniles reared in potassium fortified inland saline water and ocean water at different salinities. Aquaculture 2006, 258, 594–605. [Google Scholar] [CrossRef]
- Rahi, M.L.; Mather, P.B.; Hurwood, D.A. Do plasticity in gene expression and physiological responses in Palaemonid prawns facilitate adaptive response to different osmotic challenges? Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2021, 251, 110810. [Google Scholar] [CrossRef]
- Santhoshi, S.; Sugumar, V.; Munuswamy, N. Serotonin modulation of hemolymph glucose and crustacean hyperglycemic hormone titers in Fenneropenaeus indicus. Aquaculture 2008, 281, 106–112. [Google Scholar]
- Freire, C.A.; Onken, H.; McNamara, J.C. A structure–function analysis of ion transport in crustacean gills and excretory organs. Comp. Biochem. Physiol. 2008, 151, 272–304. [Google Scholar] [CrossRef]
- McNamara, J.C.; Freire, C.A.; Torres, A.H.; Faria, S.C. The conquest of fresh water by the palaemonid shrimps: An evolutionary history scripted in the osmoregulatory epithelia of the gills and antennal glands. Biol. J. Linn. Soc. 2015, 114, 673–688. [Google Scholar] [CrossRef]
- Rahi, M.L.; Ferdusy, T.; Ahmed, S.W.; Khan, M.N.; Aziz, D.; Salin, K.R. Impact of salinity changes on growth, oxygen consumption and expression pattern of selected candidate genes in the orange mud crab (Scylla olivacea). Aquac. Res. 2020, 51, 4290–4301. [Google Scholar] [CrossRef]
- Freire, C.A.; Maraschi, A.C.; Lara, A.F.; Amado, E.M.; Prodocimo, V. Late rise in hemolymph osmolality in Macrobrachium acanthurus (diadromous freshwater shrimp) exposed to brackish water: Early reduction in branchial Na+/K+ pump activity but stable muscle HSP70 expression. Comp. Biochem. Physiol. 2018, 216, 69–74. [Google Scholar] [CrossRef]
- Moshtaghi, A.; Rahi, M.L.; Mather, P.B.; Hurwood, D.A. An investigation of gene expression pattern that contribute to osmoregulation in Macrobrachium australiense: Assessment of adaptive responses to different osmotic niches. Gen. Rep. 2018, 13, 76–83. [Google Scholar] [CrossRef]
- Rahi, M.L.; Moshtaghi, A.; Mather, P.B.; Hurwood, D.A. Osmoregulation in decapod crustaceans: Physiological and genomic perspectives. Hydrobiologia 2018, 825, 177–188. [Google Scholar] [CrossRef]
- Vogt, G. Abbreviation of larval development and extension of brood care as key features of the evolution of freshwater Decapoda. Biol. Rev. 2013, 88, 81–116. [Google Scholar] [CrossRef]
- Rahi, M.L. Understanding the Molecular Basis of Adaptation to Freshwater Environments by Prawns in the Genus Macrobrachium. Ph.D. Thesis, Science and Engineering Faculty, Queensland University of Technology, Brisbane City, Australia, 2017. [Google Scholar] [CrossRef]
- Li, R.; Tian, J.-Z.; Zhuang, C.-H.; Zhang, Y.-C.; Geng, X.-Y.; Zhu, L.-N.; Sun, J.-S. CHHBP: A newly identified receptor of crustacean hyperglycemic hormone. J. Exp. Biol. 2016, 219, 1259–1268. [Google Scholar] [CrossRef]
- Moshtaghi, A.; Rahi, M.L.; Tuan, V.T.; Mather, P.B.; Hurwood, D.A. A transcriptomic scan for potential candidate genes involved in osmoregulation in an obligate freshwater palaemonid prawn (Macrobrachium australiense). PeerJ 2016, 4, e2520. [Google Scholar] [CrossRef]
- Rahi, M.L.; Amin, S.; Mather, P.B.; Hurwood, D.A. Candidate genes that have facilitated freshwater adaptation by palaemonid prawns in the genus Macrobrachium: Identification and expression validation in a model species (M. koombooloomba). PeerJ 2017, 5, e2977. [Google Scholar] [CrossRef][Green Version]
- Huong, D.T.T.; Yang, W.-J.; Okunu, K.; Wilder, M.N. Changes in free amino acids in the hemolymph of giant freshwater prawn Macrobrachium rosenbergii exposed to varying salinities: Relationship to osmoregulatory ability. Comp. Biochem. Physiol. 2001, 128, 317–326. [Google Scholar] [CrossRef]
- Tantulo, U.; Fotedar, R. Physiological performance and serum Na+, K+ Ca2+ and Mg2+ regulation of black tiger prawn (Penaeus monodon Fabricius 1798) reared in varying Na+/K+ ratios of inland saline water. Aquaculture 2016, 479, 52–59. [Google Scholar] [CrossRef]
- Nadal, E.D.; Ammerer, G.; Posas, F. Controlling gene expression in response to stress. Nat. Rev. Gen. 2011, 12, 833–845. [Google Scholar] [CrossRef]
- Moshtaghi, A.; Rahi, M.L.; Mather, P.B.; Hurwood, D.A. Understanding the genomic basis of adaptive response to variable osmotic niches in freshwater prawns: A comparative intraspecific RNA-seq analysis of Macrobrachium australiense. J. Hered. 2017, 108, 544–552. [Google Scholar] [CrossRef]
- Rahi, M.L.; Mather, P.B.; Tariq, E.; Hurwood, D.A. The molecular basis of freshwater adaptation in prawns: Insights from comparative transcriptomics of three Macrobrachium species. Genome Biol. Evol. 2019, 11, 1002–1018. [Google Scholar] [CrossRef]
- Rogl, K.A.; Rahi, M.L.; Royle, J.; Prentis, P.J.; Hurwood, D.A. A transcriptome-wide assessment of differentially expressed genes among two highly divergent, yet sympatric, lineages of the freshwater Atyid shrimp, Paratya australiensis. Hydrobiologia 2018, 825, 189–196. [Google Scholar] [CrossRef]
- FAO. Cultured Aquatic Species Information Programme: Penaeus monodon. Cultured Aquatic Species. Information Programme. Text by Kongkeo, H. FAO Fisheries and Aquaculture Department. 2019. Available online: https://www.fao.org/fishery/culturedspecies/Penaeus_monodon/ (accessed on 29 July 2020).
- Deris, Z.M.; Iehata, S.; Ikhwanuddin, M.; Sahimi, M.B.M.K.; Do, T.D.; Sorgeloos, P.; Sung, Y.Y.; Wong, L.L. Immune and bacterial toxin genes expression in different giant tiger prawn, Penaeus monodon post-larvae stages following AHPND-causing strain of Vibrio parahaemolyticus challenge. Aquac. Rep. 2020, 16, 100248. [Google Scholar] [CrossRef]
- Nath, R.D.; Rahi, M.L.; Hossain, G.S.; Huq, K.A. Marketing status of freshwater snail in Khulna district. Bang. Res. Pub. J. 2008, 1, 337–347. [Google Scholar]
- Duan, Y.; Huang, J.; Wang, Y.; Zhang, J. Characterization of bacterial community in intestinal and rearing water of Penaeus monodon differing growth performances in outdoor and indoor ponds. Aquac. Res. 2020, 51, 4279–4289. [Google Scholar] [CrossRef]
- FSY. Fisheries Statistical Yearbook of Bangladesh, 36th ed.; Department of Fisheries, Ministry of Fisheries and Livestock, Government of the People’s Republic of Bangladesh: Dhaka, Bangladesh, 2019; pp. 32–40. [Google Scholar]
- Afroz, K.B.; Shah, M.S.; Salin, K.R.; Rahi, M.L. Growth and survival of diploid and triploid bata, Labeo bata (Hamilton, 1822). Aquac. Fish Fish. 2021, 1, 1–9. [Google Scholar] [CrossRef]
- Sabbir, W.; Masud, M.A.A.; Islam, S.S.; Rahman, M.A.; Islam, M.R.; Rahi, M.L. Some aspects of water quality parameters of the Mouri River, Khulna: An attempt to estimate pollution status. Bang. Res. Pub. J. 2010, 4, 95–102. [Google Scholar]
- Ahmed, M.S.; Biswas, B.; Roy, D.; Raseduzzaman, M.; Rahi, M.L.; Khanom, M.; Sarower, M.G. Polymerase chain reaction technique for rapid check of virulency of Escherichia coli from shrimp farms. Int. J. Eng. Appl. Sci. 2013, 3, 8269. [Google Scholar]
- DoF (Department of Fisheries). (In Bengali) 2020; Department of Fisheries, Ministry of Fisheries and Livestock: Dhaka, Bangladesh, 2020; pp. 86–97. [Google Scholar]
- Havird, J.C.; Santos, S.R.; Henry, R.P. Osmoregulation in Hawaiian anchialine shrimp Halocaridina rubra (Crustacea: Atyidae): Expression of ion transporters, mitochondria-rich cell proliferation and hemolymph osmolality during salinity transfers. J. Exp. Biol. 2014, 217, 2309–2320. [Google Scholar] [CrossRef]
- Jung, H.; Lyons, L.E.; Hurwood, D.A.; Mather, P.B. Genes and growth performance in crustacean species: A review of relevant genomic studies in crustaceans and other taxa. Rev. Aquac. 2013, 5, 77–110. [Google Scholar] [CrossRef]
- Aziz, D.; Nguyen, V.T.; Rahi, M.L.; Hurwood, D.A.; Mather, P.B. Identification of genes that potentially affect social dominance hierarchy in adult male giant freshwater prawns (Macrobrachium rosenbergii). Aquaculture 2017, 476, 168–184. [Google Scholar] [CrossRef]
- Zokaeifar, H.; Balcazar, J.L.; Saad, C.R.; Kamarudin, M.S.; Sijam, K.; Arshad, A.; Nejat, N. Effects of Bacillus subtilis on the growth performance, digestive enzymes, immune gene expression and disease resistance of white shrimp, Litopenaeus vannamei. Fish Shellfish Immun. 2012, 33, 683–689. [Google Scholar] [CrossRef]
- Lai, A.G.; Aboobaker, A.Z. Comparative genomic analysis of innate immunity reveals novel and conserved components in crustacean food crop species. BMC Genom. 2017, 18, 389. [Google Scholar] [CrossRef]
- Qin, Z.; Babu, V.S.; Wan, Q.; Zhou, M.; Liang, R.; Muhammad, A.; Zhao, L.; Li, J.; Lan, J.; Lin, L. Transcriptome analysis of Pacific white shrimp (Litopenaeus vannamei) challenged by Vibrio parahaemolyticus reveals unique immune-related genes. Fish Shellfish Immun. 2018, 77, 164–174. [Google Scholar] [CrossRef]
- Zeynali, M.; Bahabadi, M.N.; Morshedi, V.; Ghasemi, A.; Mozanzadeh, M.T. Replacement of dietary fishmeal with Sargassum ilicifolium meal on growth, innate immunity and immune gene mRNA transcript abundance in Lates calcarifer juveniles. Aquac. Nutr. 2020, 26, 1657–1668. [Google Scholar] [CrossRef]
- Rosas, C.; Lopez, N.; Mercado, P.; Martinez, E. Effect of salinity acclimation on oxygen consumption of juvenile of the white shrimp Litopenaeus vannamei. J. Crust. Biol. 2001, 21, 912–922. [Google Scholar] [CrossRef][Green Version]
- Sang, H.M.; Fotedar, R. Growth, survival, haemolymph osmolality and organosomatic indices of the western king prawn (Penaeus latisulcatus) reared at different salinities. Aquaculture 2004, 234, 601–614. [Google Scholar] [CrossRef]
- Song, L.-Y.; Yu, C.-I.; Lien, T.-W.; Huang, C.-C.; Lin, M.-N. Haemolymph parameters of Pacific white shrimp (Litopenaeus vannamei) infected with Taura syndrome virus. Fish Shellfish. Immun. 2003, 14, 317–331. [Google Scholar] [CrossRef]
- Rodriguez, L.S.; Picones, A.; Rosete, G.C.; Islas, S.; Arechiga, H. Localization and release of 5-hydroxytryptamine in the crayfish eyestalk. J. Exp. Biol. 1997, 200, 3067–3077. [Google Scholar] [CrossRef]
- Rajendiran, S.; Iqbal, B.M.M.; Vasudevan, S. Induced thermal stress on serotonin levels in the blue swimmer crab, Portunus pelagicus. Biochem. Biophy. Rep. 2016, 5, 425–429. [Google Scholar] [CrossRef]
- Long, X.; Wu, X.; Zhao, L.; Ye, H.; Cheng, Y.; Zeng, C. Effects of salinity on gonadal development, osmoregulation and metabolism of adult male Chinese mitten crab, Eriocheir sinensis. PLoS ONE 2017, 12, e0179036. [Google Scholar] [CrossRef]
- Folch, J.; Lees, M.; Sloane-Stanley, G.H. A simple method for the isolation and purification of total lipids from animal tissues. J. Biol. Chem. 1975, 226, 497–509. [Google Scholar] [CrossRef]
- Huerlimann, R.; Wade, N.M.; Gordon, L.; Montenegro, J.D.; Goodall, J.; McWilliam, S.; Tinning, M.; Siemering, K.; Giardina, E.; Donovan, D.; et al. De novo assembly, characterization, functional annotation and expression patterns of the black tiger shrimp (Penaeus monodon) transcriptome. Sci. Rep. 2018, 8, 13553. [Google Scholar] [CrossRef]
- Sittikankaew, K.; Pootakham, W.; Sonthirod, C. Transcriptome analyses reveal the synergistic effects of feeding and eyestalk ablation on ovarian maturation in black tiger shrimp. Sci. Rep. 2020, 10, 3239. [Google Scholar] [CrossRef]
- Jeswin, J.; Thomas, A.A.P.C.; Paulton, M.P.; Vijayan, K.K. Survivability of Penaeus monodon during white spot syndrome virus infection and its correlation with immune related genes. Aquaculture 2013, 380, 84–90. [Google Scholar] [CrossRef]
- Soo, T.C.C.; Devadas, S.; Din, M.S.M.; Bhassu, S. Differential transcriptome analysis of the disease tolerant Madagascar–Malaysia crossbred black tiger shrimp, Penaeus monodon hepatopancreas in response to acute hepatopancreatic necrosis disease (AHPND) infection: Inference on immune gene response and interaction. Gut Pathog. 2019, 11, 39. [Google Scholar] [CrossRef]
- Aziz, D.; Rahi, M.L.; Hurwood, D.A.; Mather, P.B. Analysis of candidate gene expression patterns of adult male Macrobrachium rosenbergii morphotypes in response to a social dominance hierarchy. Hydrobiologia 2018, 825, 121–136. [Google Scholar] [CrossRef]
- Pfaffi, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Jolliffe, I.T.; Cadima, J. Principal component analysis: A review and recent developments. Philos. Trans. R. Soc. A 2016, 374, 20150202. [Google Scholar] [CrossRef]
- Pootakham, W.; Uengwetwanit, T.; Sonthirod, C.; Sittikankaew, K.; Karoonuthaisiri, N. A novel full-length transcriptome resource for black tiger shrimp (Penaeus monodon) developed using isoform sequencing (Iso-Seq). Front. Mar. Sci. 2020, 7, 172. [Google Scholar] [CrossRef]
- Uengwetwanit, T.; Ponza, P.; Sangsrakru, D.; Wichadakul, D.; Ingsriswang, S.; Leelatanawit, R.; Klinbunga, S.; Tangphatsornruang, S.; Karoonuthaisiri, N. Transcriptome-based discovery of pathways and genes related to reproduction of the black tiger shrimp (Penaeus monodon). Mar. Genom. 2017, 37, 69–73. [Google Scholar] [CrossRef]
- Faleiros, R.O.; Furriel, R.P.M.; McNamara, J.C. Transcriptional, translational and systemic alterations during the time course of osmoregulatory acclimation in two palaemonid shrimps from distinct osmotic niches. Comp. Biochem. Physiol. 2017, 212, 97–106. [Google Scholar] [CrossRef]
- Freire, C.A.; Souza, L.R.; Amado, E.M.; Prodocimo, V.; Souza, M.M. Regulation of muscle hydration upon hypo-or hyper-osmotic shocks: Differences related to invasion of the freshwater habitat by decapod crustaceans. J. Exp. Zool. 2013, 319, 297–309. [Google Scholar] [CrossRef]
- Loughland, I.; Seebacher, F. Differences in oxidative status explain variation in thermal acclimation capacity between individual mosquito-fish (Gambusia holbrooki). Funct. Ecol. 2020, 34, 1380–1390. [Google Scholar] [CrossRef]
- Rahi, M.L.; Mahmud, S.; Dilruba, K.J.; Sabbir, W.; Aziz, D.; Hurwood, D.A. Temperature induced changes in physiological traits and expression of selected candidate genes in black tiger shrimp (Penaeus monodon) larvae. Aquac. Rep. 2021, 19, 100620. [Google Scholar] [CrossRef]
- Betancur-R., R.; Orti, G.; Stein, A.M.; Marceniuk, A.P.; Pyron, R.A. Apparent signal of competition limiting diversification after ecological transitions from marine to freshwater habitats. Ecol. Lett. 2012, 15, 822–830. [Google Scholar] [CrossRef]
- Henry, R.P. Environmentally mediated carbonic anhydrase induction in the gills of euryhaline crustaceans. J. Exp. Biol. 2001, 204, 991–1002. [Google Scholar] [CrossRef]
- Joseph, A.; Philip, R. Acute salinity stress alters the haemolymph metabolic profile of Penaeus monodon and reduces immunocompetence to white spot syndrome virus infection. Aquaculture 2007, 272, 87–97. [Google Scholar] [CrossRef]
- Ali, M.Y.; Sabbir, W.; Rahi, M.L.; Chowdhury, M.M.R.; Faruque, M.O. Quality changes in shrimp (Penaeus monodon) stored at ambient temperature in plastic and bamboo basket. Int. J. Anim. Fish. Sci. 2008, 1, 7–13. [Google Scholar]
- Islam, S.S.; Shah, M.S.; Rahi, M.L. Assessment of genetic variability of prawn (Macrobrachium rosenbergii) post larvae (PL) from the broods stocked under different sex ratios. Int. J. Aquac. 2014, 4, 55–63. [Google Scholar] [CrossRef]
- Ali, M.R.; Rahi, M.L.; Islam, S.S.; Shah, M.S.; Shams, F.I. Genetic Variability Assay of Different Strains of Catla catla. Int. J. Life Sci. 2015, 9, 37–42. [Google Scholar] [CrossRef]
- Chang, E.S.; Chang, S.A.; Beltz, B.A.; Kravitz, E.A. Crustacean hyperglycemic hormone in the lobster nervous system: Localization and release from cells in the sub-oesophageal ganglion and thoracic second roots. J. Comp. Neurol. 1999, 414, 50–56. [Google Scholar] [CrossRef]
- Faleiros, R.O.; Goldman, M.H.; Furriel, R.P.; McNamara, J.C. Differential adjustments in gill Na+/K+- and V-ATPase activities and transporter mRNA expression during osmoregulatory acclimation in the cinnamon shrimp Macrobrachium amazonicum (Decapoda, Palaemonidae). J. Exp. Biol. 2010, 213, 3894–3905. [Google Scholar] [CrossRef]
- Havird, J.C.; Meyer, E.; Fujita, Y.; Vaught, R.C.; Henry, R.P.; Santos, S.R. Disparate responses to salinity across species and organizational levels in anchialine shrimps. J. Exp. Biol. 2019, 222, jeb211920. [Google Scholar] [CrossRef]
- Boudour-Boucheker, N.; Boulo, V.; Charmantier-Daures, M.; Anger, K.; Charmantier, G.; Lorin-Nebel, C. Osmoregulation in larvae and juveniles of two recently separated Macrobrachium species: Expression patterns of ion transporter genes. Comp. Biochem. Physiol. 2016, 195, 39–45. [Google Scholar] [CrossRef]
- Charmantier, G.; Charmantier-Daures, M.; Towle, D. Osmotic and ionic regulations in aquatic arthropods. In Osmotic and Ionic Regulation: Cells and Animals; Evans, D.H., Ed.; CRC Press: Boca Raton, FL, USA, 2009; pp. 165–230. [Google Scholar]
- Coast, G.M.; Webster, S.G.; Schegg, K.M.; Tobe, S.S.; Schooley, D.A. The Drosophila melanogaster homologue of an insect calcitonin-like diuretic peptide stimulates V-ATPase activity in fruit fly Malpighian tubules. J. Exp. Biol. 2001, 204, 1795–1804. [Google Scholar] [CrossRef]
- Fink, I.R.; Pietretti, D.; Voogdt, C.G.P.; Westphal, A.H.; Savelkoul, H.F.J.; Forlenza, M.; Wiegertjes, G.F. Molecular and functional characterization of Toll-like receptor (Tlr) 1 and Tlr2 in common carp (Cyprinus carpio). Fish Shellfish Immunol. 2016, 56, 70–83. [Google Scholar] [CrossRef]
- Rahi, M.L.; Sabbir, W.; Banerjee, P.; Sultana, I. Effect of delayed icing on the quality of tiger shrimp (Penaeus monodom Fab.). Bang. J. Fish. Res. 2008, 12, 225–234. [Google Scholar]
- Rahi, M.L.; Shah, M.S. Triploidization in rohu x mrigal hybrid and comparison of growth performance of triploid hybrid. Aquac. Res. 2012, 43, 1867–1879. [Google Scholar] [CrossRef]



| Gene with ID | Primer Sequence (5′-3′) | Size | Annealing Temperature | Reference |
|---|---|---|---|---|
| α-Amylase | F: TGGACTCGACAACTAGC | 118 | 59 °C | [55] |
| R: TAGATGCACACTCGCTC | ||||
| Crustacean Hyperglycemic Hormone (CHH) | F: TTCTGCAGTCTCTCCAACGA | 120 | 59 °C | [56] |
| R: GCGACGTATGGTTTCCTGTA | ||||
| Diuretic Hormone (DH) | F: ACAACTCGGCTCTGGTGTTC | 110 | 60 °C | [48] |
| R: CTCGGCCTAGACCCAAGTC | ||||
| Na+/H+ Exchanger (NHE) | F: TTGGAGGAGGGGTCTACCTT | 123 | 59 °C | [48] |
| R: GCTCTCTCCAAAGACAAGCA | ||||
| Na+/K+-ATPase (NKA) | F: CACCCCACCCAAACAAACT | 120 | 60 °C | [55] |
| R: TCGTGAACTCTTGCTTTCTTGA | ||||
| Na+/K+/2Cl− Co-transporter (NKCC) | F: GGGTCACCAGGGTCCAGAT | 115 | 56 °C | [49] |
| R: TAGCACCAGCAACAATTCCA | ||||
| V-type H+-ATPase (VTA) | F: GTCGATTTTCCGAAGCGAAT | 105 | 61 °C | [55] |
| R: GCCTTCCAAGTAGCGAAGC | ||||
| Toll Like Receptor (TLR) | F: CTT AGC CTT GGA GAC AAC | 118 | 53 °C | [26] |
| R: GAT GCT TAA CAG CTC CTC | ||||
| Elongation factor 1-alpha (EF1α) | F: TTC CGA CTC CAA GAA CGA CC | 122 | 60 °C | [49] |
| R: GAG CAG TGT GGC AAT CAA GC |
| 0‰ | 2.5‰ | 5‰ | 10‰ | 20‰ | 30‰ | |
|---|---|---|---|---|---|---|
| Initial Weight (BWi) (g) | 0.21 a ± 0.04 | 0.21 a ± 0.03 | 0.20 a ± 0.05 | 0.20 a ± 0.04 | 0.20 a ± 0.05 | 0.21 a ± 0.03 |
| Final Weight (BWf) (g) | 1.35 a ± 0.24 | 1.59 b ± 0.36 | 1.73 b ± 0.46 | 2.07 c ± 0.35 | 2.35 d ± 0.39 | 1.98 c ± 0.34 |
| DWG (%) | 8.56 ± 0.66 | 10.95 ± 0.86 | 12.06 ± 0.91 | 15.58 ± 0.92 | 17.92 ± 0.76 | 14.05 ± 0.79 |
| SGR (%) | 3.1 ± 0.14 | 3.37 ± 0.26 | 3.6 ± 0.16 | 3.89 ± 0.27 | 4.11 ± 0.36 | 3.74 ± 0.25 |
| Feed Intake (g g−1 day−1) | 0.162 ± 0.02 | 0.161 ± 0.03 | 0.158 ± 0.02 | 0.158 ± 0.04 | 0.157 ± 0.01 | 0.156 ± 0.02 |
| FCR | 1.89 | 1.44 | 1.31 | 1.04 | 0.89 | 1.11 |
| Survival (%) | 61 a | 64 a | 71 b | 76 b | 86 c | 83 c |
| Amino Acids | Salinity | |||||
|---|---|---|---|---|---|---|
| 0‰ | 2.5‰ | 5‰ | 10‰ | 20‰ | 30‰ | |
| Alanine * | 586.3 ± 41.2 | 561.6 ± 39.6 | 537.1 ± 40.2 | 496.5 ± 38.4 | 406.7 ± 43.8 | 453.6 ± 33.4 |
| β-Alanine | 13.8 ± 1.1 | 13.1 ± 1.2 | 11.2 ± 1.1 | 10.6 ± 0.8 | 8.76 ± 0.7 | 9.66 ± 0.9 |
| Arginine ** | 307.5 ± 3.5 | 324.8 ± 3.7 | 353.7 ± 4.6 | 388.3 ± 4.9 | 419.8 ± 5.8 | 428.6 ± 6.2 |
| Asparagine | 41.9 ± 2.8 | 43.7 ± 3.2 | 46.6 ± 3.5 | 49.5 ± 4.1 | 56.2 ± 4.3 | 54.6 ± 3.9 |
| Aspartic acid | 26.4 ± 3.8 | 26.1 ± 1.7 | 25.4 ± 2.9 | 24.6 ± 3.1 | 23.6 ± 3.6 | 24.1 ± 2.15 |
| α-AAA | 15.1 ± 2.4 | 16.3 ± 1.7 | 18.4 ± 1.9 | 20.7 ± 2.1 | 22.6 ± 1.9 | 21.9 ± 1.6 |
| α-ABA | 15.4 ± 1.1 | 10.1 ± 0.9 | 8.6 ± 0.7 | 6.6 ± 0.8 | 4.9 ± 0.4 | 5.3 ± 0.6 |
| Citrulline | 10.4 ± 0.6 | 12.6 ± 0.5 | 13.2 ± 0.6 | 13.9 ± 0.7 | 15.4 ± 0.3 | 14.8 ± 0.4 |
| Cystathionine | 11.5 ± 2.1 | 8.93 ± 1.4 | 6.98 ± 0.8 | 5.28 ± 0.6 | 3.97 ± 0.4 | 4.57 ± 0.5 |
| Cystine | 4.73 ± 0.4 | 4.96 ± 0.5 | 5.14 ± 0.3 | 6.11 ± 0.5 | 6.71 ± 0.4 | 6.02 ± 0.7 |
| Glutamic acid | 62.9 ± 4.4 | 60.2 ± 4.6 | 58.9 ± 3.9 | 56.7 ± 3.3 | 53.6 ± 3.4 | 55.4 ± 3.6 |
| Glutamine | 196.8 ± 10.4 | 206.3 ± 31.6 | 216.5 ± 26.2 | 226.4 ± 33.4 | 242.8 ± 39.1 | 230.4 ± 38.4 |
| Glycine * | 396.5 ± 7.6 | 358.6 ± 9.5 | 329.6 ± 8.6 | 356.7 ± 9.8 | 379.6 ± 6.9 | 391.5 ± 9.9 |
| Histidine ** | 44.5 ± 2.9 | 38.4 ± 2.2 | 39.4 ± 1.8 | 37.3 ± 2.1 | 33.6 ± 1.9 | 34.8 ± 1.7 |
| Isoleucine ** | 26.4 ± 1.1 | 24.1 ± 1.2 | 22.9 ± 1.5 | 22.3 ± 1.7 | 20.3 ± 1.4 | 21.2 ± 1.3 |
| Leucine ** | 38.4 ± 1.9 | 35.7 ± 2.4 | 34.2 ± 1.7 | 31.8 ± 1.4 | 29.7 ± 2.2 | 30.6 ± 1.6 |
| Lysine ** | 41.5 ± 2.7 | 47.5 ± 3.1 | 51.6 ± 2.8 | 56.2 ± 3.9 | 63.9 ± 4.3 | 63.4 ± 3.8 |
| Methionine ** | 3.11 ± 0.5 | 4.38 ± 0.4 | 4.68 ± 0.5 | 5.01 ± 0.6 | 5.91 ± 0.2 | 6.06 ± 0.7 |
| Ornithine | 39.1 ± 3.4 | 38.4 ± 4.1 | 35.6 ± 4.1 | 31.7 ± 3.6 | 26.38 ± 4.4 | 22.41 ± 2.9 |
| Phenylalanine ** | 19.9 ± 1.2 | 21.3 ± 1.1 | 24.7 ± 1.7 | 25.7 ± 1.4 | 30.6 ± 1.5 | 29.9 ± 1.8 |
| Proline * | 1086 ± 56.7 | 1068 ± 54.1 | 1026 ± 43.9 | 943.5 ± 50.9 | 930.6 ± 43.8 | 923.5 ± 60.8 |
| Serine | 55.3 ± 4.1 | 63.2 ± 5.6 | 72.4 ± 5.9 | 79.6 ± 6.2 | 87.8 ± 3.9 | 86.1 ± 5.4 |
| Taurine | 506.8 ± 23.7 | 486.7 ± 33.2 | 463.2 ± 32.6 | 408.5 ± 41.7 | 384.1 ± 10.4 | 390.4 ± 39.4 |
| Threonine ** | 47.2 ± 3.9 | 47.6 ± 3.6 | 51.6 ± 3.9 | 66.4 ± 4.1 | 86.3 ± 6.5 | 87.1 ± 5.2 |
| Tryptophan ** | 3.92 ± 0.8 | 4.02 ± 0.7 | 4.58 ± 1.2 | 5.09 ± 0.9 | 6.13 ± 0.8 | 6.27 ± 1.1 |
| Tyrosine | 3.98 ± 0.6 | 4.2 ± 0.4 | 4.65 ± 0.7 | 5.1 ± 0.6 | 6.2 ± 0.7 | 5.6 ± 0.5 |
| Valine ** | 31.2 ± 4.26 | 34.4 ± 3.9 | 41.7 ± 4.25 | 47.8 ± 4.38 | 52.3 ± 4.18 | 51.6 ± 4.39 |
| ∑EAAs | 563.63 a ± 5.3 | 582.2 a ± 5.6 | 629.1 b ± 8.6 | 685.9 c ± 9.3 | 748.5 d ± 9.6 | 759.5 d ± 6.9 |
| TFAAs | 3181.9 a ± 61 | 3605.2 b ± 60 | 3612.6 b ± 62 | 3629.3 b ± 76 | 3708.6 c ± 71 | 3669.8 bc ± 66 |
| Fatty Acids | Salinity | |||||
|---|---|---|---|---|---|---|
| 0‰ | 2.5‰ | 5‰ | 10‰ | 20‰ | 30‰ | |
| C14:0 | 0.46 ± 0.05 | 0.41 ± 0.04 | 0.38 ± 0.04 | 0.37 ± 0.03 | 0.30 ± 0.02 | 0.35 ± 0.03 |
| C15:0 | 0.42 ± 0.06 | 0.37 ± 0.05 | 0.31 ± 0.05 | 0.29 ± 0.04 | 0.27 ± 0.03 | 0.28 ± 0.02 |
| C16:0 | 15.54 ± 0.79 | 14.86 ± 0.76 | 14.66 ± 0.81 | 14.34 ± 0.61 | 13.96 ± 0.62 | 14.26 ± 0.59 |
| C17:0 | 0.53 ± 0.04 | 0.47 ± 0.03 | 0.46 ± 0.05 | 0.46 ± 0.04 | 0.44 ± 0.03 | 0.45 ± 0.03 |
| C18:0 | 7.95 ± 0.08 | 7.81 ± 0.06 | 7.75 ± 0.06 | 7.56 ± 0.08 | 6.86 ± 0.06 | 7.66 ± 0.07 |
| C20:0 | 0.98 ± 0.04 | 0.93 ± 0.03 | 0.89 ± 0.02 | 0.88 ± 0.04 | 0.76 ± 0.03 | 0.86 ± 0.02 |
| C22:0 | 0.96 ± 0.06 | 0.86 ± 0.05 | 0.78 ± 0.04 | 0.77 ± 0.03 | 0.74 ± 0.05 | 0.71 ± 0.04 |
| ∑SFAs | 26.86 a ± 0.66 | 25.71 ab ± 0.69 | 25.23 b ± 0.64 | 24.67 bc ± 0.61 | 23.33 c ± 0.56 | 24.57 bc ± 0.65 |
| C16:1n7 | 3.46 ± 0.05 | 3.26 ± 0.06 | 3.11 ± 0.09 | 2.93 ± 0.07 | 2.76 ± 0.06 | 2.89 ± 0.08 |
| C18:1n9 | 19.83 ± 0.18 | 19.46 ± 0.19 | 19.31 ± 0.21 | 19.06 ± 0.16 | 18.61 ± 0.11 | 18.99 ± 0.13 |
| C18:1n7 | 3.78 ± 0.09 | 3.62 ± 0.06 | 3.49 ± 0.07 | 3.36 ± 0.05 | 2.98 ± 0.02 | 3.18 ± 0.06 |
| C20:1n7 | 1.04 ± 0.04 | 0.99 ± 0.05 | 0.96 ± 0.03 | 0.93 ± 0.04 | 0.83 ± 0.01 | 0.89 ± 0.02 |
| ∑MUFAs | 28.11 a ± 0.32 | 27.33 ab ± 0.31 | 26.87 ab ± 0.33 | 26.26 bc ± 0.29 | 25.18 c ± 0.22 | 25.95 bc ± 0.28 |
| C18:2n6 | 7.86 ± 0.14 | 7.69 ± 0.09 | 7.66 ± 0.13 | 7.44 ± 0.11 | 6.96 ± 0.06 | 7.33 ± 0.09 |
| C18:3n3 | 0.87 ± 0.05 | 0.82 ± 0.06 | 0.74 ± 0.02 | 0.71 ± 0.04 | 0.59 ± 0.03 | 0.64 ± 0.01 |
| C20:2n6 | 2.45 ± 0.05 | 2.41 ± 0.07 | 2.39 ± 0.10 | 2.34 ± 0.06 | 2.26 ± 0.02 | 2.29 ± 0.08 |
| C20:3n6 | 0.28 ± 0.05 | 0.26 ± 0.03 | 0.25 ± 0.04 | 0.22 ± 0.02 | 0.19 ± 0.01 | 0.20 ± 0.03 |
| C20:4n6 | 17.03 ± 0.21 | 16.96 ± 0.23 | 16.76 ± 0.14 | 16.50 ± 0.09 | 16.02 ± 0.15 | 16.46 ± 0.17 |
| C20:5n3 | 10.42 ± 0.23 | 10.34 ± 0.27 | 10.23 ± 0.21 | 9.97 ± 0.16 | 9.68 ± 0.18 | 9.86 ± 0.12 |
| C22:6n3 | 5.41 ± 0.08 | 5.46 ± 0.07 | 5.56 ± 0.09 | 5.76 ± 0.08 | 5.96 ± 0.04 | 6.35 ± 0.22 |
| ∑PUFAs | 44.32 a ± 0.17 | 43.94 a ± 0.24 | 43.59 ab ± 0.28 | 42.94 b ± 0.26 | 41.66 c ± 0.12 | 43.12 b ± 0.24 |
| ∑n-3PUFAs | 15.56 a ± 0.12 | 15.36 a ± 0.13 | 15.24 ab ± 0.14 | 15.06 ab ± 0.10 | 14.35 b ± 0.11 | 14.98 ab ± 0.09 |
| ∑n-6PUFAs | 24.52 a ± 0.10 | 24.31 a ± 0.11 | 24.16 ab ± 0.13 | 23.96 ab ± 0.09 | 23.32 b ± 0.07 | 23.76 ab ± 0.08 |
| n-3/n-6 | 0.64 ± 0.03 | 0.63 ± 0.02 | 0.63 ± 0.02 | 0.62 ± 0.04 | 0.61 ± 0.01 | 0.62 ± 0.03 |
| ∑HUFAs | 35.06 a ± 0.25 | 34.76 ab ± 0.37 | 34.56 ab ± 0.34 | 34.11 b ± 0.27 | 33.60 b ± 0.32 | 34.06 b ± 0.29 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rahi, M.L.; Azad, K.N.; Tabassum, M.; Irin, H.H.; Hossain, K.S.; Aziz, D.; Moshtaghi, A.; Hurwood, D.A. Effects of Salinity on Physiological, Biochemical and Gene Expression Parameters of Black Tiger Shrimp (Penaeus monodon): Potential for Farming in Low-Salinity Environments. Biology 2021, 10, 1220. https://doi.org/10.3390/biology10121220
Rahi ML, Azad KN, Tabassum M, Irin HH, Hossain KS, Aziz D, Moshtaghi A, Hurwood DA. Effects of Salinity on Physiological, Biochemical and Gene Expression Parameters of Black Tiger Shrimp (Penaeus monodon): Potential for Farming in Low-Salinity Environments. Biology. 2021; 10(12):1220. https://doi.org/10.3390/biology10121220
Chicago/Turabian StyleRahi, Md. Lifat, Khairun Naher Azad, Maliha Tabassum, Hasna Hena Irin, Kazi Sabbir Hossain, Dania Aziz, Azam Moshtaghi, and David A Hurwood. 2021. "Effects of Salinity on Physiological, Biochemical and Gene Expression Parameters of Black Tiger Shrimp (Penaeus monodon): Potential for Farming in Low-Salinity Environments" Biology 10, no. 12: 1220. https://doi.org/10.3390/biology10121220
APA StyleRahi, M. L., Azad, K. N., Tabassum, M., Irin, H. H., Hossain, K. S., Aziz, D., Moshtaghi, A., & Hurwood, D. A. (2021). Effects of Salinity on Physiological, Biochemical and Gene Expression Parameters of Black Tiger Shrimp (Penaeus monodon): Potential for Farming in Low-Salinity Environments. Biology, 10(12), 1220. https://doi.org/10.3390/biology10121220






