Lassa Virus Circulation in Small Mammal Populations in Bo District, Sierra Leone
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
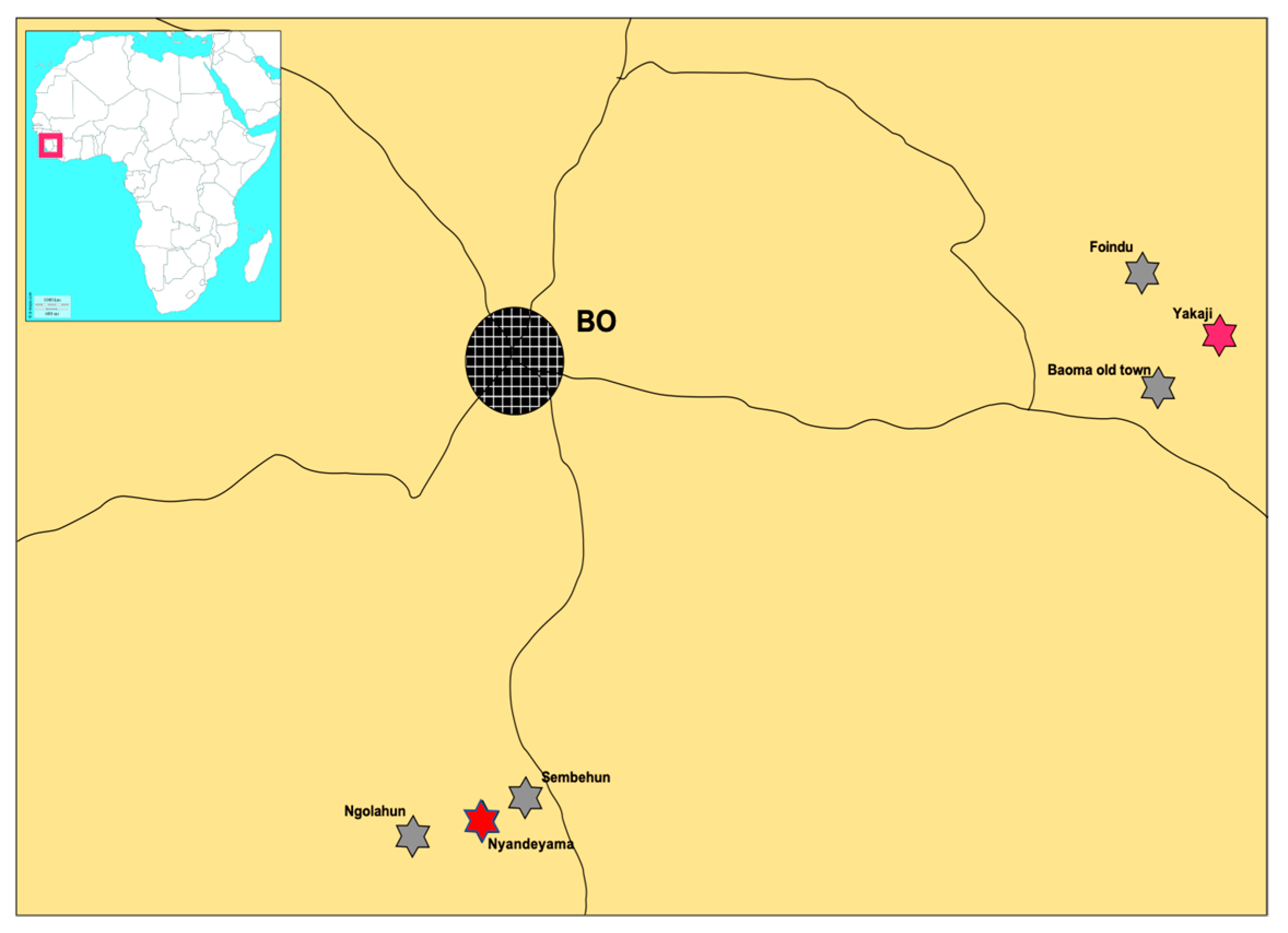
2.1. Small Mammal Sampling
2.2. Serology
2.3. RT-PCR Workflow and Sequencing
2.4. Statistical Analysis
3. Results
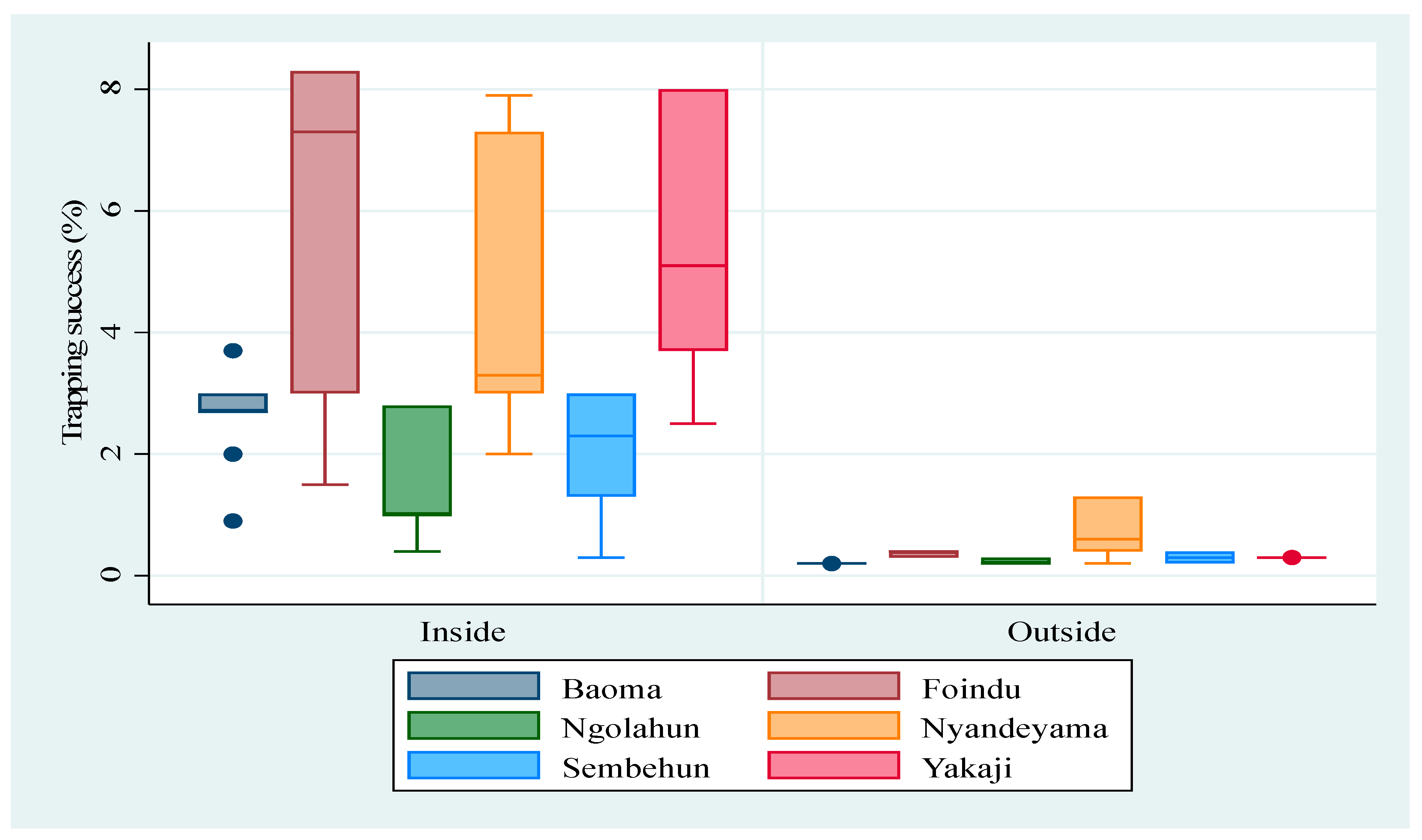
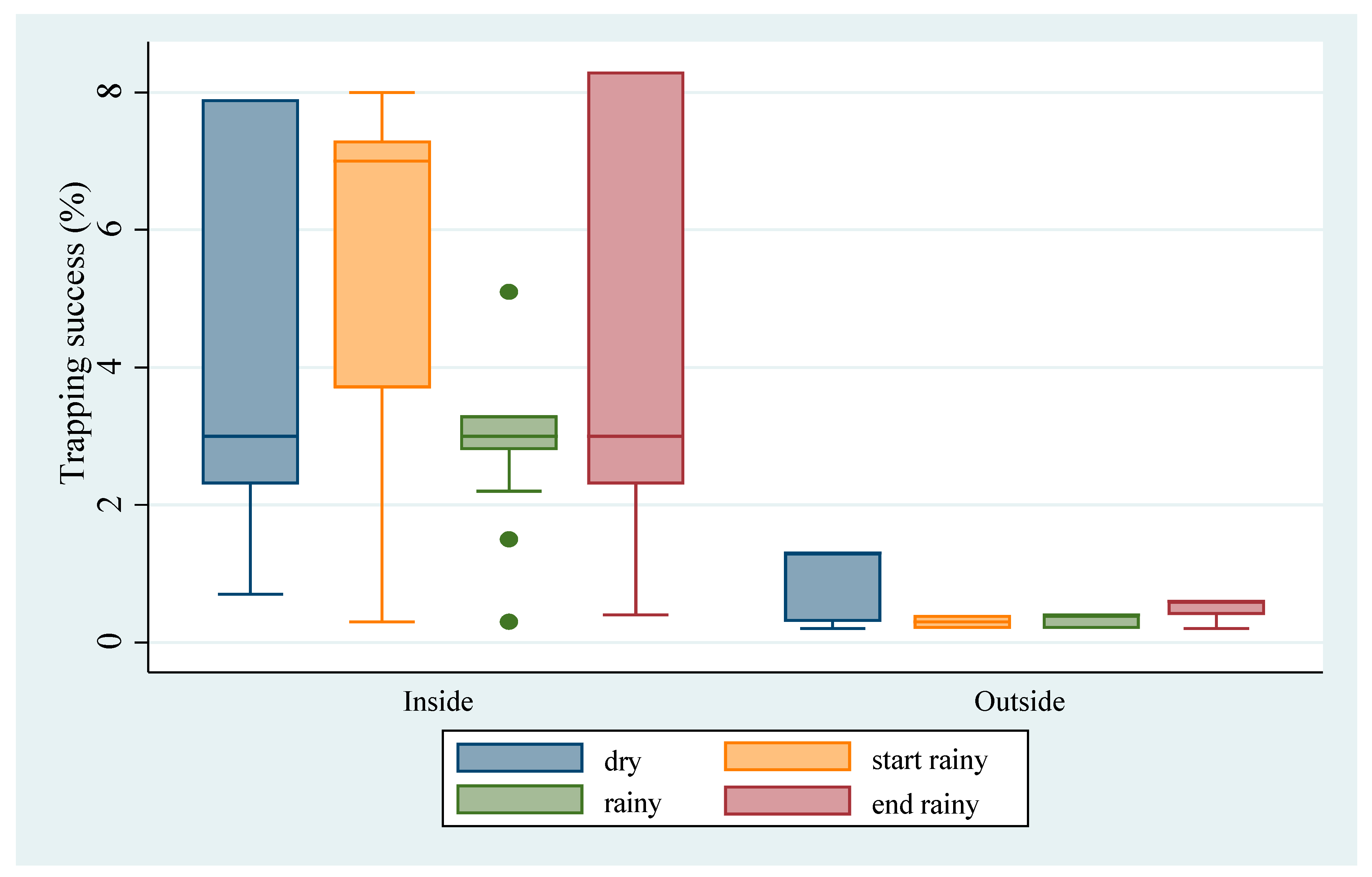
3.1. Abundance of M. natalensis
3.2. Serology
3.3. LASV Testing
3.4. Phylogenetic Analysis
4. Discussion
4.1. Abundance of M. natalensis
4.2. Arenavirus Antibodies in the Rodent Community
4.3. LASV Infection
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cummins, D.; Bennett, D.; Fisher-Hoch, S.P.; Farrar, B.; Machin, S.J.; McCormick, J.B. Lassa fever encephalopathy: Clinical and laboratory findings. J. Trop. Med. Hyg. 1992, 95, 197–201. [Google Scholar] [PubMed]
- Frame, J.D.; Baldwin, J.M., Jr.; Gocke, D.J.; Troup, J.M. Lassa fever, a new virus disease of man from West Africa. I. Clinical description and pathological findings. Am. J. Trop. Med. Hyg. 1970, 19, 670–676. [Google Scholar] [CrossRef] [PubMed]
- McCormick, J.B.; King, I.J.; Webb, P.A.; Johnson, K.M.; O’Sullivan, R.; Smith, E.S.; Trippel, S.; Tong, T.C. A case-control study of the clinical diagnosis and course of Lassa fever. J. Infect. Dis. 1987, 155, 445–455. [Google Scholar] [CrossRef] [PubMed]
- McCormick, J.B.; Webb, P.A.; Krebs, J.W.; Johnson, K.M.; Smith, E.S. A prospective study of the epidemiology and ecology of Lassa fever. J. Infect. Dis. 1987, 155, 437–444. [Google Scholar] [CrossRef]
- Fichet-Calvet, E.; Rogers, D.J. Risk maps of Lassa fever in West Africa. PLoS Negl. Trop. Dis. 2009, 3, e388. [Google Scholar] [CrossRef]
- Lecompte, E.; Fichet-Calvet, E.; Daffis, S.; Koulemou, K.; Sylla, O.; Kourouma, F.; Dore, A.; Soropogui, B.; Aniskin, V.; Allali, B.; et al. Mastomys natalensis and Lassa fever, West Africa. Emerg. Infect. Dis. 2006, 12, 1971–1974. [Google Scholar] [CrossRef]
- Monath, T.P.; Newhouse, V.F.; Kemp, G.E.; Setzer, H.W.; Cacciapuoti, A. Lassa virus isolation from Mastomys natalensis rodents during an epidemic in Sierra Leone. Science 1974, 185, 263–265. [Google Scholar] [CrossRef]
- Fichet-Calvet, E.; Lecompte, E.; Koivogui, L.; Soropogui, B.; Dore, A.; Kourouma, F.; Sylla, O.; Daffis, S.; Koulemou, K.; Ter Meulen, J. Fluctuation of abundance and Lassa virus prevalence in Mastomys natalensis in Guinea, West Africa. Vector Borne Zoonotic. Dis. 2007, 7, 119–128. [Google Scholar] [CrossRef]
- Olayemi, A.; Cadar, D.; Magassouba, N.; Obadare, A.; Kourouma, F.; Oyeyiola, A.; Fasogbon, S.; Igbokwe, J.; Rieger, T.; Bockholt, S.; et al. New Hosts of the lassa virus. Sci. Rep. 2016, 6, 25280. [Google Scholar] [CrossRef]
- Monath, T.P.; Maher, M.; Casals, J.; Kissling, R.E.; Cacciapuoti, A. Lassa fever in the Eastern Province of Sierra Leone, 1970-1972. II. Clinical observations and virological studies on selected hospital cases. Am. J. Trop. Med. Hyg. 1974, 23, 1140–1149. [Google Scholar] [CrossRef]
- Stephenson, E.H.; Larson, E.W.; Dominik, J.W. Effect of environmental factors on aerosol-induced Lassa virus infection. J. Med. Virol. 1984, 14, 295–303. [Google Scholar] [CrossRef] [PubMed]
- Ter Meulen, J.; Lukashevich, I.; Sidibe, K.; Inapogui, A.; Marx, M.; Dorlemann, A.; Yansane, M.L.; Koulemou, K.; Chang-Claude, J.; Schmitz, H. Hunting of peridomestic rodents and consumption of their meat as possible risk factors for rodent-to-human transmission of Lassa virus in the Republic of Guinea. Am. J. Trop. Med. Hyg. 1996, 55, 661–666. [Google Scholar] [CrossRef]
- Günther, S.; Lenz, O. Lassa Virus. Crit. Rev. Clin. Lab. Sci. 2004, 41, 339–390. [Google Scholar] [CrossRef]
- Fraser, D.W.; Campbell, C.C.; Monath, T.P.; Goff, P.A.; Gregg, M.B. Lassa fever in the Eastern Province of Sierra Leone, 1970-1972. I. Epidemiologic studies. Am. J. Trop. Med. Hyg. 1974, 23, 1131–1139. [Google Scholar] [CrossRef]
- Keane, E.; Gilles, H.M. Lassa fever in panguma hospital, Sierra Leone, 1973–1976. Br. Med. J. 1977, 1, 1399–1402. [Google Scholar] [CrossRef][Green Version]
- Shaffer, J.G.; Grant, D.S.; Schieffelin, J.S.; Boisen, M.L.; Goba, A.; Hartnett, J.N.; Levy, D.C.; Yenni, R.E.; Moses, L.M.; Fullah, M.; et al. Lassa fever in post-conflict sierra leone. PLoS Negl. Trop. Dis. 2014, 8, e2748. [Google Scholar] [CrossRef]
- Bonwitt, J.; Saez, A.M.; Lamin, J.; Ansumana, R.; Dawson, M.; Buanie, J.; Lamin, J.; Sondufu, D.; Borchert, M.; Sahr, F.; et al. At home with mastomys and rattus: Human-rodent interactions and potential for primary transmission of lassa virus in domestic spaces. Am. J. Trop Med. Hyg 2017, 96, 935–943. [Google Scholar] [CrossRef]
- Mills, J.N.; Centers for Disease, Control and Prevention. Methods for Trapping and sampling Small Mammals for Virologic Testing; U.S. Dept. of Health & Human Services, Public Health Service, Centers for Disease Control and Prevention: Atlanta, GA, USA, 1995. [Google Scholar]
- Fichet-Calvet, E. Chapter 5—Lassa fever: A rodent-human interaction. In The Role of Animals in Emerging Viral Diseases; Johnson, N., Ed.; Academic Press: Boston, MA, USA, 2014; pp. 89–123. [Google Scholar] [CrossRef]
- Happold, D.C.D. Rodents, Hares and Rabbits. In Mammals of Africa; Happold, D.C.D., Ed.; Bloomsbury: London, UK, 2013; Volume III, p. 784. [Google Scholar]
- Ducroz, J.F.; Granjon, L.; Chevret, P.; Duplantier, J.M.; Lombard, M.; Volobouev, V. Characterization of two distinct species of Arvicanthis (Rodentia: Muridae) in West Africa: Cytogenetic, molecular and reproductive evidence. J. Zool. 1997, 241, 709–723. [Google Scholar] [CrossRef]
- Lecompte, E.; Brouat, C.; Duplantier, J.-M.; Galan, M.; Granjon, L.; Loiseau, A.; Mouline, K.; Cosson, J.-F. Molecular identification of four cryptic species of Mastomys (Rodentia, Murinae). Biochem. Syst. Ecol. 2005, 33, 681–689. [Google Scholar] [CrossRef]
- Borremans, B. Ammonium improves elution of fixed dried blood spots without affecting immunofluorescence assay quality. Trop Med. Int. Health 2014, 19, 413–416. [Google Scholar] [CrossRef]
- Wulff, H.; Lange, J.V. Indirect immunofluorescence for the diagnosis of Lassa fever infection. Bull. World Health Organ. 1975, 52, 429–436. [Google Scholar] [PubMed]
- Olschlager, S.; Lelke, M.; Emmerich, P.; Panning, M.; Drosten, C.; Hass, M.; Asogun, D.; Ehichioya, D.; Omilabu, S.; Gunther, S. Improved detection of Lassa virus by reverse transcription-PCR targeting the 5′ region of S RNA. J. Clin. Microbiol. 2010, 48, 2009–2013. [Google Scholar] [CrossRef] [PubMed]
- Vieth, S.; Drosten, C.; Lenz, O.; Vincent, M.; Omilabu, S.; Hass, M.; Becker-Ziaja, B.; ter Meulen, J.; Nichol, S.T.; Schmitz, H.; et al. RT-PCR assay for detection of Lassa virus and related Old World arenaviruses targeting the L gene. Trans. R Soc. Trop. Med. Hyg. 2007, 101, 1253–1264. [Google Scholar] [CrossRef] [PubMed]
- Ehichioya, D.U.; Hass, M.; Becker-Ziaja, B.; Ehimuan, J.; Asogun, D.A.; Fichet-Calvet, E.; Kleinsteuber, K.; Lelke, M.; ter Meulen, J.; Akpede, G.O.; et al. Current molecular epidemiology of Lassa virus in Nigeria. J. Clin. Microbiol. 2011, 49, 1157–1161. [Google Scholar] [CrossRef]
- Fichet-Calvet, E.; Olschlager, S.; Strecker, T.; Koivogui, L.; Becker-Ziaja, B.; Camara, A.B.; Soropogui, B.; Magassouba, N.; Gunther, S. Spatial and temporal evolution of Lassa virus in the natural host population in Upper Guinea. Sci. Rep. 2016, 6, 21977. [Google Scholar] [CrossRef]
- Thomson, E.; Ip, C.L.; Badhan, A.; Christiansen, M.T.; Adamson, W.; Ansari, M.A.; Bibby, D.; Breuer, J.; Brown, A.; Bowden, R.; et al. Comparison of next-generation sequencing technologies for comprehensive assessment of full-length hepatitis c viral genomes. J. Clin. Microbiol. 2016, 54, 2470–2484. [Google Scholar] [CrossRef]
- Davis, C.; Mgomella, G.S.; da Silva Filipe, A.; Frost, E.H.; Giroux, G.; Hughes, J.; Hogan, C.; Kaleebu, P.; Asiki, G.; McLauchlan, J.; et al. Highly diverse hepatitis c strains detected in sub-saharan Africa have unknown susceptibility to direct-acting antiviral treatments. Hepatology 2019, 69, 1426–1441. [Google Scholar] [CrossRef]
- Drummond, A.J.; Rambaut, A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 2007, 7, 214. [Google Scholar] [CrossRef]
- Baele, G.; Lemey, P.; Bedford, T.; Rambaut, A.; Suchard, M.A.; Alekseyenko, A.V. Improving the accuracy of demographic and molecular clock model comparison while accommodating phylogenetic uncertainty. Mol. Biol. Evol. 2012, 29, 2157–2167. [Google Scholar] [CrossRef]
- Bonwitt, J.; Kelly, A.H.; Ansumana, R.; Agbla, S.; Sahr, F.; Saez, A.M.; Borchert, M.; Kock, R.; Fichet-Calvet, E. Rat-atouille: A mixed method study to characterize rodent hunting and consumption in the context of lassa fever. Ecohealth 2016, 13, 234–247. [Google Scholar] [CrossRef]
- Fichet-Calvet, E.; Becker-Ziaja, B.; Koivogui, L.; Gunther, S. Lassa serology in natural populations of rodents and horizontal transmission. Vector. Borne Zoonotic. Dis. 2014, 14, 665–674. [Google Scholar] [CrossRef] [PubMed]
- Olayemi, A.; Oyeyiola, A.; Obadare, A.; Igbokwe, J.; Adesina, A.S.; Onwe, F.; Ukwaja, K.N.; Ajayi, N.A.; Rieger, T.; Gunther, S.; et al. Widespread arenavirus occurrence and seroprevalence in small mammals, Nigeria. Parasites Vectors 2018, 11, 416. [Google Scholar] [CrossRef] [PubMed]
- Coulibaly-N’Golo, D.; Allali, B.; Kouassi, S.K.; Fichet-Calvet, E.; Becker-Ziaja, B.; Rieger, T.; Olschlager, S.; Dosso, H.; Denys, C.; Ter Meulen, J.; et al. Novel arenavirus sequences in Hylomyscus sp. and Mus (Nannomys) setulosus from Cote d’Ivoire: Implications for evolution of arenaviruses in Africa. PLoS ONE 2011, 6, e20893. [Google Scholar] [CrossRef] [PubMed]
- Borremans, B.; Vossen, R.; Becker-Ziaja, B.; Gryseels, S.; Hughes, N.; Van Gestel, M.; Van Houtte, N.; Gunther, S.; Leirs, H. Shedding dynamics of Morogoro virus, an African arenavirus closely related to Lassa virus, in its natural reservoir host Mastomys natalensis. Sci. Rep. 2015, 5, 10445. [Google Scholar] [CrossRef] [PubMed]
- Walker, D.H.; Wulff, H.; Lange, J.V.; Murphy, F.A. Comparative pathology of Lassa virus infection in monkeys, guinea-pigs, and Mastomys natalensis. Bull. World Health Organ. 1975, 52, 523–534. [Google Scholar]
- Leski, T.A.; Stockelman, M.G.; Moses, L.M.; Park, M.; Stenger, D.A.; Ansumana, R.; Bausch, D.G.; Lin, B. Sequence variability and geographic distribution of Lassa virus, Sierra Leone. Emerg. Infect. Dis. 2015, 21, 609–618. [Google Scholar] [CrossRef]
- Paff, M.L.; Stolte, S.P.; Bull, J.J. Lethal mutagenesis failure may augment viral adaptation. Mol. Biol. Evol. 2014, 31, 96–105. [Google Scholar] [CrossRef][Green Version]
- Jankhöfer, C.H.; Marí Sáez, A.; Magassouba, N.; Soropogui, B.; Camara, A.; Günther, S.; Gabriel, M.; Fichet-Calvet, E.; Borchert, M. Lassa-seroprevalence and its predictors in Faranah prefecture, Guinea: A cross-sectional survey. In Proceedings of the 11th Europen Congress of Tropical Medicine and International Health, Liverpool, UK, 16–20 September 2019; p. S111. [Google Scholar]
| Species | Baoma | Foindu | Ngolahun | Nyandeyama | Sembehun | Yakaji | Total |
|---|---|---|---|---|---|---|---|
| Crocidura spp. | 1 | 1 | 3 | 2 | 7 | ||
| Hylomyscus simus | 1 | 1 | |||||
| Mastomys erythroleucus | 1 | 1 | 1 | 1 | 1 | 1 | 6 |
| Mastomys natalensis | 57 | 93 | 13 | 86 | 30 | 49 | 328 |
| Praomys rostratus | 2 | 2 | 4 | 1 | 9 | ||
| Rattus rattus | 61 | 25 | 38 | 55 | 39 | 30 | 248 |
| Total | 121 | 122 | 53 | 149 | 71 | 83 | 599 |
| % Mastomys natalensis | 47 | 76 | 25 | 58 | 42 | 59 | 54 |
| % Rattus rattus | 50 | 20 | 72 | 37 | 55 | 36 | 41 |
| Species | Village | Total | |||||
|---|---|---|---|---|---|---|---|
| Baoma | Foindu | Ngolahun | Nyandeyama | Sembehun | Yakaji | ||
| Crocidura spp. | 42 | 24 | 27 | 54 | 35 | 28 | 0/210 |
| Dasymys rufulus | 2 | 4 | 1 | 0/7 | |||
| Grammomys buntingi | 1 | 1 | 0/2 | ||||
| Graphiurus sp. | 1 | 0/1 | |||||
| Hybomys sp. | 3 | 4 | 3 | 1 | 5 | 0/16 | |
| Hylomyscus simus | 8 | 5 | 2 | 1 | 1 | 6 | 0/23 |
| Lemniscomys striatus | 1 | 1 | 1 | 1 | 0/4 | ||
| Lophuromys sikapusi | 1/41 | 14 | 33 | 1/25 | 31 | 30 | 2/174 |
| Malacomys edwardsi | 1 | 1/3 | 4 | 6 | 9 | 1/23 | |
| Mastomys erythroleucus | 4 | 1 | 2 | 2 | 2/3 | 2 | 2/14 |
| Mastomys natalensis | 5/60 | 2/96 | 2/16 | 12/100 | 3/34 | 5/51 | 29/357 |
| Nannomys minutoides | 4 | 3 | 2 | 1 | 1 | 0/11 | |
| Nannomys setulosus | 7 | 2 | 1 | 3 | 11 | 0/24 | |
| Praomys rostratus | 44 | 1/39 | 3/21 | 1/102 | 48 | 92 | 5/346 |
| Rattus rattus | 1/61 | 1/26 | 40 | 58 | 42 | 32 | 2/261 |
| Uranomys ruddi | 2 | 0/2 | |||||
| Total | 7/276 | 4/213 | 6/154 | 14/356 | 5/204 | 5/270 | 41/1473 (2.8%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bangura, U.; Buanie, J.; Lamin, J.; Davis, C.; Bongo, G.N.; Dawson, M.; Ansumana, R.; Sondufu, D.; Thomson, E.C.; Sahr, F.; et al. Lassa Virus Circulation in Small Mammal Populations in Bo District, Sierra Leone. Biology 2021, 10, 28. https://doi.org/10.3390/biology10010028
Bangura U, Buanie J, Lamin J, Davis C, Bongo GN, Dawson M, Ansumana R, Sondufu D, Thomson EC, Sahr F, et al. Lassa Virus Circulation in Small Mammal Populations in Bo District, Sierra Leone. Biology. 2021; 10(1):28. https://doi.org/10.3390/biology10010028
Chicago/Turabian StyleBangura, Umaru, Jacob Buanie, Joyce Lamin, Christopher Davis, Gédéon Ngiala Bongo, Michael Dawson, Rashid Ansumana, Dianah Sondufu, Emma C. Thomson, Foday Sahr, and et al. 2021. "Lassa Virus Circulation in Small Mammal Populations in Bo District, Sierra Leone" Biology 10, no. 1: 28. https://doi.org/10.3390/biology10010028
APA StyleBangura, U., Buanie, J., Lamin, J., Davis, C., Bongo, G. N., Dawson, M., Ansumana, R., Sondufu, D., Thomson, E. C., Sahr, F., & Fichet-Calvet, E. (2021). Lassa Virus Circulation in Small Mammal Populations in Bo District, Sierra Leone. Biology, 10(1), 28. https://doi.org/10.3390/biology10010028







