Genome-Wide Identification and Evolution of Receptor-Like Kinases (RLKs) and Receptor like Proteins (RLPs) in Brassica juncea
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Brassica Reference Genomes
2.2. Genome Wide Identification of RLK and RLP Genes
2.3. Genomic Distribution of RLK and RLP Genes
2.4. Gene Duplication Analysis of RLK and RLP Genes
2.5. Ortholog and Paralog Analysis
2.6. Multiple Alignment and Phylogenetic Analysis
3. Results
3.1. Genome-Wide Identification of RLK and RLP Genes in B. juncea
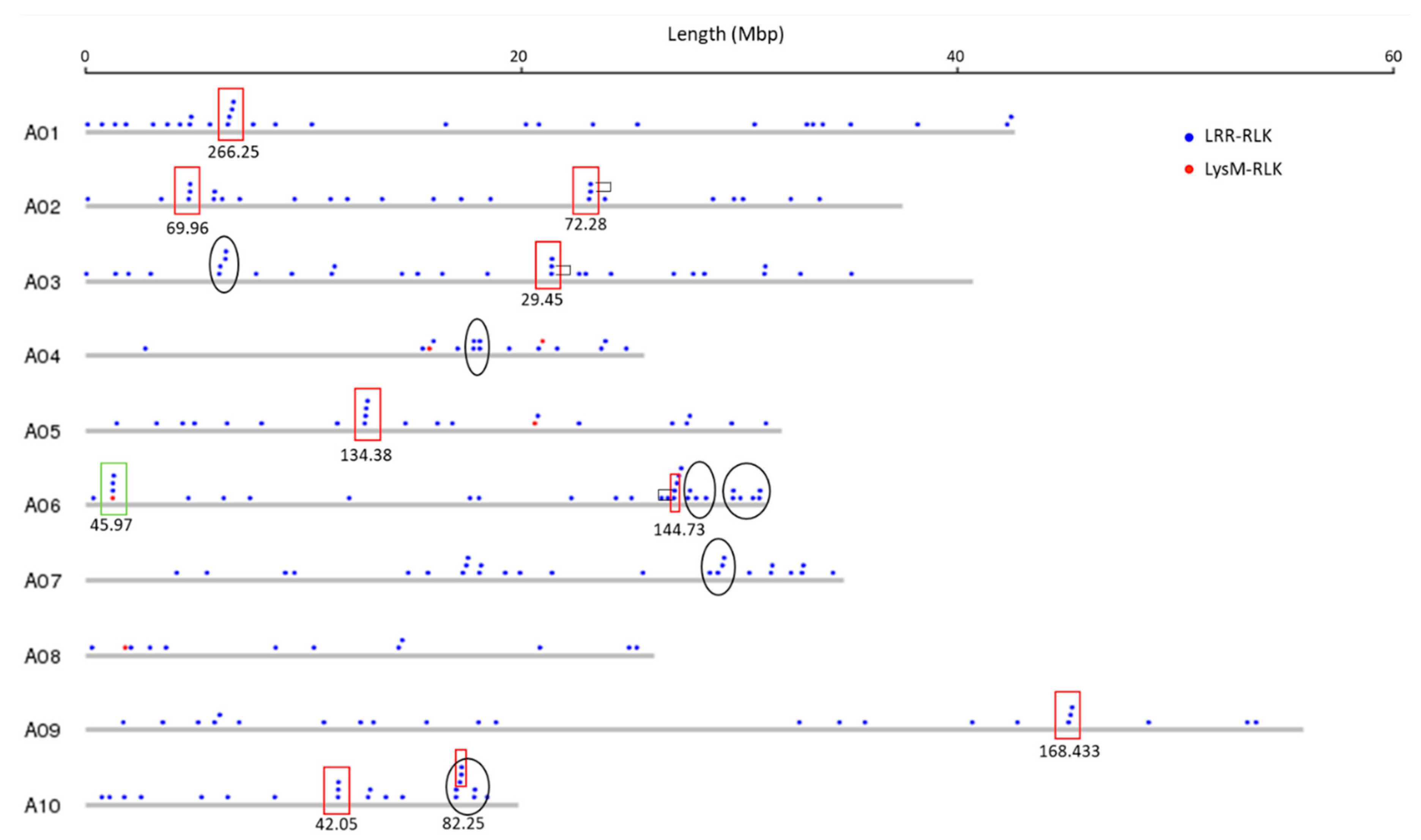
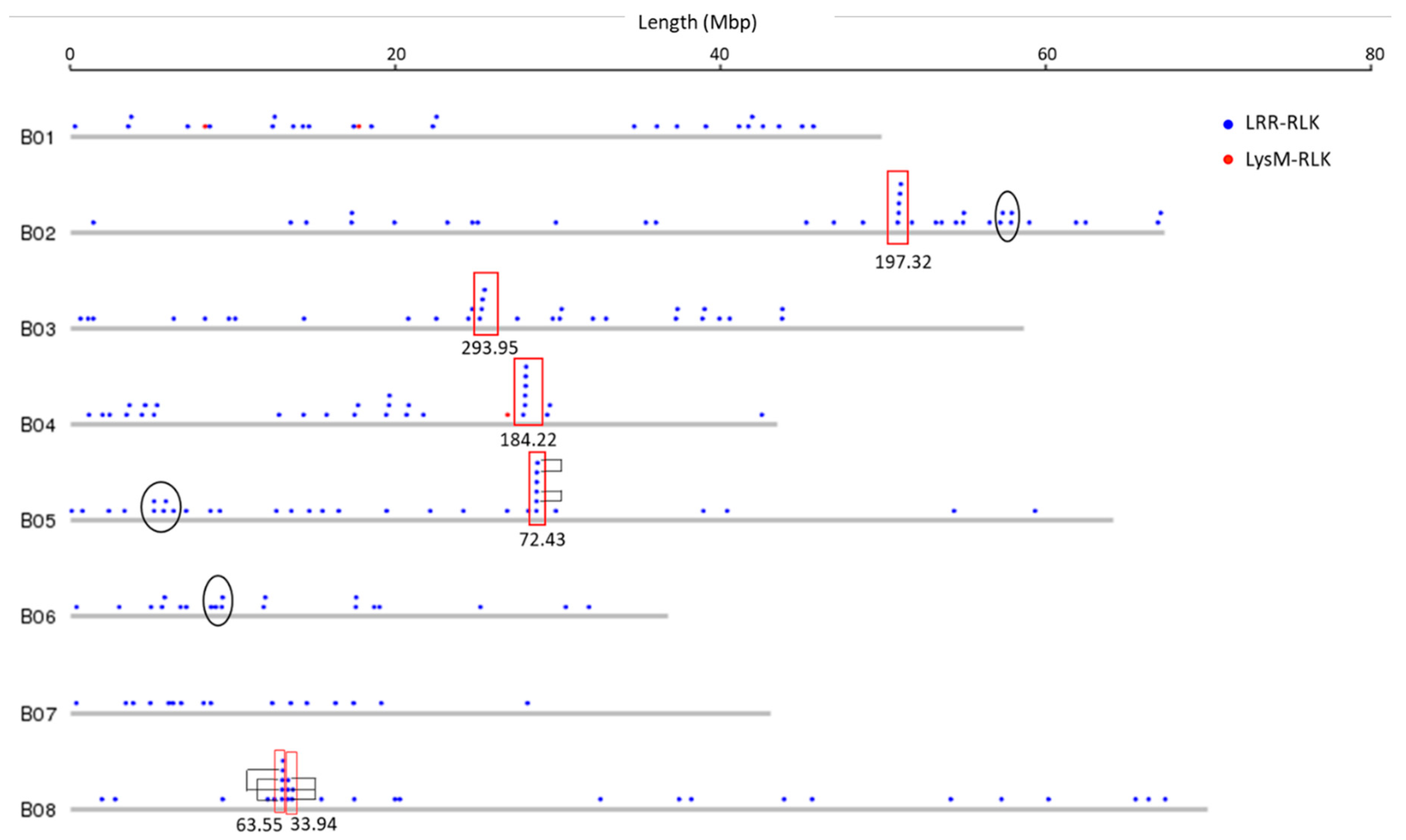
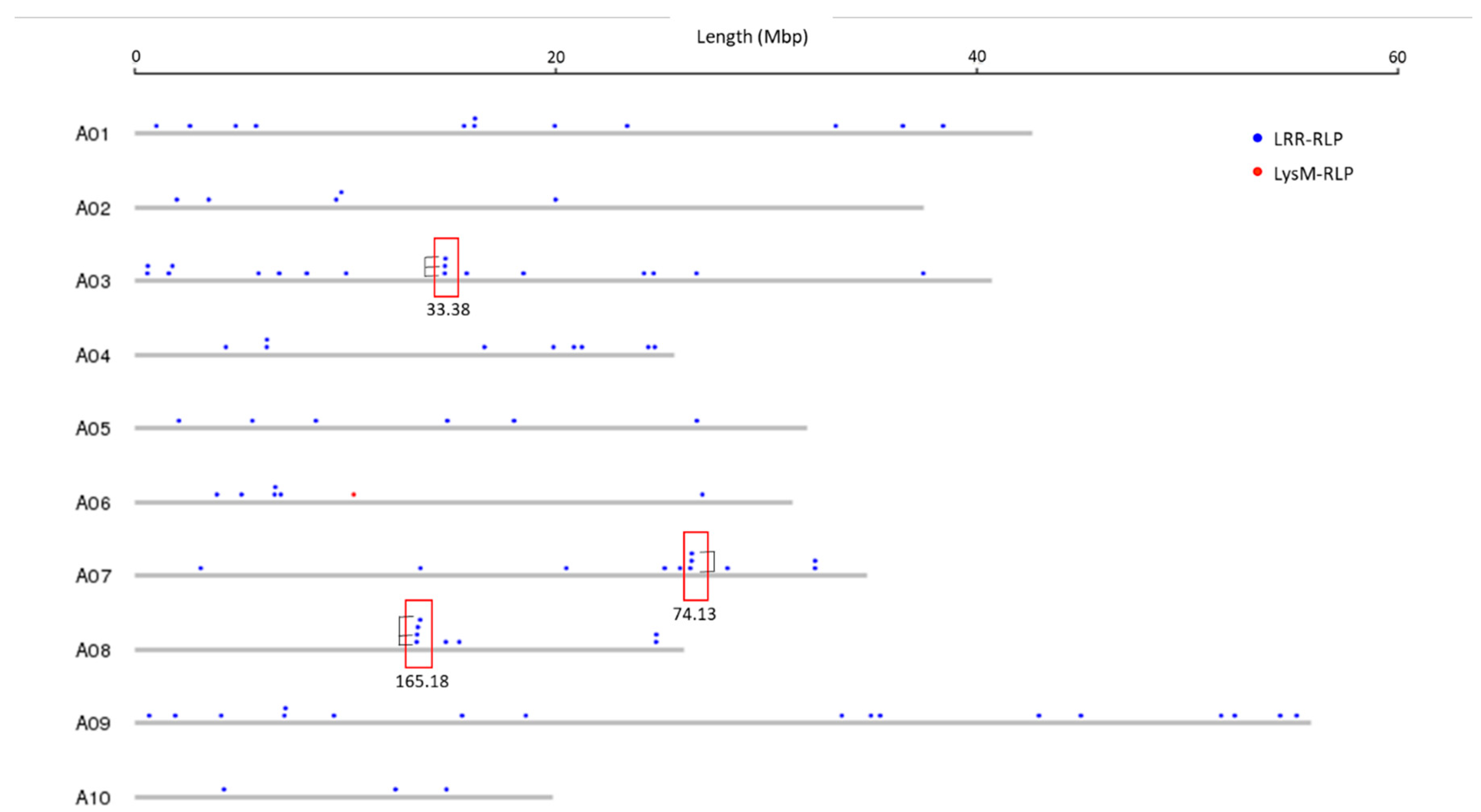
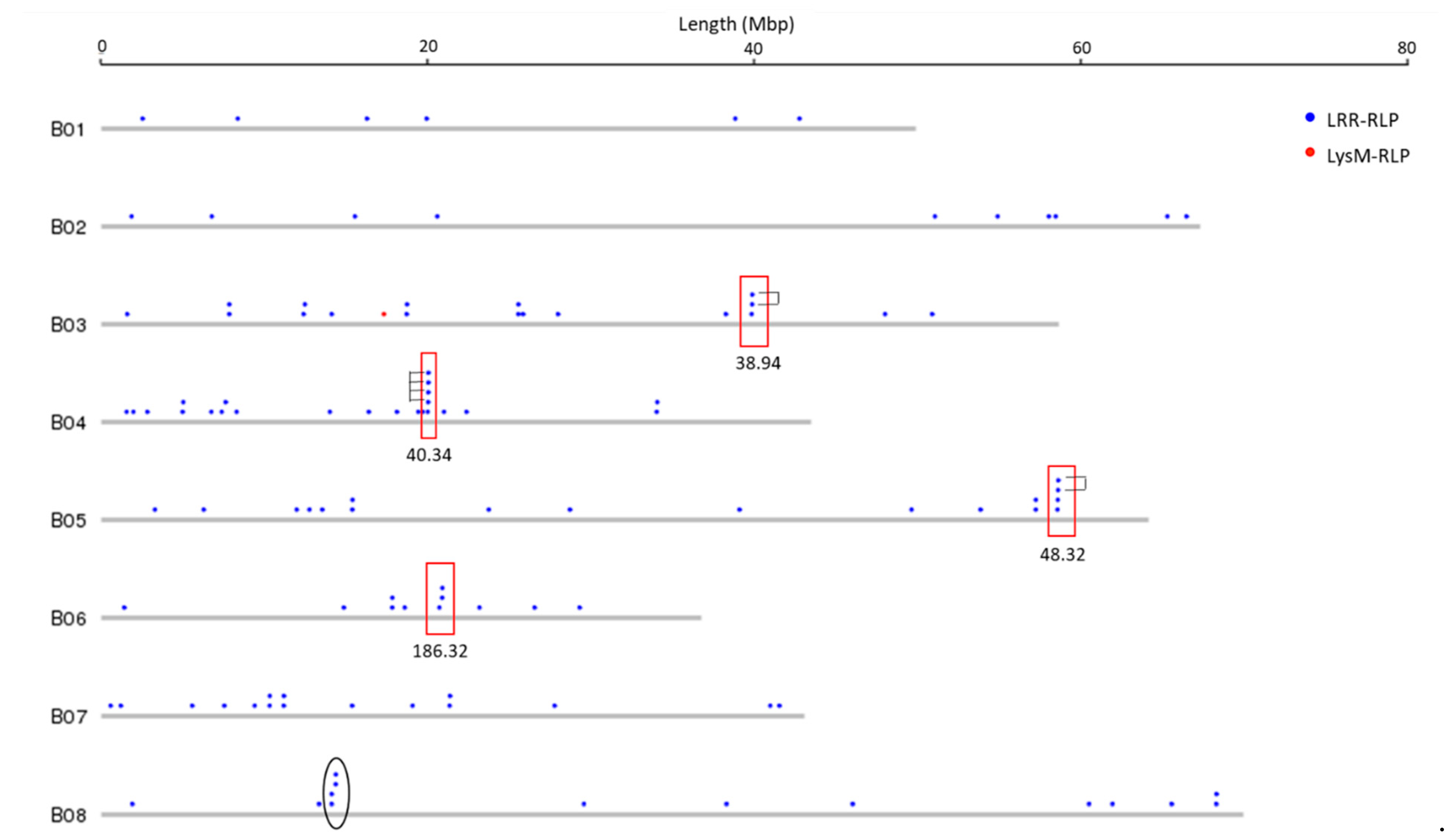
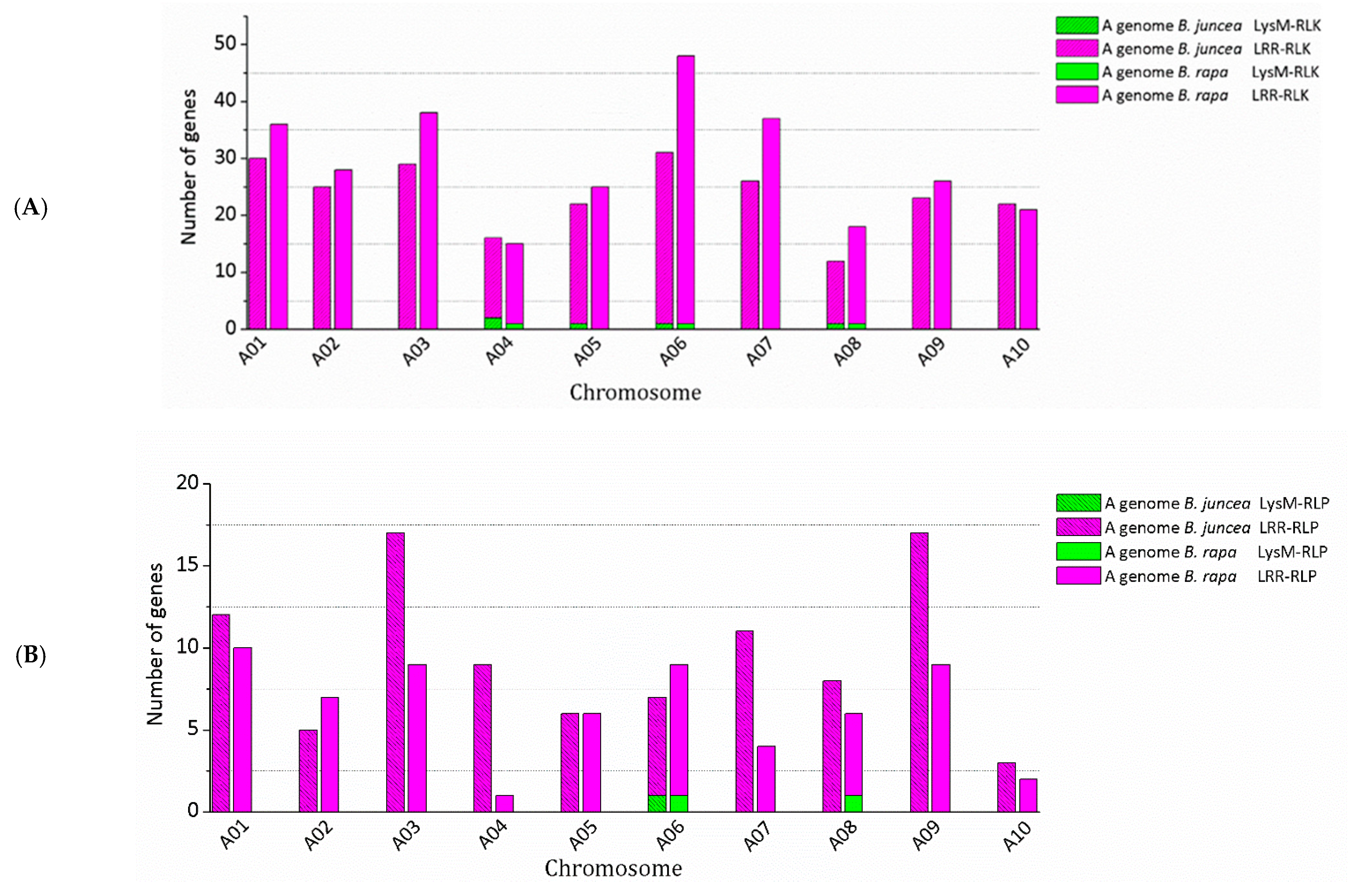
3.2. Genomic Distribution of RLK and RLP Genes in B. juncea
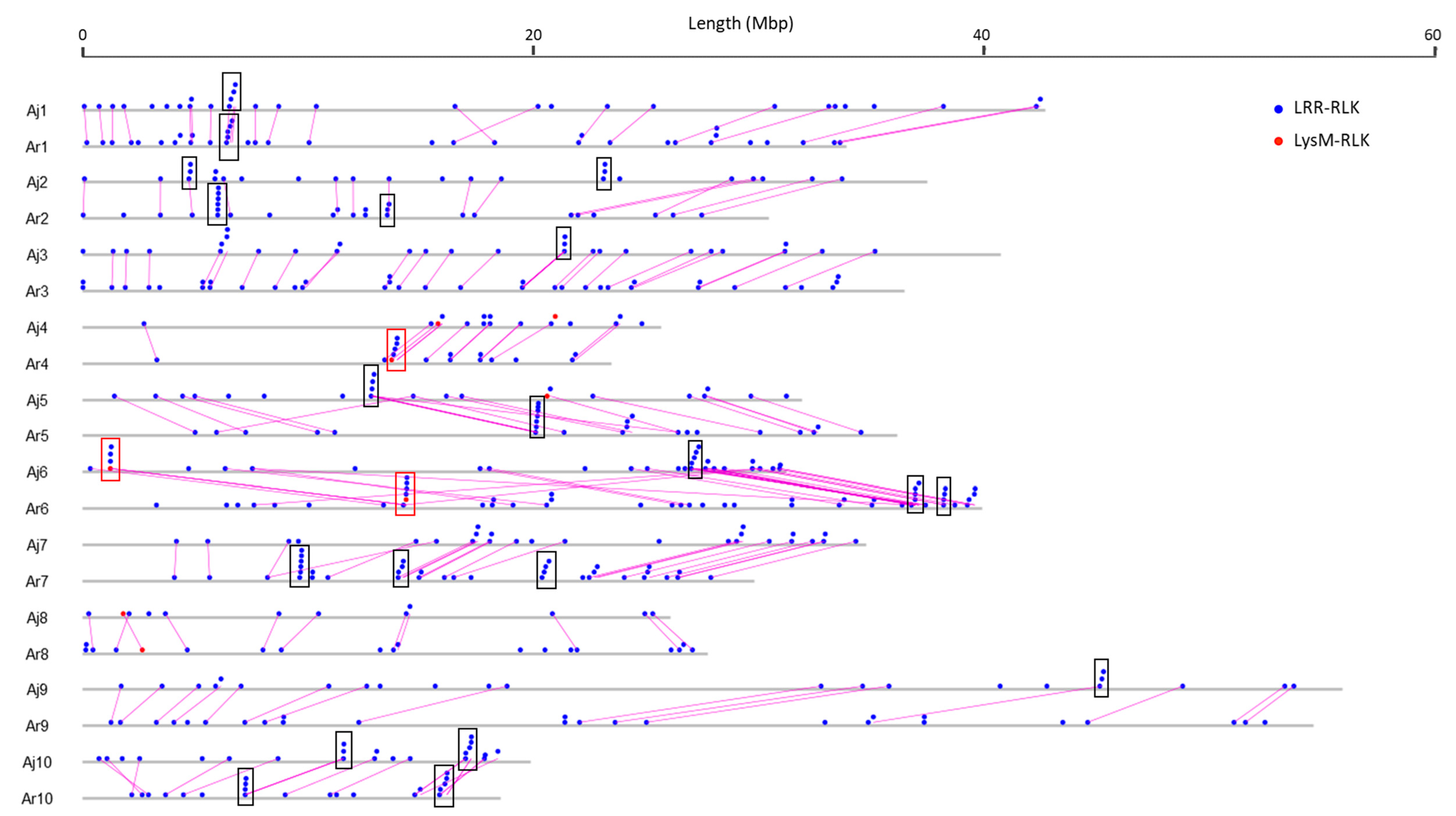
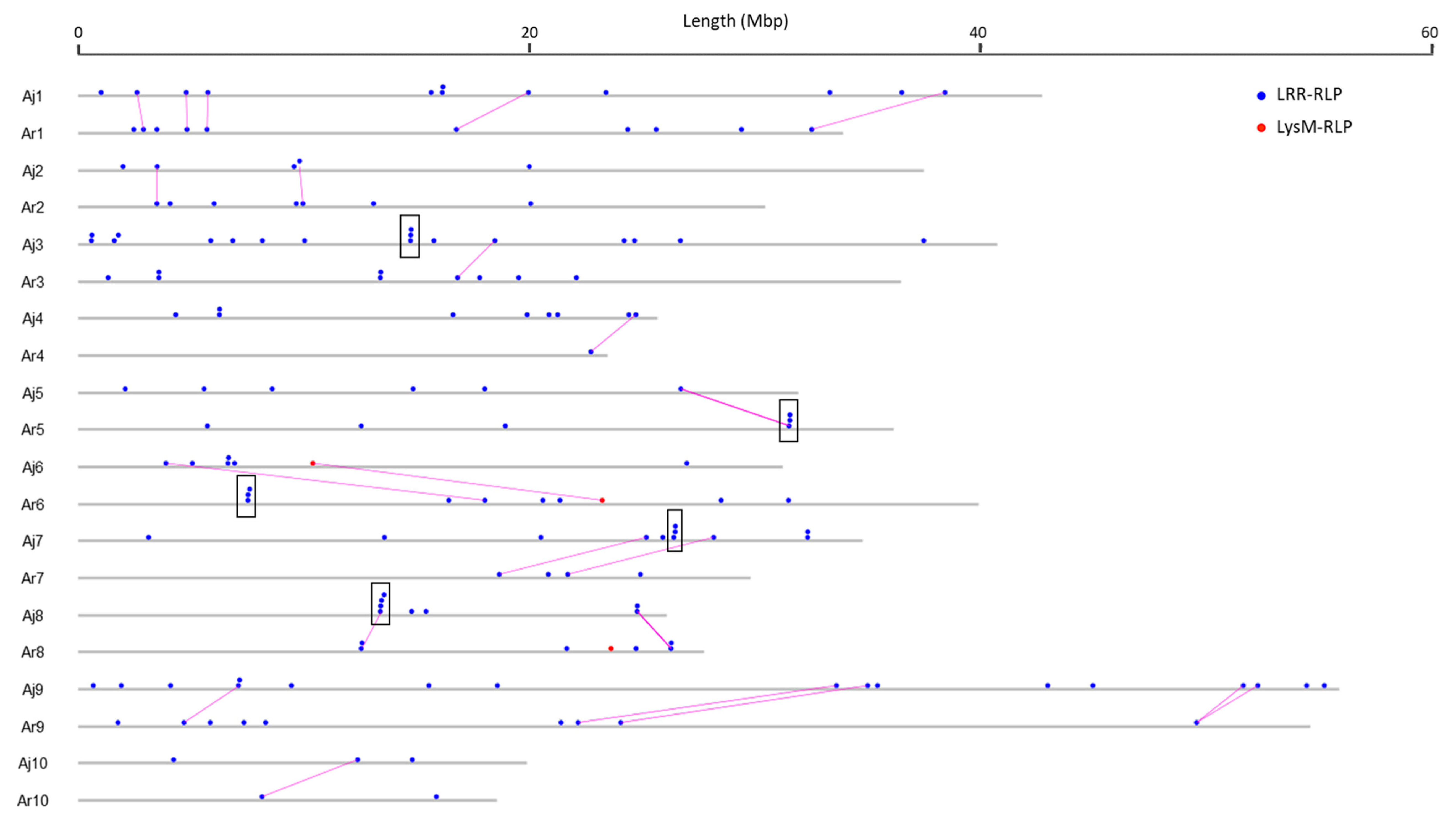
3.3. RLK and RLP Gene Clustering in B. juncea
3.4. Analysis of Duplications and Paralogues of RLK and RLP Genes in B. juncea
3.5. Phylogenetic Analysis of RLK and RLP Genes in B. juncea
3.6. Comparison and Conservation Analysis of RLK and RLP Genes between B. juncea and Its Diploid Progenitor Species B. rapa and B. nigra
4. Discussion
4.1. Genome Wide Identification of RLK and RLP Genes in B. juncea
4.2. RLK and RLP Gene Duplication in B. juncea
4.3. Phylogenetic Analysis
4.4. Comparative Analysis of RLK and RLP Genes in B. juncea to Its Related Diploid Progenitor Species B. rapa and B. nigra
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Monaghan, J.; Zipfel, C. Plant pattern recognition receptor complexes at the plasma membrane. Curr. Opin. Plant Biol. 2012, 15, 349–357. [Google Scholar] [CrossRef] [PubMed]
- Wolf, S. Plant cell wall signalling and receptor-like kinases. Biochem. J. 2017, 474, 471–492. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Zhou, J.M. Receptor-like kinases in plant innate immunity. J. Integr. Plant Biol. 2013, 55, 1271–1286. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Thomma, B.P.H.J. Structure-function aspects of extracellular leucine-rich repeat-containing cell surface receptors in plants. J. Integr. Plant Biol. 2013, 55, 1212–1223. [Google Scholar] [CrossRef]
- Cai, W.; Zhang, D. The role of receptor-like kinases in regulating plant male reproduction. Plant Reprod. 2018, 31, 77–87. [Google Scholar] [CrossRef]
- Ye, Y.; Ding, Y.; Jiang, Q.; Wang, F.; Sun, J.; Zhu, C. The role of receptor-like protein kinases (RLKs) in abiotic stress response in plants. Plant Cell Rep. 2017, 36, 235–242. [Google Scholar] [CrossRef]
- Tang, D.; Wang, G.; Zhou, J.M. Receptor kinases in plant-pathogen interactions: More than pattern recognition. Plant Cell 2017, 29, 618–637. [Google Scholar] [CrossRef]
- Kruijt, M.; de Kock, M.J.; de Wit, P.J. Receptor-like proteins involved in plant disease resistance. Mol. Plant Pathol. 2005, 6, 85–97. [Google Scholar] [CrossRef]
- Wang, G.; Fiers, M.; Ellendorff, U.; Wang, Z.; de Wit, P.J.; Angenent, G.C.; Thomma, B.P. The diverse roles of extracellular leucine-rich repeat-containing receptor-like proteins in plants. Crit. Rev. Plant Sci. 2010, 29, 285–299. [Google Scholar] [CrossRef]
- Shiu, S.H.; Bleecker, A.B. Plant receptor-like kinase gene family: Diversity, function, and signaling. Sci. Stke 2001, 2001, 22. [Google Scholar] [CrossRef]
- Shiu, S.H.; Karlowski, W.M.; Pan, R.; Tzeng, Y.H.; Mayer, K.F.; Li, W.H. Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 2004, 16, 1220–1234. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Ellendorff, U.; Kemp, B.; Mansfield, J.W.; Forsyth, A.; Mitchell, K.; Bastas, K.; Liu, C.M.; Woods-Tör, A.; Zipfel, C.; et al. A genome-wide functional investigation into the roles of receptor-like proteins in Arabidopsis. Plant Physiol. 2008, 147, 503. [Google Scholar] [CrossRef] [PubMed]
- Liebrand, T.W.; van den Berg, G.C.; Zhang, Z.; Smit, P.; Cordewener, J.H.; America, A.H.; Sklenar, J.; Jones, A.M.; Tameling, W.I.; Robatzek, S. Receptor-like kinase SOBIR1/EVR interacts with receptor-like proteins in plant immunity against fungal infection. Proc. Natl. Acad. Sci. USA 2013, 110, 10010–10015. [Google Scholar] [CrossRef]
- Liebrand, T.W.; van den Burg, H.A.; Joosten, M.H. Two for all: Receptor-associated kinases SOBIR1 and BAK1. Trends Plant Sci. 2014, 19, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Gust, A.A.; Felix, G. Receptor like proteins associate with SOBIR1-type of adaptors to form bimolecular receptor kinases. Curr. Opin. Plant Biol. 2014, 21, 104–111. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.A.; Thomas, C.M.; Hammond-Kosack, K.E.; Balint-Kurti, P.J.; Jones, J.D.G. Isolation of the tomato Cf-9 gene for resistance to Cladosporium fulvum by transposon tagging. Science 1994, 266, 789–793. [Google Scholar] [CrossRef] [PubMed]
- Walker, J.C.; Zhang, R. Relationship of a putative receptor protein kinase from maize to the S-locus glycoproteins of Brassica. Nature 1990, 345, 743. [Google Scholar] [CrossRef]
- Shiu, S.H.; Bleecker, A.B. Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc. Natl. Acad. Sci. USA 2001, 98, 10763–10768. [Google Scholar] [CrossRef]
- Fischer, I.; Diévart, A.; Droc, G.; Dufayard, J.F.; Chantret, N. Evolutionary dynamics of the leucine-rich repeat receptor-like kinase (LRR-RLK) subfamily in Angiosperms. Plant Physiol. 2016, 170, 1595–1610. [Google Scholar] [CrossRef]
- Petre, B.; Hacquard, S.; Duplessis, S.; Rouhier, N. Genome analysis of poplar LRR-RLP gene clusters reveals RISP, a defense-related gene coding a candidate endogenous peptide elicitor. Front. Plant Sci. 2014, 5, 111. [Google Scholar] [CrossRef]
- Fritz-Laylin, L.K.; Krishnamurthy, N.; Tör, M.; Sjölander, K.V.; Jones, J.D.G. Phylogenomic analysis of the receptor-like proteins of rice and Arabidopsis. Plant Physiol. 2005, 138, 611–623. [Google Scholar] [CrossRef] [PubMed]
- Shiu, S.H.; Bleecker, A.B. Expansion of the receptor-like kinase/pelle gene family and receptor-like proteins in Arabidopsis. Plant Physiol. 2003, 132, 530–543. [Google Scholar] [CrossRef] [PubMed]
- Miya, A.; Albert, P.; Shinya, T.; Desaki, Y.; Ichimura, K.; Shirasu, K.; Narusaka, Y.; Kawakami, N.; Kaku, H.; Shibuya, N. CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 2007, 104, 19613–19618. [Google Scholar] [CrossRef] [PubMed]
- Erwig, J. Analysis of the Subcellular Behavior of Arabidopsis thaliana LysM-Proteins and Their Role in Plant Innate Immunity. Ph.D. Thesis, Georg-August-Universität Göttingen, Göttingen, Germany, April 2016. [Google Scholar]
- Wan, J.; Zhang, X.C.; Neece, D.; Ramonell, K.M.; Clough, S.; Kim, S.Y.; Stacey, M.G.; Stacey, G. A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell 2008, 20, 471–481. [Google Scholar] [CrossRef]
- Petutschnig, E.K.; Jones, A.M.; Serazetdinova, L.; Lipka, U.; Lipka, V. The lysin motif receptor-like kinase (LysM-RLK) CERK1 is a major chitin-binding protein in Arabidopsis thaliana and subject to chitin-induced phosphorylation. J. Biol. Chem. 2010, 285, 28902–28911. [Google Scholar] [CrossRef]
- Kelly, S.; Radutoiu, S.; Stougaard, J. Legume LysM receptors mediate symbiotic and pathogenic signalling. Curr. Opin. Plant Biol. 2017, 39, 152–158. [Google Scholar] [CrossRef]
- Gust, A.A.; Willmann, R.; Desaki, Y.; Grabherr, H.M.; Nürnberger, T. Plant LysM proteins: Modules mediating symbiosis and immunity. Trends Plant Sci. 2012, 17, 495–502. [Google Scholar] [CrossRef]
- Limpens, E.; Franken, C.; Smit, P.; Willemse, J.; Bisseling, T.; Geurts, R. LysM domain receptor kinases regulating rhizobial nod factor-induced infection. Science 2003, 302, 630–633. [Google Scholar] [CrossRef]
- Madsen, E.B.; Madsen, L.H.; Radutoiu, S.; Olbryt, M.; Rakwalska, M.; Szczyglowski, K.; Sato, S.; Kaneko, T.; Tabata, S.; Sandal, N. A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 2003, 425, 637. [Google Scholar] [CrossRef]
- Radutoiu, S.; Madsen, L.H.; Madsen, E.B.; Felle, H.H.; Umehara, Y.; Grønlund, M.; Sato, S.; Nakamura, Y.; Tabata, S.; Sandal, N. Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 2003, 425, 585. [Google Scholar] [CrossRef]
- Nagaharu, U.; Nagaharu, N. Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Jap. J. Bot. 1935, 7, 389–452. [Google Scholar]
- Yang, J.; Liu, D.; Wang, X.; Ji, C.; Cheng, F.; Liu, B.; Hu, Z.; Chen, S.; Pental, D.; Ju, Y.; et al. The genome sequence of allopolyploid Brassica juncea and analysis of differential homoeolog gene expression influencing selection. Nat. Genet. 2016, 48, 1225. Available online: http://www.nature.com/ng/journal/vaop/ncurrent/abs/ng.3657.html#supplementary-information (accessed on 1 January 2017). [CrossRef] [PubMed]
- Oram, R.N.; Kirk, J.T.O.; Veness, P.E.; Hurlstone, C.J.; Edlington, J.P.; Halsall, D.M. Breeding Indian mustard [Brassica juncea (L.) Czern.] for cold-pressed, edible oil production—A review. Aust. J. Agric. Res. 2005, 56, 581–596. [Google Scholar] [CrossRef]
- Chèvre, A.M.; Barret, P.; Eber, F.; Dupuy, P.; Brun, H.; Tanguy, X.; Renard, M. Selection of stable Brassica napus-B. juncea recombinant lines resistant to blackleg (Leptosphaeria maculans). 1. Identification of molecular markers, hromosomal and genomic origin of the introgression. Theor. Appl. Genet. 1997, 95, 1104–1111. [Google Scholar]
- Bhardwaj, A.R.; Joshi, G.; Kukreja, B.; Malik, V.; Arora, P.; Pandey, R.; Shukla, R.N.; Bankar, K.G.; Katiyar-Agarwal, S.; Goel, S.; et al. Global insights into high temperature and drought stress regulated genes by RNA-Seq in economically important oilseed crop Brassica juncea. BMC Plant Biol. 2015, 1, 1–15. [Google Scholar] [CrossRef]
- Van de Wouw, A.P.; Lowe, R.G.; Elliott, C.E.; Dubois, D.J.; Howlett, B.J. An avirulence gene, AvrLmJ1, from the blackleg fungus, Leptosphaeria maculans, confers avirulence to Brassica juncea cultivars. Mol. Plant Pathol. 2014, 15, 523–530. [Google Scholar] [CrossRef]
- Potter, T. Development of Brassica juncea as a biodiesel feedstock in low rainfall areas of Australia. In Proceedings of the 13th International Rapeseed Congress, Prague, Crech Republic, 5–9 June 2011. [Google Scholar]
- Elliott, V.L.; Marcroft, S.J.; Norton, R.M.; Salisbury, P.A. Reaction of Brassica juncea to Australian isolates of Leptosphaeria maculans and Leptosphaeria biglobosa ‘canadensis’. Can. J. Plant Pathol. 2011, 33, 38–48. [Google Scholar] [CrossRef]
- Inturrisi, F.; Barbetti, M.; Tirnaz, S.; Patel, D.; Edwards, D.; Batley, J. Molecular characterisation of disease resistance in Brassica juncea–the current status and the way forward. Plant Pathol. 2021, 70, 13–34. [Google Scholar] [CrossRef]
- Inturrisi, F.; Bayer, P.E.; Yang, H.; Tirnaz, S.; Edwards, D.; Batley, J. Genome-wide identification and comparative analysis of resistance genes in Brassica juncea. Mol. Breed. 2020, 40, 78. [Google Scholar] [CrossRef]
- Cai, C.; Wang, X.; Liu, B.; Wu, J.; Liang, J.; Cui, Y.; Cheng, F.; Wang, X. Brassica rapa genome 2.0: A reference upgrade through sequence re-assembly and gene re-annotation. Mol. Plant 2017, 10, 649–651. [Google Scholar] [CrossRef]
- Li, P.; Quan, X.; Jia, G.; Xiao, J.; Cloutier, S.; You, F.M. RGAugury: A pipeline for genome-wide prediction of resistance gene analogs (RGAs) in plants. BMC Genom. 2016, 17, 852. [Google Scholar] [CrossRef]
- Holub, E.B. The arms race is ancient history in Arabidopsis, the wildflower. Nat. Rev. Genet. 2001, 2, 516–527. [Google Scholar] [CrossRef] [PubMed]
- Alamery, S.; Tirnaz, S.; Bayer, P.; Tollenaere, R.; Chaloub, B.; Edwards, D.; Batley, J. Genome-wide identification and comparative analysis of NBS-LRR resistance genes in Brassica napus. Crop. Pasture Sci. 2018, 69, 72–93. [Google Scholar] [CrossRef]
- Wei, Z.; Wang, J.; Yang, S.; Song, Y. Identification and expression analysis of the LRR-RLK gene family in tomato (Solanum lycopersicum) Heinz 1706. Genome 2015, 58, 121–134. [Google Scholar] [CrossRef]
- Zhou, T.; Wang, Y.; Chen, J.Q.; Araki, H.; Jing, Z.; Jiang, K.; Shen, J.; Tian, D. Genome-wide identification of NBS genes in japonica rice reveals significant expansion of divergent non-TIR NBS-LRR genes. Mol. Genet. Genom. 2004, 271, 402–415. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Kumar, R.; Mendu, V.; Singh, K.; Upadhyay, S.K. Genomic dissection and expression profiling revealed functional divergence in Triticum aestivum leucine rich repeat receptor like kinases (TaLRRKs). Front. Plant Sci. 2016, 7, 1374. [Google Scholar]
- Sharma, S.; Pandey, A.K.; Singh, K.; Upadhyay, S.K. Molecular characterization and global expression analysis of lectin receptor kinases in bread wheat (Triticum aestivum). PLoS ONE 2016, 11, e0153925. [Google Scholar]
- Agarwal, G.; Garg, V.; Kudapa, H.; Doddamani, D.; Pazhamala, L.T.; Khan, A.W.; Thudi, M.; Lee, S.H.; Varshney, R.K. Genome-wide dissection of AP2/ERF and HSP90 gene families in five legumes and expression profiles in chickpea and pigeonpea. Plant Biotechnol. J. 2016, 14, 1563–1577. [Google Scholar] [CrossRef]
- Sonnhammer, E.L.; Koonin, E.V. Orthology, paralogy and proposed classification for paralog subtypes. Trends Genet. 2002, 18, 619–620. [Google Scholar] [CrossRef]
- Rameneni, J.J.; Lee, Y.; Dhandapani, V.; Yu, X.; Choi, S.R.; Oh, M.H.; Lim, Y.P. Genomic and post-translational modification analysis of leucine-rich-repeat receptor-like kinases in Brassica rapa. PLoS ONE 2015, 10, e0142255. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Diévart, A.; Gilbert, N.; Droc, G.; Attard, A.; Gourgues, M.; Guiderdoni, E.; Périn, C. Leucine-rich repeat receptor kinases are sporadically distributed in eukaryotic genomes. BMC Evol. Biol. 2011, 11, 367. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Wang, G.L. Genome-wide identification, characterization and phylogenetic analysis of the rice LRR-kinases. PLoS ONE 2011, 6, e16079. [Google Scholar] [CrossRef] [PubMed]
- Zan, Y.; Ji, Y.; Zhang, Y.; Yang, S.; Song, Y.; Wang, J. Genome-wide identification, characterization and expression analysis of populus leucine-rich repeat receptor-like protein kinase genes. BMC Genom. 2013, 14, 318. [Google Scholar] [CrossRef]
- Zhou, F.; Guo, Y.; Qiu, L.J. Genome-wide identification and evolutionary analysis of leucine-rich repeat receptor-like protein kinase genes in soybean. BMC Plant Biol. 2016, 16, 58. [Google Scholar] [CrossRef] [PubMed]
- International Wheat Genome Sequencing Consortium (IWGSC). A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 2014, 345, 1251788. [Google Scholar] [CrossRef] [PubMed]
- Schmutz, J.; Cannon, S.B.; Schlueter, J.; Ma, J.; Mitros, T.; Nelson, W.; Hyten, D.L.; Song, Q.; Thelen, J.J.; Cheng, J. Genome sequence of the palaeopolyploid soybean. Nature 2010, 463, 178. [Google Scholar] [CrossRef]
- Tirnaz, S.; Bayer, P.; Inturrisi, F.; Zhang, F.; Yang, H.; Dolatabadian, A.; Neik, T.X.; Severn-Ellis, A.; Patel, D.; Ibrahim, M.I.; et al. Resistance gene analogs in the Brassicaceae: Identification, characterization, distribution, and evolution. Plant Physiol. 2020. [Google Scholar] [CrossRef]
- Lohmann, G.V.; Shimoda, Y.; Nielsen, M.W.; Jørgensen, F.G.; Grossmann, C.; Sandal, N.; Sørensen, K.; Thirup, S.; Madsen, L.H.; Tabata, S. Evolution and regulation of the Lotus japonicus LysM receptor gene family. Mol. Plant Microbe Interact. 2010, 23, 510–521. [Google Scholar] [CrossRef]
- Arrighi, J.F.; Barre, A.; Amor, B.B.; Bersoult, A.; Soriano, L.C.; Mirabella, R.; de Carvalho-Niebel, F.; Journet, E.P.; Ghérardi, M.; Huguet, T. The Medicago truncatula lysine motif-receptor-like kinase gene family includes NFP and new nodule-expressed genes. Plant Physiol. 2006, 142, 265–279. [Google Scholar] [CrossRef]
- Zhang, X.C.; Wu, X.; Findley, S.; Wan, J.; Libault, M.; Nguyen, H.T.; Cannon, S.B.; Stacey, G. Molecular evolution of lysin motif-type receptor-like kinases in plants. Plant Physiol. 2007, 144, 623–636. [Google Scholar] [CrossRef]
- Kouzai, Y.; Mochizuki, S.; Nakajima, K.; Desaki, Y.; Hayafune, M.; Miyazaki, H.; Yokotani, N.; Ozawa, K.; Minami, E.; Kaku, H.; et al. Targeted gene disruption of OsCERK1 reveals its indispensable role in chitin perception and involvement in the peptidoglycan response and immunity in rice. Mol. Plant Microbe Interact. 2014, 27, 975–982. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.C.; Cannon, S.B.; Stacey, G. Evolutionary genomics of LysM genes in land plants. BMC Evol. Biol. 2009, 9, 183. [Google Scholar] [CrossRef]
- Liu, Q.; Chang, S.; Hartman, G.L.; Domier, L.L. Assembly and annotation of a draft genome sequence for Glycine latifolia, a perennial wild relative of soybean. Plant J. Cell Mol. Biol. 2018, 95, 71–85. [Google Scholar] [CrossRef] [PubMed]
- Magalhães, D.M.; Scholte, L.L.; Silva, N.V.; Oliveira, G.C.; Zipfel, C.; Takita, M.A.; De Souza, A.A. LRR-RLK family from two Citrus species: Genome-wide identification and evolutionary aspects. BMC Genom. 2016, 17, 623. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.; Shao, Z.Q.; Wu, X.Z.; Wang, Q.; Wang, B.; Chen, J.Q.; Hang, Y.Y.; Xue, J.Y. Loss/retention and evolution of NBS-encoding genes upon whole genome triplication of Brassica rapa. Gene 2014, 540, 54–61. [Google Scholar] [CrossRef]
- Chen, J.; Piao, Y.; Liu, Y.; Li, X.; Piao, Z. Genome-wide identification and expression analysis of chitinase gene family in Brassica rapa reveals its role in clubroot resistance. Plant Sci. 2018, 270, 257–267. [Google Scholar] [CrossRef] [PubMed]
- Friedman, A.R.; Baker, B.J. The evolution of resistance genes in multi-protein plant resistance systems. Curr. Opin. Genet. & Dev. 2007, 17, 493–499. [Google Scholar]
- Zhang, J. Evolution by gene duplication: An update. Trends Ecol. & Evol. 2003, 18, 292–298. [Google Scholar]
- Flagel, L.E.; Wendel, J.F. Gene duplication and evolutionary novelty in plants. New Phytol. 2009, 183, 557–564. [Google Scholar] [CrossRef]
- Moore, R.C.; Purugganan, M.D. The early stages of duplicate gene evolution. Proc. Natl. Acad. Sci. USA 2003, 100, 15682–15687. [Google Scholar] [CrossRef]
- Hwang, S.-G.; Kim, D.S.; Jang, C.S. Comparative analysis of evolutionary dynamics of genes encoding leucine-rich repeat receptor-like kinase between rice and Arabidopsis. Genetica 2011, 139, 1023–1032. [Google Scholar] [CrossRef]
- Sun, J.; Li, L.; Wang, P.; Zhang, S.; Wu, J. Genome-wide characterization, evolution, and expression analysis of the leucine-rich repeat receptor-like protein kinase (LRR-RLK) gene family in Rosaceae genomes. BMC Genom. 2017, 18, 763. [Google Scholar] [CrossRef]
- Bai, J.; Pennill, L.A.; Ning, J.; Lee, S.W.; Ramalingam, J.; Webb, C.A.; Zhao, B.; Sun, Q.; Nelson, J.C.; Leach, J.E. Diversity in nucleotide binding site-leucine-rich repeat genes in cereals. Genome Res. 2002, 12, 1871–1884. [Google Scholar] [CrossRef]
- Liu, S.; Liu, Y.; Yang, X.; Tong, C.; Edwards, D.; Parkin, I.A.; Zhao, M.; Ma, J.; Yu, J.; Huang, S. The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat. Commun. 2014, 5, 3930. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, H.; Wang, J.; Sun, R.; Wu, J.; Liu, S.; Bai, Y.; Mun, J.-H.; Bancroft, I.; Cheng, F. The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 2011, 43, 1035. [Google Scholar] [CrossRef]
- Panjabi, P.; Jagannath, A.; Bisht, N.C.; Padmaja, K.L.; Sharma, S.; Gupta, V.; Pradhan, A.K.; Pental, D. Comparative mapping of Brassica juncea and Arabidopsis thaliana using Intron Polymorphism (IP) markers: Homoeologous relationships, diversification and evolution of the A, B and C Brassica genomes. BMC Genom. 2008, 9, 113. [Google Scholar] [CrossRef] [PubMed]
- Lysak, M.A.; Koch, M.A.; Pecinka, A.; Schubert, I. Chromosome triplication found across the tribe Brassiceae. Genome Res. 2005, 15, 516–525. [Google Scholar] [CrossRef] [PubMed]
- Axelsson, T.; Bowman, C.; Sharpe, A.; Lydiate, D.; Lagercrantz, U. Amphidiploid Brassica juncea contains conserved progenitor genomes. Genome 2000, 43, 679–688. [Google Scholar] [CrossRef]
- Cheung, W.; Friesen, L.; Rakow, G.; Seguin-Swartz, G.; Landry, B. A RFLP-based linkage map of mustard [Brassica juncea (L.) Czern. and Coss.]. Theor. Appl. Genet. 1997, 94, 841–851. [Google Scholar] [CrossRef]
- Paritosh, K.; Gupta, V.; Yadava, S.K.; Singh, P.; Pradhan, A.K.; Pental, D. RNA-seq based SNPs for mapping in Brassica juncea (AABB): Synteny analysis between the two constituent genomes A (from B. rapa) and B (from B. nigra) shows highly divergent gene block arrangement and unique block fragmentation patterns. BMC Genom. 2014, 15, 396. [Google Scholar] [CrossRef][Green Version]
- Liu, P.L.; Xie, L.L.; Li, P.W.; Mao, J.F.; Liu, H.; Gao, S.M.; Shi, P.H.; Gong, J.Q. Duplication and divergence of leucine-rich repeat receptor-like protein kinase (LRR-RLK) genes in basal angiosperm Amborella trichopoda. Front. Plant Sci. 2016, 7, 1952. [Google Scholar] [CrossRef] [PubMed]
- Cannon, S.B.; Zhu, H.; Baumgarten, A.M.; Spangler, R.; May, G.; Cook, D.R.; Young, N.D. Diversity, distribution, and ancient taxonomic relationships within the TIR and non-TIR NBS-LRR resistance gene subfamilies. J. Mol. Evol. 2002, 54, 548–562. [Google Scholar] [CrossRef] [PubMed]
- Wan, H.; Yuan, W.; Bo, K.L.; Shen, J.; Pang, X.; Chen, J.F. Genome-wide analysis of NBS-encoding disease resistance genes in Cucumis sativus and phylogenetic study of NBS-encoding genes in Cucurbitaceae crops. BMC Genom. 2013, 14, 109. [Google Scholar] [CrossRef] [PubMed]
- Marcussen, T.; Sandve, S.R.; Heier, L.; Spannagl, M.; Pfeifer, M.; Jakobsen, K.S.; Wulff, B.B.; Steuernagel, B.; Mayer, K.F.; Olsen, O.A. Ancient hybridizations among the ancestral genomes of bread wheat. Science 2014, 345, 1250092. [Google Scholar] [CrossRef]
- Petersen, G.; Seberg, O.; Yde, M.; Berthelsen, K. Phylogenetic relationships of Triticum and Aegilops and evidence for the origin of the A, B, and D genomes of common wheat (Triticum aestivum). Mol. Phylogenetics Evol. 2006, 39, 70–82. [Google Scholar] [CrossRef]
- Salamini, F.; Özkan, H.; Brandolini, A.; Schäfer-Pregl, R.; Martin, W. Genetics and geography of wild cereal domestication in the Near East. Nat. Rev. Genet. 2002, 3, 429. [Google Scholar] [CrossRef]
- Liu, P.L.; Du, L.; Huang, Y.; Gao, S.M.; Yu, M. Origin and diversification of leucine-rich repeat receptor-like protein kinase (LRR-RLK) genes in plants. BMC Evol. Biol. 2017, 17, 47. [Google Scholar] [CrossRef]
- Porter, B.W.; Paidi, M.; Ming, R.; Alam, M.; Nishijima, W.T.; Zhu, Y.J. Genome-wide analysis of Carica papaya reveals a small NBS resistance gene family. Mol. Genet. Genom. 2009, 281, 609–626. [Google Scholar] [CrossRef]
- Lagercrantz, U. Comparative mapping between Arabidopsis thaliana and Brassica nigra indicates that Brassica genomes have evolved through extensive genome replication accompanied by chromosome fusions and frequent rearrangements. Genetics 1998, 150, 1217–1228. [Google Scholar]
- Grant, D.; Cregan, P.; Shoemaker, R.C. Genome organization in dicots: Genome duplication in Arabidopsis and synteny between soybean and Arabidopsis. Proc. Natl. Acad. Sci. USA 2000, 97, 4168–4173. [Google Scholar] [CrossRef]
- McCouch, S.R. Genomics and synteny. Plant Physiol. 2001, 125, 152–155. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Bowers, J.E.; Wang, X.; Ming, R.; Alam, M.; Paterson, A.H. Synteny and collinearity in plant genomes. Science 2008, 320, 486–488. [Google Scholar] [CrossRef] [PubMed]
- Frary, A.; Doganlar, S.; Frary, A. Synteny among Solanaceae Genomes. In The Tomato Genome; Causse, M., Giovannoni, J., Bouzayen, M., Zouine, M., Eds.; Springer: Berlin/Heidelberg, Germany, 2016; pp. 217–243. [Google Scholar]
- Ling, H.Q.; Ma, B.; Shi, X.; Liu, H.; Dong, L.; Sun, H.; Cao, Y.; Gao, Q.; Zheng, S.; Li, Y. Genome sequence of the progenitor of wheat A subgenome Triticum urartu. Nature 2018, 557, 424–428. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Li, X.; Lim, Y.P. Comparative genomics of Brassicaceae crops. Breed. Sci. 2014, 64, 3–13. [Google Scholar] [CrossRef]
- Parkin, I. Chasing ghosts: Comparative mapping in the Brassicaceae. In Genetics and Genomics of the Brassicaceae; Schmidt, R., Bancroft, I., Eds.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 153–170. [Google Scholar]
- Leister, D. Tandem and segmental gene duplication and recombination in the evolution of plant disease resistance genes. Trends Genet. 2004, 20, 116–122. [Google Scholar] [CrossRef]
- Lehti-Shiu, M.D.; Zou, C.; Hanada, K.; Shiu, S.H. Evolutionary history and stress regulation of plant receptor-like kinase/pelle genes. Plant Physiol. 2009, 150, 12–26. [Google Scholar] [CrossRef]
- Nobuta, K.; Ashfield, T.; Kim, S.; Innes, R.W. Diversification of non-TIR class NB-LRR genes in relation to whole-genome duplication events in Arabidopsis. Mol. Plant Microbe Interact. 2005, 18, 103–109. [Google Scholar] [CrossRef]
- Zeng, L.; Deng, R.; Guo, Z.; Yang, S.; Deng, X. Genome-wide identification and characterization of Glyceraldehyde-3-phosphate dehydrogenase genes family in wheat (Triticum aestivum). BMC Genom. 2016, 17, 240. [Google Scholar] [CrossRef]

| Species | B. juncea | B. rapa | B. nigra |
|---|---|---|---|
| Gene content | 80,430 | 46,098 | 49,826 |
| RLKs | 493 (0.613%) | 300 (0.651%) | 317 (0.636%) |
| LRR-RLKs | 484 (0.602%) | 297 (0.644%) | 312 (0.626%) |
| LysM-RLKs | 9 (0.011%) | 3 (0.007%) | 5 (0.010%) |
| RLPs | 228 (0.283%) | 65 (0.141%) | 176 (0.353%) |
| LRR-RLPs | 226 (0.281%) | 63 (0.137%) | 175 (0.351%) |
| LysM-RLPs | 2 (0.002%) | 2 (0.004%) | 1 (0.002%) |
| Duplication Type | RLK | RLP | ||||||
|---|---|---|---|---|---|---|---|---|
| Total | Intra Sub-Family | Inter Sub-Family | Total | Intra Sub-Family | Inter Sub-Family | |||
| LysM- | LRR- | LysM- | LRR- | |||||
| Intra A genomic | 83 | 1 | 82 | 0 | 20 | 0 | 20 | 0 |
| Intra B genomic | 80 | 0 | 80 | 0 | 32 | 0 | 32 | 0 |
| Total of intra genome | 163 | 1 | 162 | 0 | 52 | 0 | 52 | 0 |
| Inter genome | 308 | 4 | 304 | 0 | 58 | 1 | 57 | 0 |
| Total | 471 | 5 | 466 | 0 | 110 | 1 | 109 | 0 |
| Chromosome | RLK | RLP | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | LysM-RLK | LRR-RLK | Gene Cluster | Conserved | Total | LysM-RLP | LRR-RLP | Gene Cluster | Conserved | ||||||||
| No. of Cluster | No. of Gene in Cluster | Total | LysM-RLK | LRR-RLK | No. of Cluster | No. of Gene in Cluster | Total | LysM-RLP | LRR-RLP | ||||||||
| A genome B. rapa | A01 | 36 | 0 | 36 | 1 | 5 | 29 | 0 | 29 | 10 | 0 | 10 | 0 | 0 | 5 | 0 | 5 |
| A02 | 28 | 0 | 28 | 2 | 9 | 17 | 0 | 17 | 7 | 0 | 7 | 0 | 0 | 2 | 0 | 2 | |
| A03 | 38 | 0 | 38 | 0 | 0 | 30 | 0 | 30 | 9 | 0 | 9 | 0 | 0 | 2 | 0 | 2 | |
| A04 | 16 | 1 | 15 | 1 | 5 | 14 | 1 | 13 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | |
| A05 | 25 | 0 | 25 | 1 | 7 | 19 | 0 | 19 | 6 | 0 | 6 | 1 | 3 | 4 | 0 | 4 | |
| A06 | 49 | 1 | 48 | 3 | 15 | 36 | 1 | 35 | 10 | 1 | 9 | 1 | 3 | 2 | 1 | 1 | |
| A07 | 37 | 0 | 37 | 3 | 12 | 21 | 0 | 21 | 4 | 0 | 4 | 0 | 0 | 2 | 0 | 2 | |
| A08 | 19 | 1 | 18 | 0 | 0 | 15 | 1 | 14 | 7 | 1 | 6 | 0 | 0 | 4 | 1 | 3 | |
| A09 | 26 | 0 | 26 | 0 | 0 | 16 | 0 | 16 | 9 | 0 | 9 | 0 | 0 | 4 | 0 | 4 | |
| A10 | 21 | 0 | 21 | 2 | 7 | 17 | 0 | 17 | 2 | 0 | 2 | 0 | 0 | 1 | 0 | 1 | |
| Total mapped genes | 295 | 3, 1.02% 1 | 292, 98.98% 1 | 13 | 60, 20.34% 1 | 214, 72.54% 1 | 3, 100.00% 2 | 211, 72.26% 3 | 65 | 2, 3.08% 1 | 63, 96.92% 1 | 2 | 6, 9.23% 1 | 27, 41.54% 1 | 2, 100.00% 2 | 25, 39.68% 3 | |
| Unassigned | 5 | 0 | 5 | - | - | - | - | - | 0 | 0 | 0 | - | - | - | - | - | |
| Total | 300 | 3, 1.00% 4 | 297, 99.00% 4 | 13 | - | - | - | - | 65 | 2, 3.08% 4 | 63, 96.92% 4 | 2 | - | - | - | - | |
| Chromosome | RLK | RLP | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | LysM- | LRR- | Conserved | Syntenic Genes | Total | LysM | LRR- | Conserved | Syntenic Genes | ||||||
| Total | LysM- | LRR- | Total | LysM- | LRR- | ||||||||||
| A genome B. juncea | A01 | 30 | 0 | 30 | 24 | 0 | 24 | 22 | 12 | 0 | 12 | 6 | 0 | 6 | 5 |
| A02 | 25 | 0 | 25 | 17 | 0 | 17 | 14 | 5 | 0 | 5 | 2 | 0 | 2 | 2 | |
| A03 | 29 | 0 | 29 | 26 | 0 | 26 | 28 | 17 | 0 | 17 | 1 | 0 | 1 | 1 | |
| A04 | 16 | 2 | 14 | 13 | 1 | 12 | 13 | 9 | 0 | 9 | 1 | 0 | 1 | 1 | |
| A05 | 22 | 1 | 21 | 18 | 0 | 18 | 19 | 6 | 0 | 6 | 1 | 0 | 1 | 3 | |
| A06 | 31 | 1 | 30 | 27 | 1 | 26 | 32 | 7 | 1 | 6 | 2 | 1 | 1 | 2 | |
| A07 | 26 | 0 | 26 | 22 | 0 | 22 | 21 | 11 | 0 | 11 | 2 | 0 | 2 | 2 | |
| A08 | 12 | 1 | 11 | 12 | 1 | 11 | 11 | 8 | 0 | 8 | 3 | 0 | 3 | 5 | |
| A09 | 23 | 0 | 23 | 17 | 0 | 17 | 15 | 17 | 0 | 17 | 5 | 0 | 5 | 5 | |
| A10 | 22 | 0 | 22 | 18 | 0 | 18 | 17 | 3 | 0 | 3 | 1 | 0 | 1 | 1 | |
| Total in A genome | 236, 51.53% 1 | 5, 2.12% 2 | 231, 97.88% 2 | 194, 82.20% 2 | 3, 60.00% 3 | 191, 82.68% 4 | 192, 81.36% 2 | 95, 44.81% 1 | 1, 1.05% 2 | 94, 98.95% 2 | 24, 25.26% 2 | 1, 100.00% 3 | 23, 24.47% 4 | 27, 28.42% 2 | |
| B genome B. juncea | B01 | 27 | 2 | 25 | 24 | 2 | 22 | - | 6 | 0 | 6 | 6 | 0 | 6 | - |
| B02 | 36 | 0 | 36 | 30 | 0 | 30 | - | 10 | 0 | 10 | 3 | 0 | 3 | - | |
| B03 | 30 | 0 | 30 | 27 | 0 | 27 | - | 19 | 1 | 18 | 13 | 1 | 12 | - | |
| B04 | 30 | 1 | 29 | 23 | 1 | 22 | - | 23 | 0 | 23 | 11 | 0 | 11 | - | |
| B05 | 33 | 0 | 33 | 28 | 0 | 28 | - | 18 | 0 | 18 | 11 | 0 | 11 | - | |
| B06 | 20 | 0 | 20 | 17 | 0 | 17 | - | 11 | 0 | 11 | 6 | 0 | 6 | - | |
| B07 | 16 | 0 | 16 | 16 | 0 | 16 | - | 16 | 0 | 16 | 9 | 0 | 9 | - | |
| B08 | 30 | 0 | 30 | 24 | 0 | 24 | - | 14 | 0 | 14 | 6 | 0 | 6 | - | |
| Total in B genome | 222, 48.47% 1 | 3, 1.35% 2 | 219, 98.65% 2 | 189, 85.14% 2 | 3, 100.00% 3 | 186, 84.93% 4 | - | 117, 55.19% 1 | 1, 0.85% 2 | 116, 99.15% 2 | 65, 55.56% 2 | 1, 100.00% 3 | 64, 55.17% 4 | - | |
| Total mapped genes | 458 | 8, 1.75% 1 | 450, 98.25% 1 | 383, 83.62% 1 | 6, 75.00% 3 | 377, 83.78% 4 | - | 212 | 2, 0.94% 1 | 210, 99.06% 1 | 89, 41.98% 1 | 2, 100.00% 3 | 87, 41.43% 4 | - | |
| RLK | Total | 317 | |
| LysM-RLK | 5 (1.58% 1) | ||
| LRR-RLK | 312 (98.42% 1) | ||
| Conserved | Total | 227 (71.61% 1) | |
| LysM-RLK | 4 | ||
| LRR-RLK | 223 | ||
| RLP | Total | 176 | |
| LysM-RLP | 1 (0.57% 2) | ||
| LRR-RLP | 175 (99.43% 2) | ||
| Conserved | Total | 72 (40.91% 2) | |
| LysM-RLP | 1 | ||
| LRR-RLP | 71 | ||
| Gene Family | Genome | B. juncea A Genome | B. rapa A Genome | B. juncea B Genome | B. nigra B Genome |
|---|---|---|---|---|---|
| RLK family | Total (LysM, LRR) | 236 (5, 231) | 300 (3, 297) | 222 (3, 219) | 317 (5, 315) |
| Total conserved | 194 (82.20%) | 214 (71.33%) | 189 (85.14%) | 227 (71.61%) | |
| Conserved LysM-RLK | 3 (60.00%) | 3 (100.00%) | 3 (100.00%) | 4 (80.00%) | |
| Conserved LRR-RLK | 191 (82.68%) | 211 (71.04%) | 186 (84.93%) | 223 (70.79%) | |
| Not conserved | 42 (17.80%) | 86 (28.67%) | 33 (14.86%) | 90 (28.39%) | |
| Lost | 0 | 86 | 0 | 90 | |
| Gained | 42 | 0 | 33 | 0 | |
| RLP family | Total (LysM, LRR) | 95 (1, 94) | 65 (2, 63) | 117 (1, 116) | 176 (1, 175) |
| Total conserved | 24 (25.26%) | 27 (41.54%) | 65 (55.56%) | 72 (40.91%) | |
| Conserved LysM-RLP | 1 (100.00%) | 2 (100.00%) | 1 (100.00%) | 1 (100.00%) | |
| Conserved LRR-RLP | 23 (24.47%) | 25 (39.68%) | 64 (55.17%) | 71 (40.57%) | |
| Not conserved | 71 (74.74%) | 38 (58.46%) | 52 (44.44%) | 104 (59.09%) | |
| Lost | 0 | 38 | 0 | 104 | |
| Gained | 71 | 0 | 52 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, H.; Bayer, P.E.; Tirnaz, S.; Edwards, D.; Batley, J. Genome-Wide Identification and Evolution of Receptor-Like Kinases (RLKs) and Receptor like Proteins (RLPs) in Brassica juncea. Biology 2021, 10, 17. https://doi.org/10.3390/biology10010017
Yang H, Bayer PE, Tirnaz S, Edwards D, Batley J. Genome-Wide Identification and Evolution of Receptor-Like Kinases (RLKs) and Receptor like Proteins (RLPs) in Brassica juncea. Biology. 2021; 10(1):17. https://doi.org/10.3390/biology10010017
Chicago/Turabian StyleYang, Hua, Philipp E. Bayer, Soodeh Tirnaz, David Edwards, and Jacqueline Batley. 2021. "Genome-Wide Identification and Evolution of Receptor-Like Kinases (RLKs) and Receptor like Proteins (RLPs) in Brassica juncea" Biology 10, no. 1: 17. https://doi.org/10.3390/biology10010017
APA StyleYang, H., Bayer, P. E., Tirnaz, S., Edwards, D., & Batley, J. (2021). Genome-Wide Identification and Evolution of Receptor-Like Kinases (RLKs) and Receptor like Proteins (RLPs) in Brassica juncea. Biology, 10(1), 17. https://doi.org/10.3390/biology10010017








