Disappearance of Temporal Collinearity in Vertebrates and Its Eventual Reappearance
Abstract
:Simple Summary
Abstract
1. Introduction
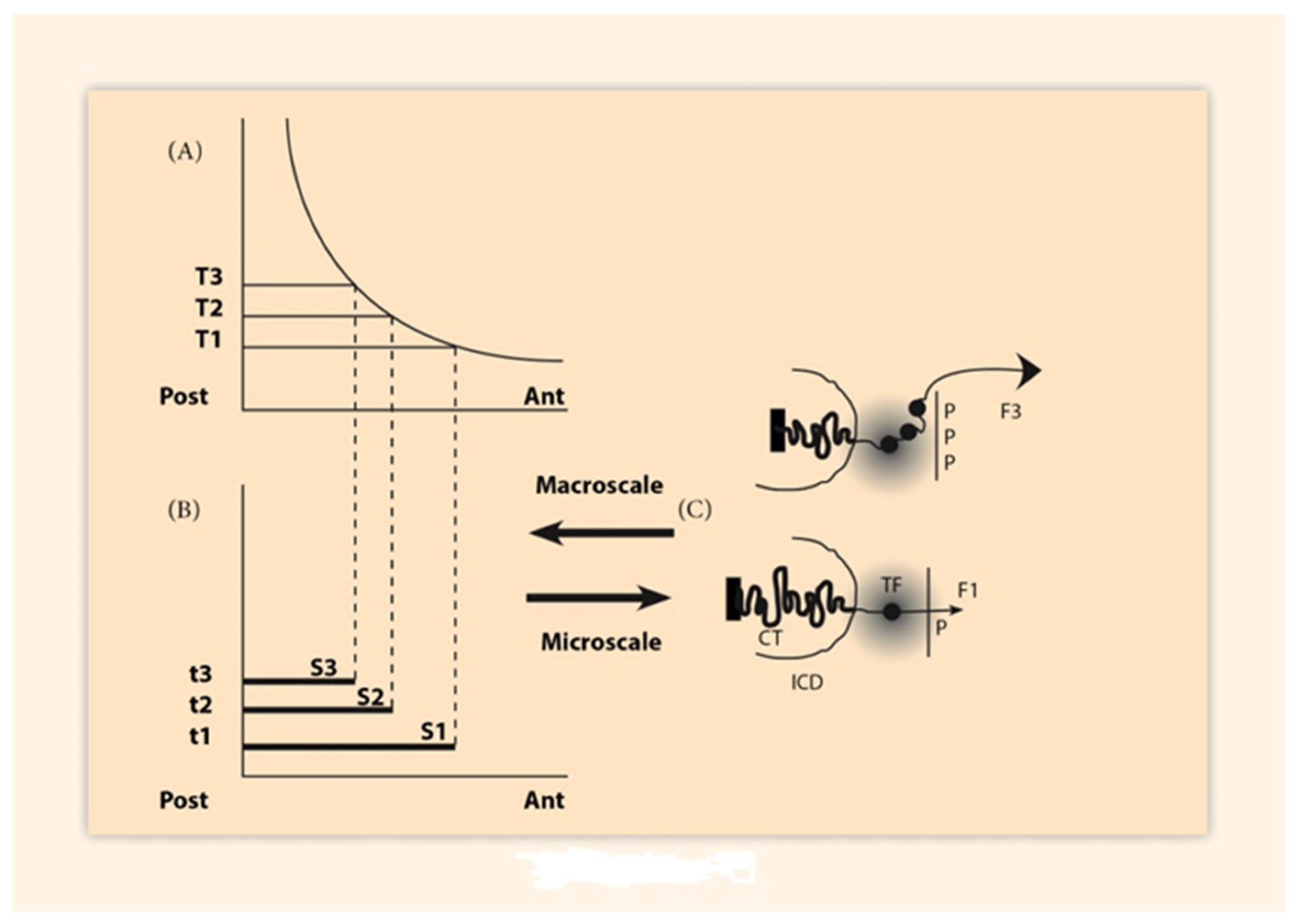
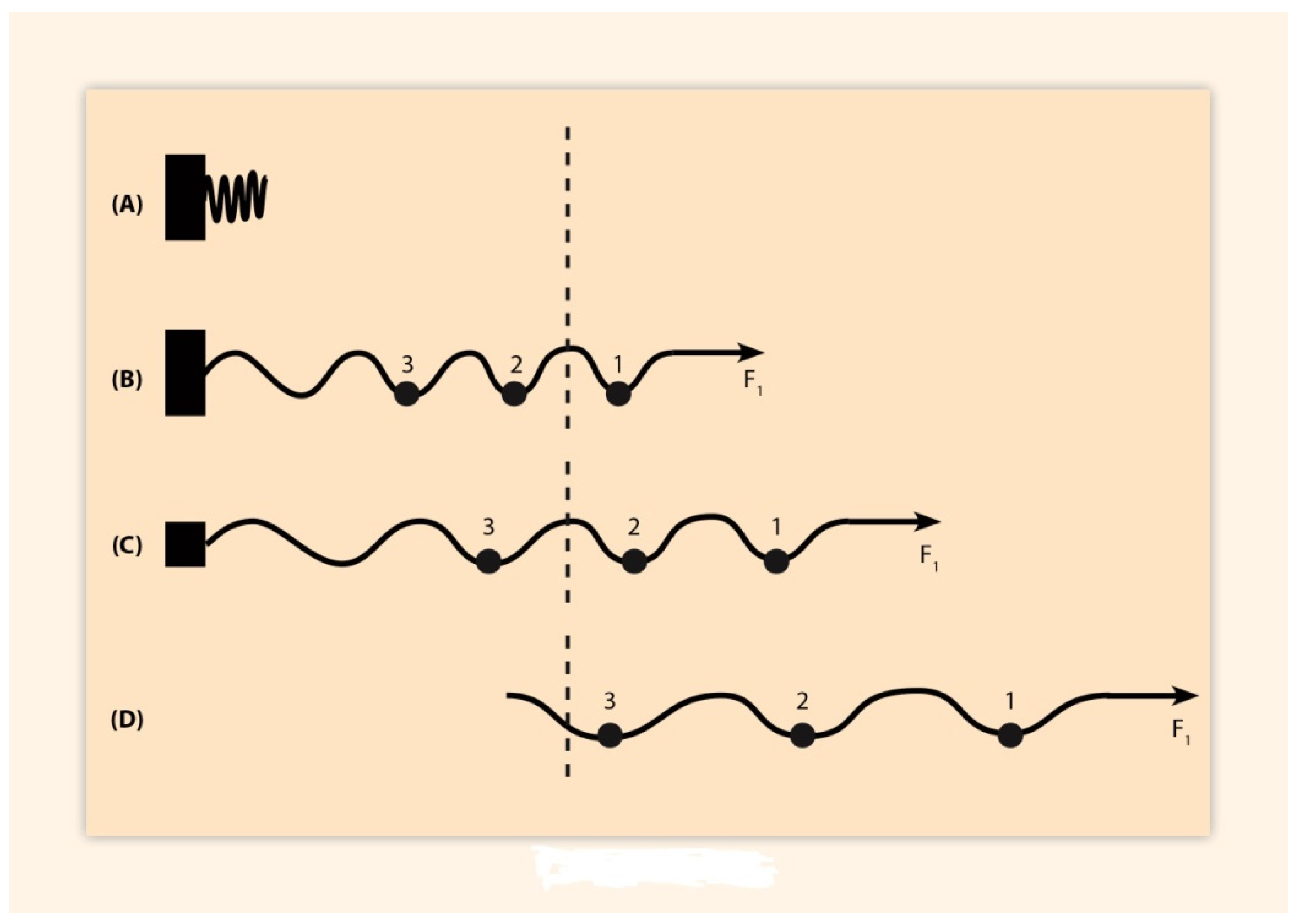
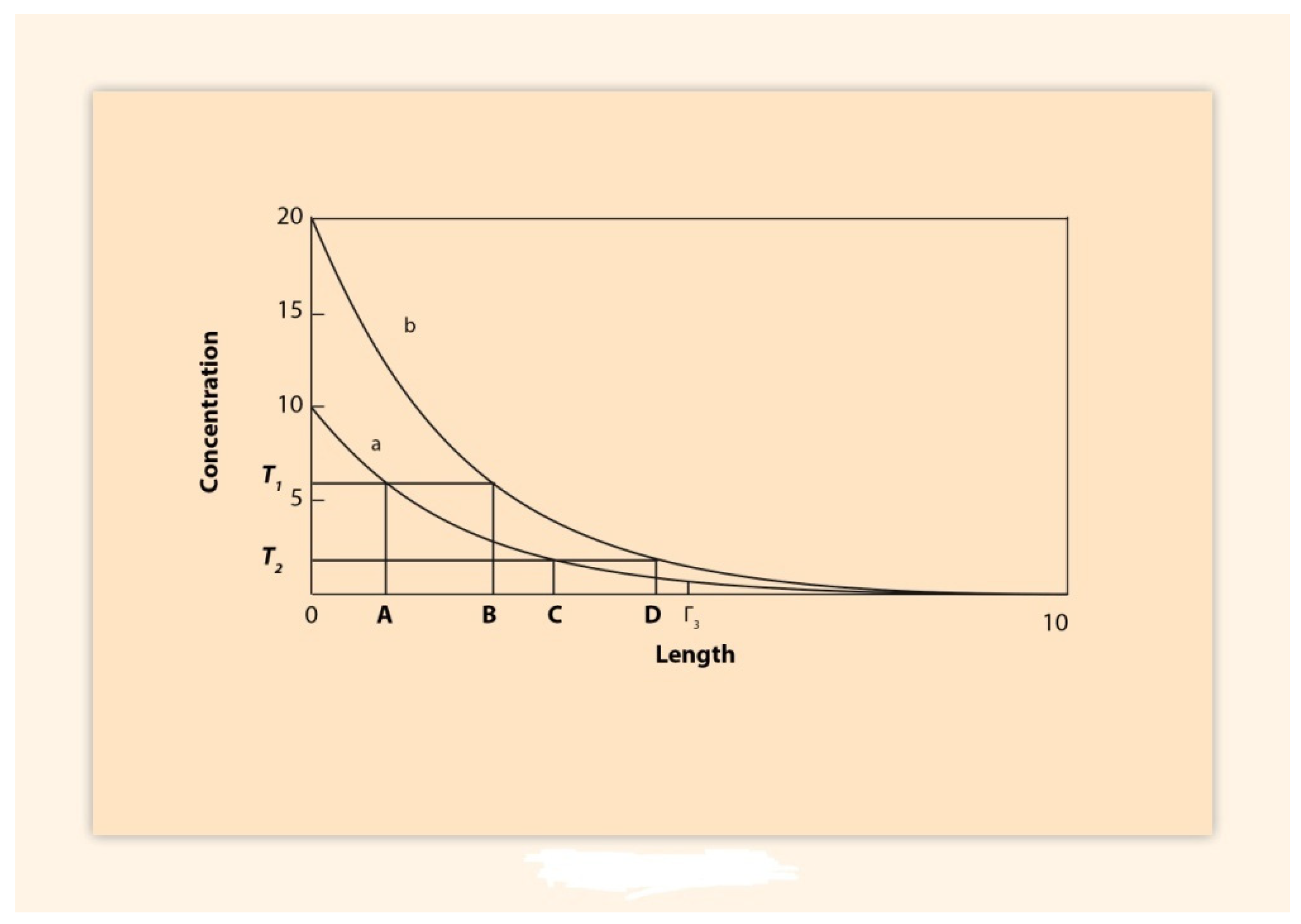
2. BM Formulation and Its Elastic Spring Approximation
3. Spatial and Temporal Collinearities in the Vertebrates
3.1. Paradigm of the HoxA Expressions in the Chick Limb Bud
- After the AER excision, HoxA13 is the first gene that rapidly switches off.
- Upon continuous exposure of the limb bud to an FGF soaked bead, HoxA13 is rescued after at least 6 h.
| Exp II. | (a) → (b) → (c) → (d) | (direct step) |
| (af) ← (b) ← (c) ← (d) | (reverse step) |
3.2. Paradigm of Gene Activation in the Mouse Embryo
| (Exp. I) | (a) → (b) → (c) → (d) | (direct step) |
| (a?) ← (b?) ← (c?) ← (d) | (reverse step) |
4. Discussion
- A.
- In the case of paradigm I the chip insertion operates in terra incognita and any prediction would be hazardous. Only Experiment can confirm a theory and fix the parameters involved in the data description. Note that an ideal Model is a ‘theory’ that predicts all experimental results in the realm of its implementation. Usually, a model can predict a limited set of results and all efforts aim at enlarging this domain by experimental trials. In the case of a microarray chip insertion the possible results one can anticipate are the following:
- no response at all—no difference observed with or without pulling forces.
- a partial restoration of HoxA13 expression (b?, c?)
- a full rescue of HoxA13 (a?)
- B.
- A very important consequence of the above TC disappearance is its connection to TC loss as an evolutionary instrument applied to many animal phyla where TC is absent as, for instance, in Drosophila [8].
- C.
- TC disappearance is a very striking event. A less striking effect is TC violation as observed by M. Kondo et al. in Xenopus leavis [16].
5. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Lewis, E.B. A gene complex controlling segmentation in Drosophila. Nature 1978, 276, 565–570. [Google Scholar] [CrossRef] [PubMed]
- Krumlauf, R. Hox genes, clusters and collinearity. Int. J. Dev. Biol. 2018, 62, 659–663. [Google Scholar] [CrossRef] [PubMed]
- Lesne, A. Systems biology: The multiscale organization of living systems. Med.-Sci. 2009, 25, 585–587. [Google Scholar]
- Izpisua-Belmonte, J.C.; Duboule, D. Murine genes related to the Drosophila AbdB homeotic genes are sequentially expressed. EMBO J. 1991, 10, 2279–2289. [Google Scholar] [CrossRef] [PubMed]
- Kondo, T.; Duboule, D. Breaking Collinearity in the mouse HoxD Complex. Cell 1999, 97, 407–417. [Google Scholar] [CrossRef]
- Papageorgiou, S. A physical force may expose Hox genes to express in a morphogenetic density gradient. Bull. Math. Biol. 2001, 63, 185–200. [Google Scholar] [CrossRef] [PubMed]
- Papageorgiou, S. Physical Forces May Cause the HoxD Gene Cluster Elongation. Biology 2017, 6, 32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papageorgiou, S. Physical Laws Shape Up HOX Gene Collinearity. J. Dev. Biol. 2021, 9, 17. [Google Scholar] [CrossRef] [PubMed]
- Vargesson, N.; Kostakopoulou, K.; Drossopoulou, G.; Papageorgiou, S.; Tickle, C. Characterisation of Hoxa gene expression in the chick limb bud in response to FGF. Dev. Dyn. 2001, 220, 87–90. [Google Scholar] [CrossRef]
- Towers, M.; Wolpert, L.; Tickle, C. Gradients of signaling in the developing limb. Curr. Opin. Cell Biol. 2012, 24, 181–187. [Google Scholar] [CrossRef]
- Papageorgiou, S. Cooperating morphogens control hoxd gene expression in the developing vertebrate limb. J. Theor. Biol. 1998, 192. [Google Scholar]
- Ye, Z.; Kimelman, D. Identification of in vivo Hox13-binding sites reveals an essential locus controlling zebrafish brachyury expression. Development 2021, 148, dev199408. [Google Scholar] [CrossRef]
- Rodaway, A.; Takeda, H.; Koshida, S.; Broadbent, J.; Price, B.; Smith, J.; Patient, R.; Holder, N. Induction of the mesendoderm in the zebrafish germ ring by yolk cell-derived TGF-beta family signals and discrimination of mesoderm and endoderm by FGF. Development 1999, 126, 3067–3078. [Google Scholar] [CrossRef]
- Gentsch, G.E.; Owens, N.D.L.; Martin, S.R.; Piccinelli, P.; Faial, T.; Trotter, M.W.B.; Gilchrist, M.J.; Smith, J.C. In vivo T-box transcription factor profiling reveals joint regulation of embryonic neuromesodermalbipotency. Cell Rep. 2013, 4, 1185–1196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Evans, A.L.; Faial, T.; Gilchrist, M.J.; Down, T.; Vaillier, L.; Pedersen, R.A.; Wardle, F.C.; Smith, J.C. Genomic targets of Brachyury (T) in differentiating mouse embryonic stem cells. PLoS ONE 2012, 7, e33346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kondo, M.; Yamamoto, T.; Takahashi, S.; Taira, M. Comprehensive analysis of hox gene expression in Xenopus laevis embryos and adult tissues. Dev. Growth Differ. 2017, 59, 526–539. [Google Scholar] [CrossRef] [Green Version]
- Gordon, N.K.; Chen, Z.; Gordon, R.; Zou, Y. French flag gradients and Turing reaction-diffusion versus differentiation waves as models of morphogenesis. Biosystems 2020, 196, 104–169. [Google Scholar] [CrossRef] [PubMed]
- Tompkins, N.; Li, N.; Girabawe, C.; Heymann, M.; Ermentrout, G.B.; Epstein, I.R.; Fraden, S. Testing Turing’s theory of Morphogenesis in chemical cells. Proc. Nat. Acad. Sci. USA 2014, 111, 4397–4402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kmita, M.; van der Hoeven, F.; Zakany, J.; Krumlauf, R.; Duboule, D. Mechanisms of Hox gene collinearity: Transposition of the anterior Hoxb1 gene into the posterior HoxD complex. Genes Dev. 2000, 14, 198–211. [Google Scholar] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Papageorgiou, S. Disappearance of Temporal Collinearity in Vertebrates and Its Eventual Reappearance. Biology 2021, 10, 1018. https://doi.org/10.3390/biology10101018
Papageorgiou S. Disappearance of Temporal Collinearity in Vertebrates and Its Eventual Reappearance. Biology. 2021; 10(10):1018. https://doi.org/10.3390/biology10101018
Chicago/Turabian StylePapageorgiou, Spyros. 2021. "Disappearance of Temporal Collinearity in Vertebrates and Its Eventual Reappearance" Biology 10, no. 10: 1018. https://doi.org/10.3390/biology10101018
APA StylePapageorgiou, S. (2021). Disappearance of Temporal Collinearity in Vertebrates and Its Eventual Reappearance. Biology, 10(10), 1018. https://doi.org/10.3390/biology10101018




