3D Liquid Marble Microbioreactors Support In Vitro Maturation of Prepubertal Ovine Oocytes and Affect Expression of Oocyte-Specific Factors
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Approval
2.2. Source of Oocytes and In Vitro Maturation
2.2.1. Control Group (4W)
2.2.2. Liquid Marble Group (LM)
2.3. In Vitro Fertilization
2.4. In Vitro Embryo Development
2.5. Gene Expression Analysis
2.6. Sample Collection for Gene Expression Analysis
2.7. RNA Isolation and Reverse Transcription
2.8. Real-Time Polymerase Chain Reaction
2.9. Statistical Analysis
3. Results
3.1. In Vitro Maturation and Development
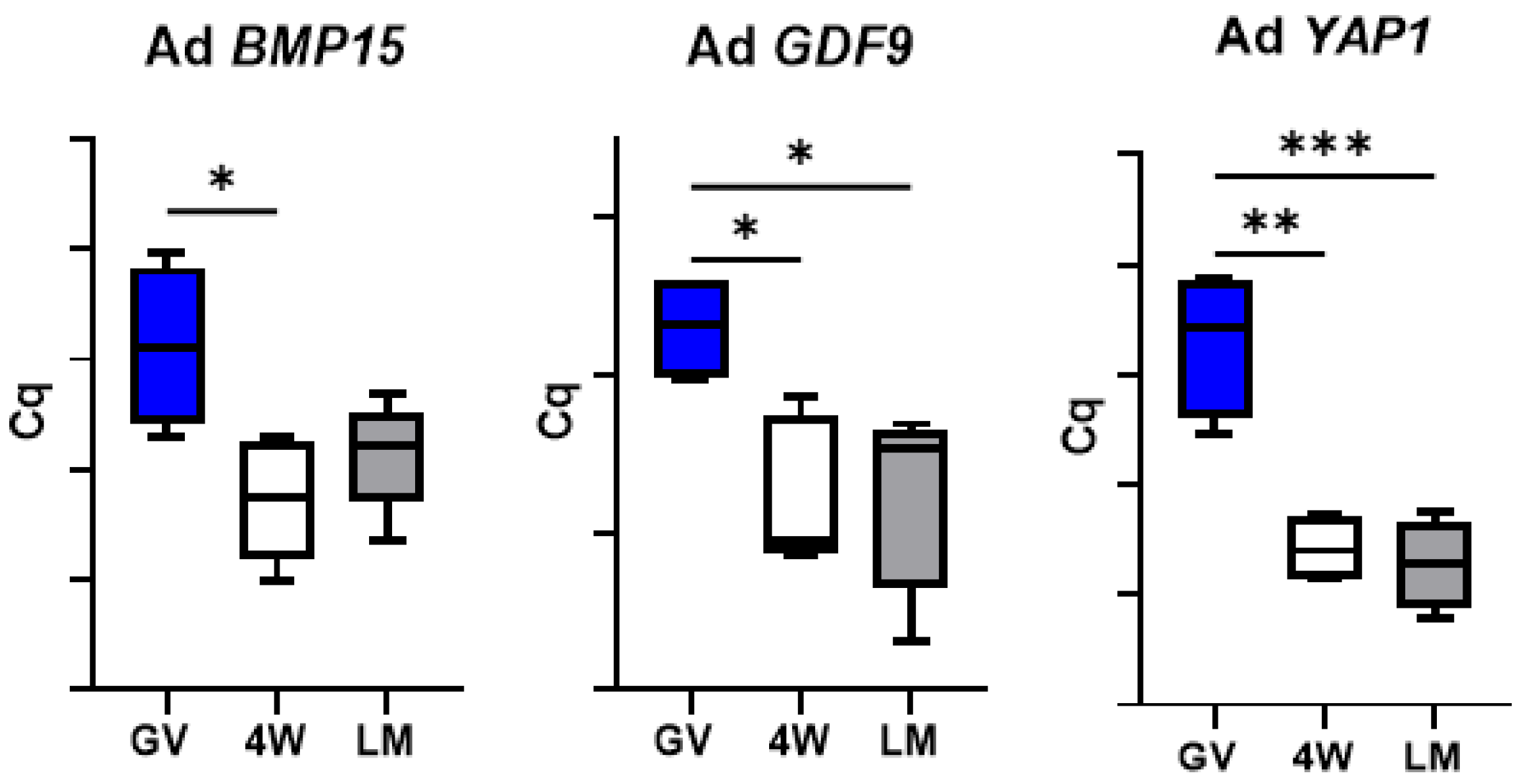
3.2. Gene Expression Analysis
3.3. Expression of the SCMC Components
3.4. Expression of Genes Involved in Cell Stress Response
3.5. Expression of Genes Encoding Oocyte-Secreted Factors (OSF)
3.6. Expression of Enzymes Involved in DNA Methylation Reprogramming
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sánchez, F.; Lolicato, F.; Romero, S.; De Vos, M.; Van Ranst, H.; Verheyen, G.; Anckaert, E.; Smitz, J.E.J. An improved IVM method for cumulus-oocyte complexes from small follicles in polycystic ovary syndrome patients enhances oocyte competence and embryo yield. Hum. Reprod. 2017, 32, 2056–2068. [Google Scholar] [CrossRef]
- De Vos, M.; Grynberg, M.; Ho, T.M.; Yuan, Y.; Albertini, D.F.; Gilchrist, R.B. Perspectives on the development and future of oocyte IVM in clinical practice. J. Assist. Reprod. Genet. 2021, 38, 1265–1280. [Google Scholar] [CrossRef] [PubMed]
- Gilchrist, R.B.; Thompson, J.G. Oocyte maturation: Emerging concepts and technologies to improve developmental potential in vitro. Theriogenology 2007, 67, 6–15. [Google Scholar] [CrossRef]
- Farsi, M.M.; Kamali, N.; Pourghasem, M. Embryological aspects of oocyte in vitro maturation. Int. J. Mol. Cell. Med. 2013, 2, 99–109. [Google Scholar] [PubMed]
- Ledda, S.; Idda, A.; Kelly, J.; Ariu, F.; Bogliolo, L.; Bebbere, D. A novel technique for in vitro maturation of sheep oocytes in a liquid marble microbioreactor. J. Assist. Reprod. Genet. 2016, 33, 513–518. [Google Scholar] [CrossRef][Green Version]
- Uhm, S.J.; Gupta, M.K.; Yang, J.H.; Chung, H.J.; Min, T.S.; Lee, H.T. Epidermal growth factor can be used in lieu of follicle-stimulating hormone for nuclear maturation of porcine oocytes in vitro. Theriogenology 2010, 73, 1024–1036. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.H.; Hwang, J.L.; Seow, K.M.; Huang, L.W.; Chen, H.J.; Tzeng, C.R. Effects of growth factors and granulosa cell co-culture on in-vitro maturation of oocytes. Reprod. BioMed. Online 2009, 19, 165–170. [Google Scholar] [CrossRef]
- You, J.; Lee, E.; Bonilla, L.; Francis, J.; Koh, J.; Block, J.; Chen, S.; Hansen, P.J. Treatment with the proteasome inhibitor MG132 during the end of oocyte maturation improves oocyte competence for development after fertilization in cattle. PLoS ONE 2012, 7, e48613. [Google Scholar] [CrossRef]
- Donnay, I.; Faerge, I.; Grøndahl, C.; Verhaeghe, B.; Sayoud, H.; Ponderato, N.; Galli, C.; Lazzari, G. Effect of prematuration, meiosis activating sterol and enriched maturation medium on the nuclear maturation and competence to development of calf oocytes. Theriogenology 2004, 62, 1093–1107. [Google Scholar] [CrossRef] [PubMed]
- Antoni, D.; Burckel, H.; Josset, E.; Noel, G. Three-dimensional cell culture: A breakthrough in vivo. Int. J. Mol. Sci. 2015, 16, 5517–5527. [Google Scholar] [CrossRef]
- Mastrorocco, A.; Cacopardo, L.; Martino, N.A.; Fanelli, D.; Camillo, F.; Ciani, E.; Roelen, B.A.J.; Ahluwalia, A.; Dell’Aquila, M.E. One-step automated bioprinting-based method for cumulus-oocyte complex microencapsulation for 3D in vitro maturation. PLoS ONE 2020, 15, e0238812. [Google Scholar] [CrossRef]
- Park, J.E.; Kim, M.S.; Lee, E.; Lee, S.T. In vitro maturation using an agarose matrix with incorporated extracellular matrix proteins improves porcine oocyte developmental competence by enhancing cytoplasmic maturation. J. Tissue Eng. Regen. Med. 2021, 15, 807–817. [Google Scholar] [CrossRef]
- Shen, P.; Xu, J.; Wang, P.; Zhao, X.; Huang, B.; Wu, F.; Wang, L.; Chen, W.; Feng, Y.; Guo, Z.; et al. A new three-dimensional glass scaffold increases the in vitro maturation efficiency of buffalo (Bubalus bubalis) oocyte via remodelling the extracellular matrix and cell connection of cumulus cells. Reprod. Domest. Anim. 2020, 55, 170–180. [Google Scholar] [CrossRef]
- Ledda, S.; Bebbere, D.; Ariu, F.; Pescatori, M.; Pau, S.; Zedda, M.T.; Bogliolo, L. Unveiling mRNA changes during meiotic progression and pre-implantation development: Help from large animal models. Curr. Pharm. Des. 2012, 18, 256–263. [Google Scholar] [CrossRef] [PubMed]
- Bebbere, D.; Bogliolo, L.; Ariu, F.; Fois, S.; Leoni, G.G.; Tore, S.; Succu, S.; Berlinguer, F.; Naitana, S.; Ledda, S. Expression pattern of zygote arrest 1 (ZAR1), maternal antigen that embryo requires (MATER), growth differentiation factor 9 (GDF9) and bone morphogenetic protein 15 (BMP15) genes in ovine oocytes and in vitro-produced preimplantation embryos. Reprod. Fertil. Dev. 2008, 20, 908–915. [Google Scholar] [CrossRef]
- Tervit, H.R.; Whittingham, D.G.; Rowson, L.E. Successful culture in vitro of sheep and cattle ova. J. Reprod. Fertil. 1972, 30, 493–497. [Google Scholar] [CrossRef]
- Walker, S.K.; Hill, J.L.; Kleemann, D.O.; Nancarrow, C.D. Development of ovine embryos in synthetic oviductal fluid containing amino acids at oviductal fluid concentrations. Biol. Reprod. 1996, 55, 703–708. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Taylor, S.C.; Nadeau, K.; Abbasi, M.; Lachance, C.; Nguyen, M.; Fenrich, J. The ultimate qPCR experiment: Producing publication quality, reproducible data the first time. Trends Biotechnol. 2019, 37, 761–774. [Google Scholar] [CrossRef]
- Ohsugi, M.; Zheng, P.; Baibakov, B.; Li, L.; Dean, J. Maternally derived FILIAMATER complex localizes asymmetrically in cleavage-stage mouse embryos. Development 2008, 135, 259–269. [Google Scholar] [CrossRef]
- Evsikov, A.V.; Marín de Evsikova, C. Gene expression during the oocyte-to embryo transition in mammals. Mol. Reprod. Dev. 2009, 76, 805–818. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.Q.; Sugiura, K.; Woo, Y.; Wigglesworth, K.; Kamdar, S.; Affourtit, J.; Eppig, J.J. Selective degradation of transcripts during meiotic maturation of mouse oocytes. Dev. Biol. 2007, 302, 104–117. [Google Scholar] [CrossRef] [PubMed]
- Bebbere, D.; Masala, L.; Albertini, D.F.; Ledda, S. The subcortical maternal complex: Multiple functions for one biological structure? J. Assist. Reprod. Genet. 2016, 33, 1431–1438. [Google Scholar] [CrossRef] [PubMed]
- Bebbere, D.; Albertini, D.F.; Coticchio, G.; Borini, A.; Ledda, S. The subcortical maternal complex: Emerging roles and novel perspectives. Mol. Hum. Reprod. 2021, 27, gaab043. [Google Scholar] [CrossRef]
- Gilchrist, R.B.; Lane, M.; Thompson, J.G. Oocyte-secreted factors: Regulators of cumulus cell function and oocyte quality. Hum. Reprod. Update 2008, 14, 159–177. [Google Scholar] [CrossRef]
- Haase, M.; Fitze, G. HSP90AB1: Helping the good and the bad. Gene 2016, 575 Pt 1, 171–186. [Google Scholar] [CrossRef]
- Ito, S.; D’Alessio, A.C.; Taranova, O.V.; Hong, K.; Sowers, L.C.; Zhang, Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 2010, 466, 1129–1133. [Google Scholar] [CrossRef]
- Okano, M.; Bell, D.W.; Haber, D.A.; Li, E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 1999, 99, 247–257. [Google Scholar] [CrossRef]
- Morselli, M.G.; Luvoni, G.C.; Comizzoli, P. The nuclear and developmental competence of cumulus-oocyte complexes is enhanced by three-dimensional coculture with conspecific denuded oocytes during in vitro maturation in the domestic cat model. Reprod. Domest. Anim. 2017, 52 (Suppl. 2), 82–87. [Google Scholar] [CrossRef]
- Eppig, J.J.; Schroeder, A.C. Capacity of mouse oocytes from preantral follicles to undergo embryogenesis and development to live young after growth, maturation, and fertilization in vitro. Biol. Reprod. 1989, 41, 268–276. [Google Scholar] [CrossRef]
- Ma, J.; Flemr, M.; Strnad, H.; Svoboda, P.; Schultz, R.M. Maternally recruited DCP1A and DCP2 contribute to messenger RNA degradation during oocyte maturation and genome activation in mouse. Biol. Reprod. 2013, 88, 11. [Google Scholar] [CrossRef] [PubMed]
- Ivanova, I.; Much, C.; Di Giacomo, M.; Azzi, C.; Morgan, M.; Moreira, P.N.; Monahan, J.; Carrieri, C.; Enright, A.J.; O’Carroll, D. The RNA m(6)A Reader YTHDF2 Is Essential for the Post-transcriptional Regulation of the Maternal Transcriptome and Oocyte Competence. Mol. Cell 2017, 67, 1059–1067.e4. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zheng, P.; Dean, J. Maternal control of early mouse development. Development 2010, 137, 859–870. [Google Scholar] [CrossRef] [PubMed]
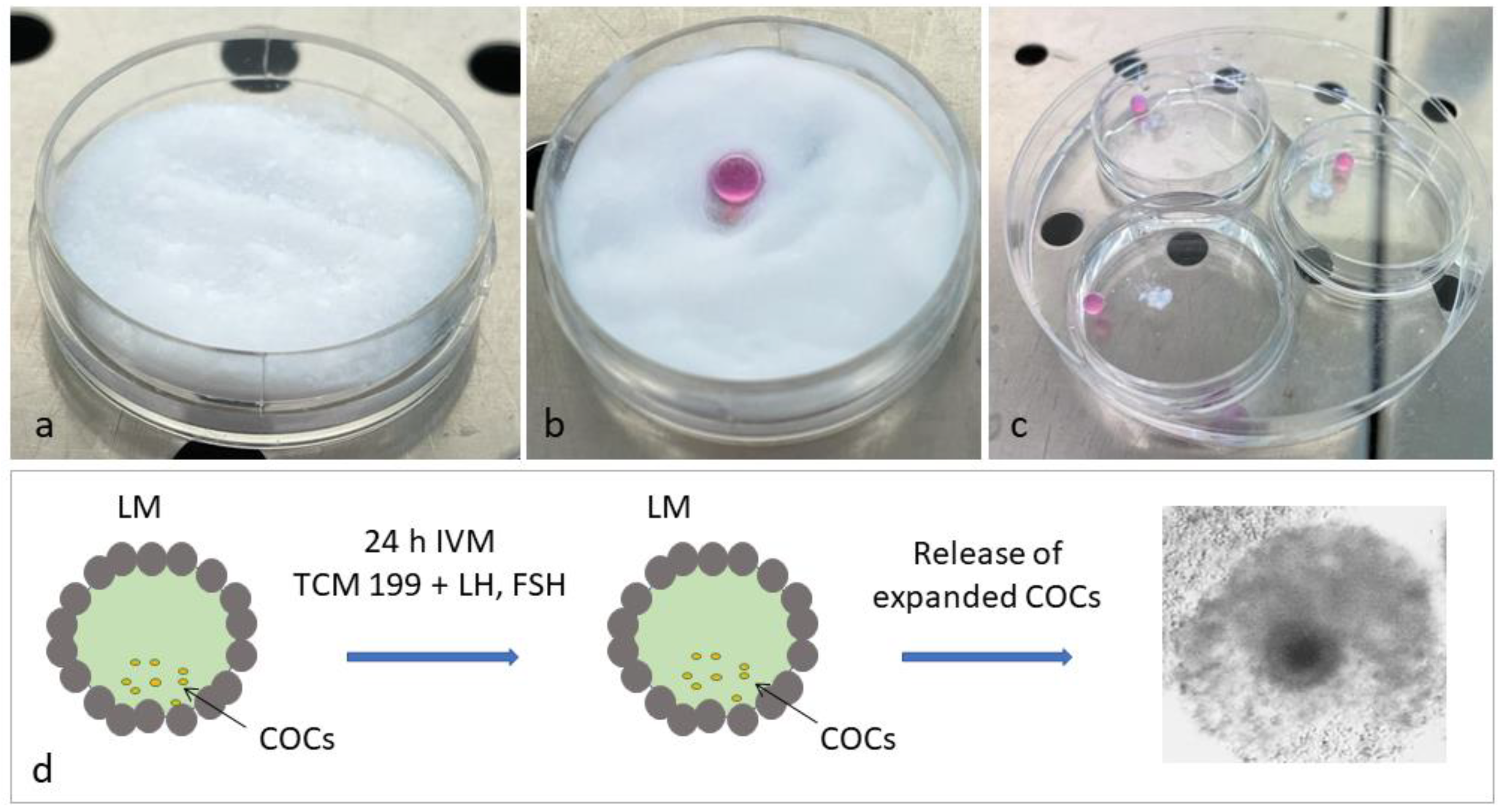







| GV | LM—MII | 4W—MII | |
|---|---|---|---|
| Adult | 4 | 5 | 5 |
| Prepubertal | 5 | 6 | 5 |
| Symbol | Gene Name | Accession Number | Primer Sequence | Ta | bps |
|---|---|---|---|---|---|
| BAX | BCL2 associated X protein | XM_004015363 | 5′ ctccccgagaggtctttttc 3′ 5′ tcgaaggaagtccaatgtcc 3′ | 58 °C | 176 |
| BMP15 | Bone morphogenetic protein 15 | NM_001114767 | 5′ gggttctacgactccgcttc 3′ 5′ ggttactttcaggcccatcat 3′ | 59 °C | 173 |
| DNMT1 | DNA methylation transferase 1 | NM_001009473 | 5′ cagctctcgtacatccacag 3′ 5′ aatctcgcgtagtcttggtc 3′ | 60 °C | 158 |
| DNMT3A | DNA methylation transferase 3A | XM_015094252 | 5′ gtgatgattgatgccaaaga 3′ 5′ ggtcctcactttgctgaact 3′ | 60 °C | 165 |
| DNMT3B | DNA methylation transferase 3B | XM_012189044 | 5′ attgcaacagggtacttggt 3′ 5′ atatttgatgttgccctcgt 3′ | 60 °C | 122 |
| GDF9 | Growth differentiation factor 9 | NM_001142888 | 5′ cagacgccacctctacaaca 3′ 5′caggaaagggaaaagaaatgg 3′ | 58 °C | 198 |
| HSP90b | Heat shock protein 90b | XM_004018854 | 5′ tggagatcaaccctgacca 3′ 5′ gggatcctcaagcgagaag 3′ | 58 °C | 143 |
| KHDC3L | KH domain containing 3 like | XM_027973471 | 5′ cagaccctgcttcacgttca 3′ 5′ cttctcagagcttcgcgcc 3′ | 60 °C | 150 |
| LUC | Luciferase reporter vector pXP2 *SA *PS | AF093685 | 5′ gctgggcgttaatcagagag 3′ 5′ gtgttcgtcttcgtcccagt 3′ | 58 °C | 151 |
| NLRP2 | NLR family pyrin domain containing 2 | XM_027977986 | 5′ gcatgtgttgctcattctgg 3′ 5′ agcactgtggaaacttgcag 3′ | 60 °C | 120 |
| NLRP5 | NLR family pyrin domain containing 5 | XM_027978862 | 5′ cagcctccaggagttctttg 3′ 5′ gacagcctaggagggtttcc 3′ | 59 °C | 212 |
| OOEP1 | Oocyte expressed protein isoform 1 | KF218578 | 5′ atccgctggtgttcttcctg 3′ 5′ gaacacggtgacttcgacc 3′ | 60 °C | 149 |
| PADI6 | Peptidyl arginine deiminase, type VI | XM_012153966 | 5′ acggctgtactccacctcac 3′ 5′ cccagacccaggttctctta 3′ | 60 °C | 109 |
| SOD1 | Superoxide dismutase 1 | NM_001145185 | 5′caactcccgccagcagat 3′ 5′ ccgggaatggacagtcaca 3′ | 58 °C | 130 |
| TET3 | Ten-eleven translocation 3 | XM_015094461 | 5′tggagcatgtacttcaatgg 3′ 5′ ggtcacctggttctgatagg 3′ | 60 °C | 173 |
| TLE6 | TLE family member 6 | XM_004009373 | 5′ gctgcaggtctccatcatct 3′ 5′ ggatcagctcaagcagcatt 3′ | 60 °C | 134 |
| YAP1 | Yes associated protein 1 | XM_015100723 | 5′ ttcctttgagatccctgacg 3′ 5′ gtcctgccaggttgttgtct 3′ | 60 °C | 115 |
| ZBED3 | Zinc finger BED-type containing 3 | XM_027971476 | 5′ cccagggtagagtgtgcatt 3′ 5′ ggcaagggctactcatcaaa 3′ | 60 °C | 97 |
| GV Oocytes | Matured Oocytes | Cleaved Embryos | Blastocysts | |
|---|---|---|---|---|
| 4W | 89 | 82 (92.13%) | 64 (78.13%) | 9 (14.06%) * |
| LM | 75 | 66 (88%) | 46 (69.7%) | 13 (28.26%) * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bebbere, D.; Nieddu, S.M.; Ariu, F.; Piras, D.; Ledda, S. 3D Liquid Marble Microbioreactors Support In Vitro Maturation of Prepubertal Ovine Oocytes and Affect Expression of Oocyte-Specific Factors. Biology 2021, 10, 1101. https://doi.org/10.3390/biology10111101
Bebbere D, Nieddu SM, Ariu F, Piras D, Ledda S. 3D Liquid Marble Microbioreactors Support In Vitro Maturation of Prepubertal Ovine Oocytes and Affect Expression of Oocyte-Specific Factors. Biology. 2021; 10(11):1101. https://doi.org/10.3390/biology10111101
Chicago/Turabian StyleBebbere, Daniela, Stefano Mario Nieddu, Federica Ariu, Davide Piras, and Sergio Ledda. 2021. "3D Liquid Marble Microbioreactors Support In Vitro Maturation of Prepubertal Ovine Oocytes and Affect Expression of Oocyte-Specific Factors" Biology 10, no. 11: 1101. https://doi.org/10.3390/biology10111101
APA StyleBebbere, D., Nieddu, S. M., Ariu, F., Piras, D., & Ledda, S. (2021). 3D Liquid Marble Microbioreactors Support In Vitro Maturation of Prepubertal Ovine Oocytes and Affect Expression of Oocyte-Specific Factors. Biology, 10(11), 1101. https://doi.org/10.3390/biology10111101






