Functionalization of Biotinylated Polyethylene Glycol on Live Magnetotactic Bacteria Carriers for Improved Stealth Properties
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Method
2.1. Reagents and Chemicals
2.2. Magnetospirillum Magneticum AMB-1 Strain Culture Conditions
2.3. Attachment of PEG–Biotin to MTB
2.4. Characterization of Magnetotactic Bacteria and Magnetosomes
2.5. Assessment of MTB/PEG–Biotin Complex Formation
2.6. Evaluation of Bacterial Viability with PEG–Biotin Attachment
2.6.1. THP-1 Cell Culture
2.6.2. CCK-8-Based Cell Viability Assay
2.6.3. THP-1 Cell Association
2.7. Statistical Analysis
3. Results and Discussion
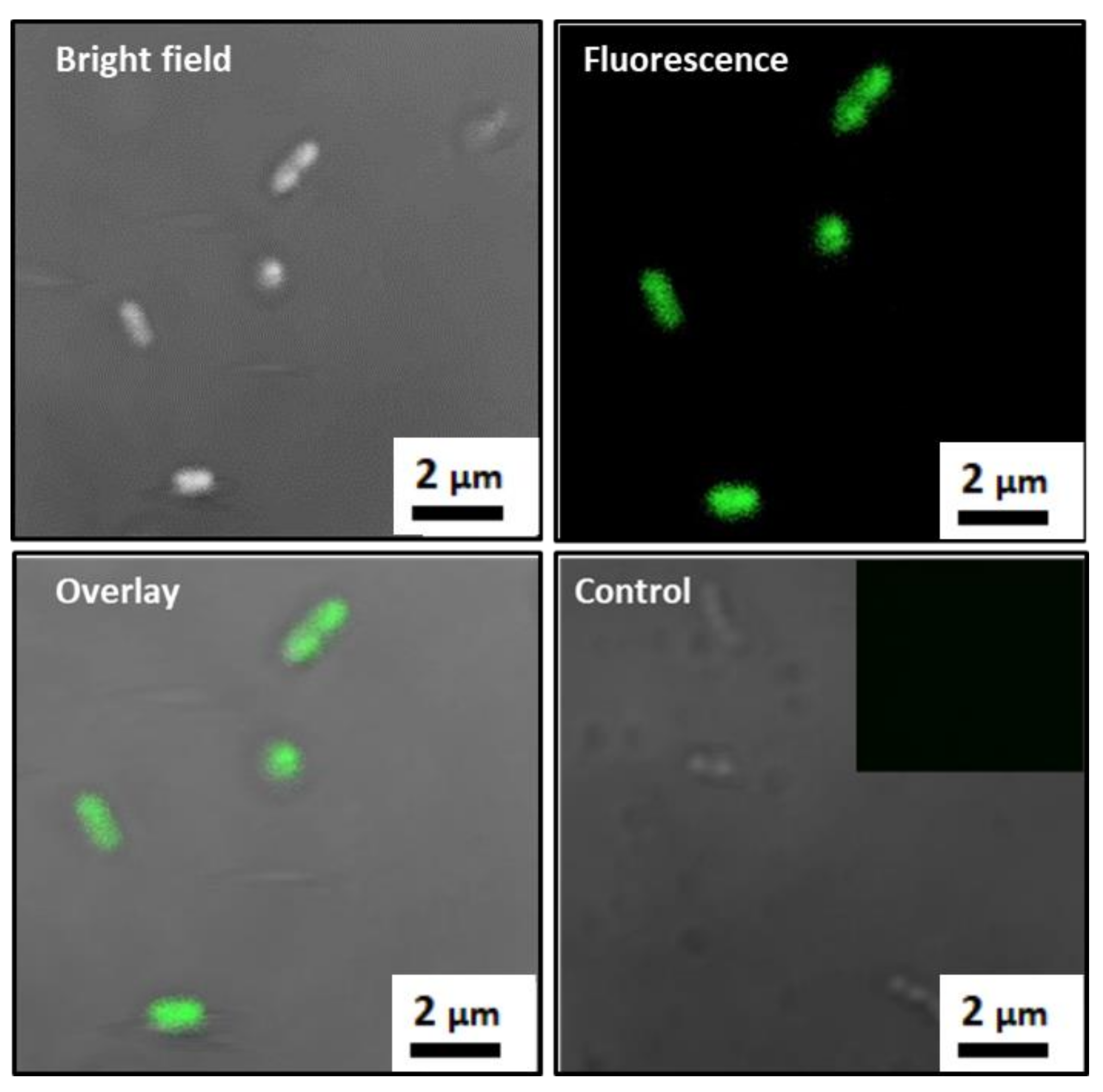
3.1. Characterization of MTB
3.2. Formation of MTB/PEG–Biotin Complex
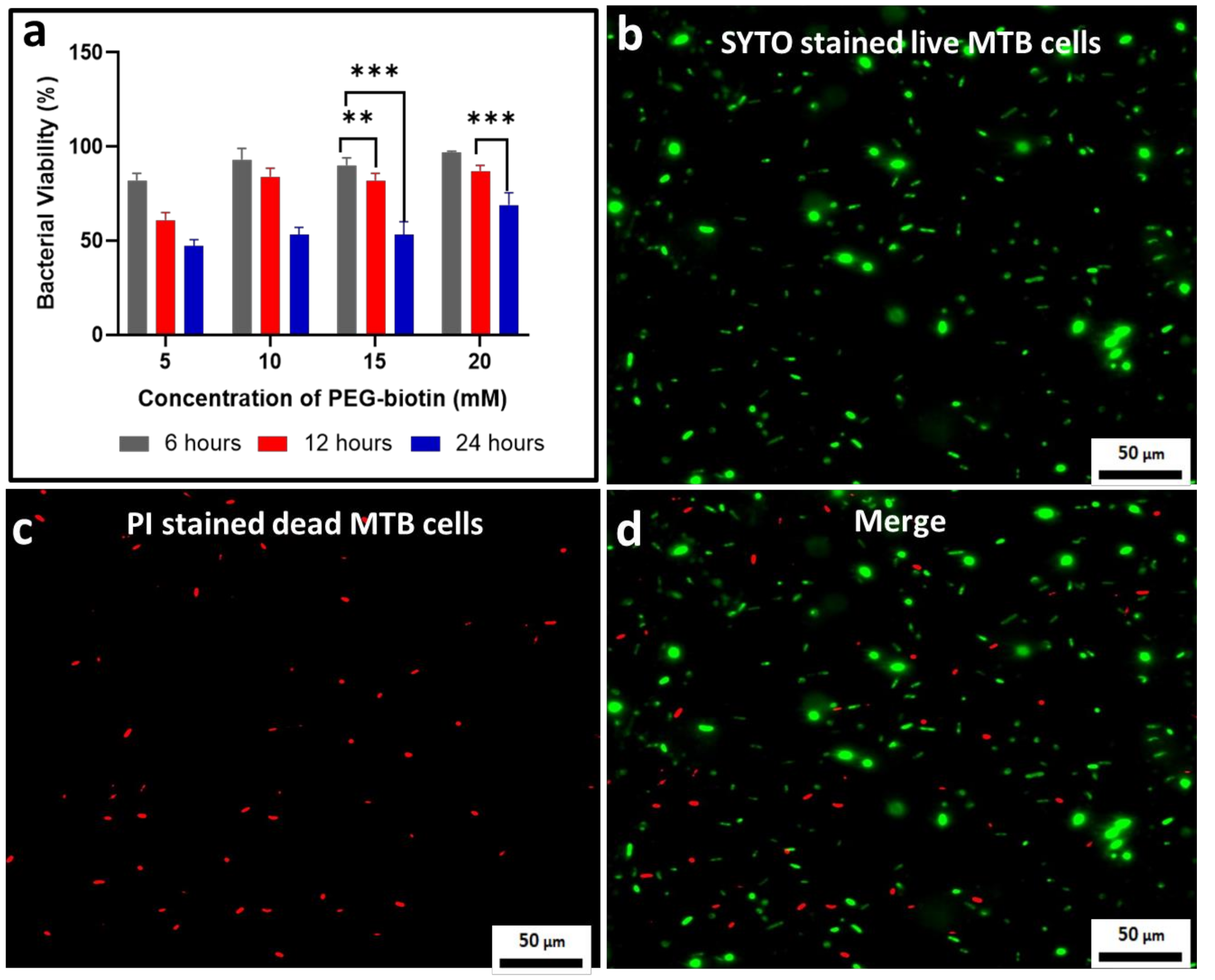
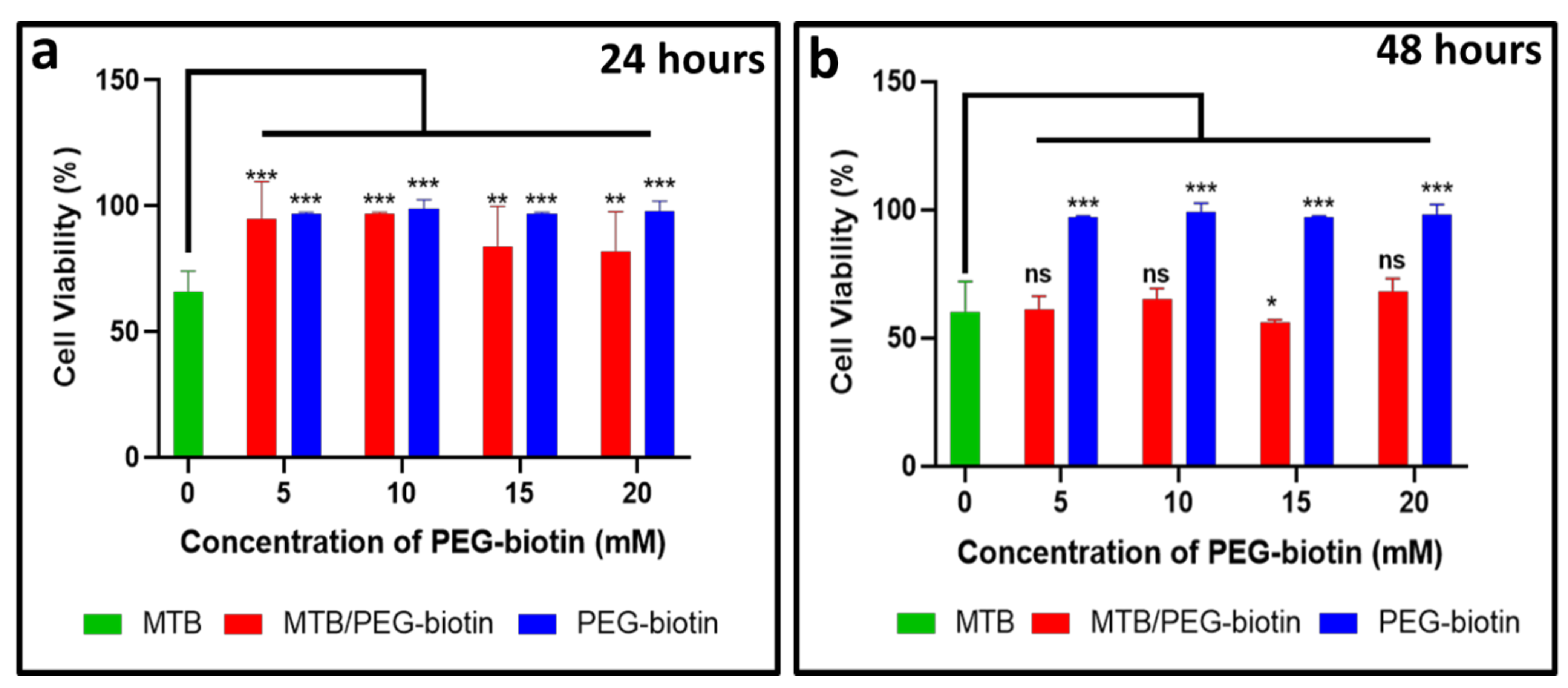
3.3. Assessing the Bacterial Viability for the MTB/PEG–Biotin Complex
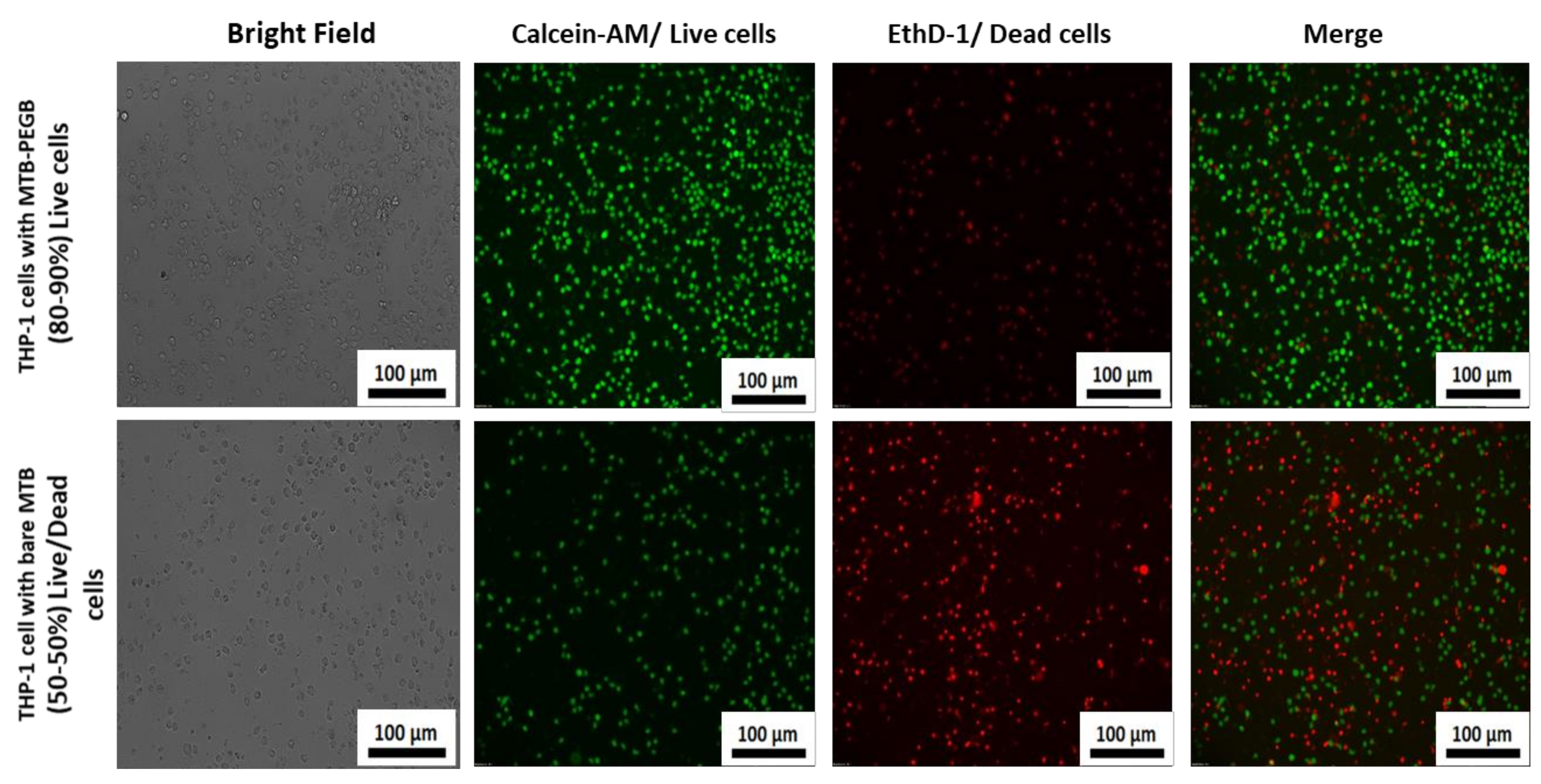
3.4. Biological Application of the MTB/PEG–Biotin Complex
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Al-Jamal, W.T.; Kostarelos, K. Liposomes: From a clinically established drug delivery system to a nanoparticle platform for theranostic nanomedicine. Acc. Chem. Res. 2011, 44, 1094–1104. [Google Scholar] [CrossRef]
- Pouponneau, P.; Leroux, J.C.; Soulez, G.; Gaboury, L.; Martel, S. Co-encapsulation of magnetic nanoparticles and doxorubicin into biodegradable microcarriers for deep tissue targeting by vascular MRI navigation. Biomaterials 2011, 32, 3481–3486. [Google Scholar] [CrossRef]
- Mikhaylov, G.; Mikac, U.; Magaeva, A.A.; Itin, V.I.; Naiden, E.P.; Psakhye, I.; Babes, L.; Reinheckel, T.; Peters, C.; Zeiser, R.; et al. Ferri-liposomes as an MRI-visible drug-delivery system for targeting tumours and their microenvironment. Nat. Nanotechnol. 2011, 6, 594–602. [Google Scholar] [CrossRef] [PubMed]
- Brannon-Peppas, L.; Blanchette, J.O. Nanoparticle and targeted systems for cancer therapy. Adv. Drug Deliv. Rev. 2004, 56, 1649–1659. [Google Scholar] [CrossRef] [PubMed]
- Torchilin, V.P. Passive and active drug targeting: Drug delivery to tumors as an example. Handb. Exp. Pharmacol. 2010, 197, 3–53. [Google Scholar]
- Albanese, A.; Tang, P.S.; Chan, W.C.W. The effect of nanoparticle size, shape, and surface chemistry on biological systems. Annu. Rev. Biomed. Eng. 2012, 14, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.Y.; Cho, S.H.; Lee, Y.M.; Chu, L.Y. Biotin-conjugated block copolymeric nanoparticles as tumor-targeted drug delivery systems. Macromol. Res. 2007, 15, 646–655. [Google Scholar] [CrossRef]
- Chen, S.; Yang, K.; Tuguntaev, R.G.; Mozhi, A.; Zhang, J.; Wang, P.C.; Liang, X.J. Targeting tumor microenvironment with PEG-based amphiphilic nanoparticles to overcome chemoresistance. Nanomed. Nanotechnol. Biol. Med. 2016, 12, 269–286. [Google Scholar] [CrossRef] [Green Version]
- Broquist, H.P.; Snell, E.E. Biotin and bacterial growth. I. Relation to aspartate, oleate, and carbon dioxide. J. Biol. Chem. 1951, 188, 431–444. [Google Scholar] [CrossRef]
- Park, S.; Kim, E.; Kim, W.Y.; Kang, C.; Kim, J.S. Biotin-guided anticancer drug delivery with acidity-triggered drug release. Chem. Commun. 2015, 51, 9343–9345. [Google Scholar] [CrossRef]
- Suk, J.S.; Xu, Q.; Kim, N.; Hanes, J.; Ensign, L.M. PEGylation as a strategy for improving nanoparticle-based drug and gene delivery. Adv. Drug Deliv. Rev. 2016, 99, 28–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wenande, E.; Garvey, L.H. Immediate-type hypersensitivity to polyethylene glycols: A review. Clin. Exp. Allergy 2016, 46, 907–922. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; De Rose, R.; Alt, K.; Alcantara, S.; Paterson, B.M.; Liang, K.; Hu, M.; Richardson, J.J.; Yan, Y.; Jeffery, C.M.; et al. Engineering poly(ethylene glycol) particles for improved biodistribution. ACS Nano 2015, 9, 1571–1580. [Google Scholar] [CrossRef] [Green Version]
- Matoba, T.; Koga, J.-I.; Nakano, K.; Egashira, K.; Tsutsui, H. Nanoparticle-mediated drug delivery system for atherosclerotic cardiovascular disease. J. Cardiol. 2017, 70, 206–211. [Google Scholar] [CrossRef] [Green Version]
- Stanton, M.M.; Park, B.W.; Vilela, D.; Bente, K.; Faivre, D.; Sitti, M.; Sánchez, S. Magnetotactic bacteria powered biohybrids target e. Coli biofilms. ACS Nano 2017, 11, 9968–9978. [Google Scholar] [CrossRef]
- Minchinton, A.I.; Tannock, I.F. Drug penetration in solid tumours. Nat. Rev. Cancer 2006, 6, 583–592. [Google Scholar] [CrossRef]
- Carlsen, R.W.; Sitti, M. Bio-hybrid cell-based actuators for microsystems. Small 2014, 10, 3831–3851. [Google Scholar] [CrossRef]
- Feinberg, A.W.; Feigel, A.; Shevkoplyas, S.S.; Sheehy, S.; Whitesides, G.M.; Parker, K.K. Muscular thin films for building actuators and powering devices. Science 2007, 317, 1366–1370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, Z.; Liu, J. Bacteria and bacterial derivatives as drug carriers for cancer therapy. J. Control. Release 2020, 326, 396–407. [Google Scholar] [CrossRef] [PubMed]
- Forbes, N.S. Engineering the perfect (bacterial) cancer therapy. Nat. Rev. Cancer 2010, 10, 785–794. [Google Scholar] [CrossRef] [Green Version]
- Williams, B.J.; Anand, S.V.; Rajagopalan, J.; Saif, M.T.A. A self-propelled biohybrid swimmer at low Reynolds number. Nat. Commun. 2014, 5, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Medina-Sánchez, M.; Schwarz, L.; Meyer, A.K.; Hebenstreit, F.; Schmidt, O.G. Cellular cargo delivery: Toward assisted fertilization by sperm-carrying micromotors. Nano Lett. 2016, 16, 555–561. [Google Scholar] [CrossRef]
- Kuzajewska, D.; Wszołek, A.; Żwierełło, W.; Kirczuk, L.; Maruszewska, A. Magnetotactic bacteria and magnetosomes as smart drug delivery systems: A new weapon on the battlefield with cancer? Biology 2020, 9, 102. [Google Scholar] [CrossRef] [PubMed]
- Ebrahimi, N.; Bi, C.; Cappelleri, D.J.; Ciuti, G.; Conn, A.T.; Faivre, D.; Habibi, N.; Hošovský, A.; Iacovacci, V.; Khalil, I.S.M.; et al. Magnetic Actuation Methods in Bio/Soft Robotics. Adv. Funct. Mater. 2021, 31, 1–40. [Google Scholar] [CrossRef]
- Gareev, K.G.; Grouzdev, D.S.; Kharitonskii, P.V.; Kosterov, A.; Koziaeva, V.V.; Sergienko, E.S.; Shevtsov, M.A. Magnetotactic bacteria and magnetosomes: Basic properties and applications. Magnetochemistry 2021, 7, 86. [Google Scholar] [CrossRef]
- Martel, S.; Tremblay, C.C.; Ngakeng, S.; Langlois, G. Controlled manipulation and actuation of micro-objects with magnetotactic bacteria. Appl. Phys. Lett. 2006, 89, 3–5. [Google Scholar] [CrossRef] [Green Version]
- Blakemore, R.P.; Maratea, D.; Wolfe, R.S. Isolation and pure culture of a freshwater magnetic spirillum in chemically defined medium. J. Bacteriol. 1979, 140, 720–729. [Google Scholar] [CrossRef] [Green Version]
- Martel, S.; Mohammadi, M.; Felfoul, O.; Zhao, L.; Pouponneau, P. Flagellated magnetotactic bacteria as controlled MRI-trackable propulsion and steering systems for medical nanorobots operating in the human microvasculature. Int. J. Rob. Res. 2009, 28, 571–582. [Google Scholar] [CrossRef] [Green Version]
- Felfoul, O.; Mohammadi, M.; Taherkhani, S.; De Lanauze, D.; Zhong Xu, Y.; Loghin, D.; Essa, S.; Jancik, S.; Houle, D.; Lafleur, M.; et al. Magneto-aerotactic bacteria deliver drug-containing nanoliposomes to tumour hypoxic regions. Nat. Nanotechnol. 2016, 11, 941–947. [Google Scholar] [CrossRef] [PubMed]
- Matsunaga, T.; Sakaguchi, T.; Tadakoro, F. Magnetite formation by a magnetic bacterium capable of growing aerobically. Appl. Microbiol. Biotechnol. 1991, 35, 651–655. [Google Scholar] [CrossRef]
- Hua, J.; Gengsheng, J. Hydrothermal synthesis and characterization of monodisperse α-Fe2O3 nanoparticles. Mater. Lett. 2009, 63, 2725–2727. [Google Scholar] [CrossRef]
- Hermanson, G.T. Chapter 3-The Reactions of Bioconjugation, Bioconjugate Techniques, 3rd ed.; Academic Press: Cambridge, MA, USA, 2013; pp. 229–258. ISBN 9780123822390. [Google Scholar]
- Perrault, S.D.; Walkey, C.; Jennings, T.; Fischer, H.C.; Chan, W.C.W. Mediating tumor targeting efficiency of nanoparticles through design. Nano Lett. 2009, 9, 1909–1915. [Google Scholar] [CrossRef]
- Nuttelman, C.R.; Tripodi, M.C.; Anseth, K.S. Synthetic hydrogel niches that promote hMSC viability. Matrix Biol. 2005, 24, 208–218. [Google Scholar] [CrossRef]
- Li, L.; Kantor, A.; Warne, N. Application of a PEG precipitation method for solubility screening: A tool for developing high protein concentration formulations. Protein Sci. 2013, 22, 1118–1123. [Google Scholar] [CrossRef] [Green Version]
- Veronese, F.M. Peptide and protein PEGylation. Biomaterials 2001, 22, 405–417. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chaturvedi, R.; Kang, Y.; Eom, Y.; Torati, S.R.; Kim, C. Functionalization of Biotinylated Polyethylene Glycol on Live Magnetotactic Bacteria Carriers for Improved Stealth Properties. Biology 2021, 10, 993. https://doi.org/10.3390/biology10100993
Chaturvedi R, Kang Y, Eom Y, Torati SR, Kim C. Functionalization of Biotinylated Polyethylene Glycol on Live Magnetotactic Bacteria Carriers for Improved Stealth Properties. Biology. 2021; 10(10):993. https://doi.org/10.3390/biology10100993
Chicago/Turabian StyleChaturvedi, Richa, Yumin Kang, Yunji Eom, Sri Ramulu Torati, and CheolGi Kim. 2021. "Functionalization of Biotinylated Polyethylene Glycol on Live Magnetotactic Bacteria Carriers for Improved Stealth Properties" Biology 10, no. 10: 993. https://doi.org/10.3390/biology10100993
APA StyleChaturvedi, R., Kang, Y., Eom, Y., Torati, S. R., & Kim, C. (2021). Functionalization of Biotinylated Polyethylene Glycol on Live Magnetotactic Bacteria Carriers for Improved Stealth Properties. Biology, 10(10), 993. https://doi.org/10.3390/biology10100993






