Abstract
The TLRs and IL-1 receptors have evolved to coordinate the innate immune response following pathogen invasion. Receptors and signalling intermediates of these systems are generally characterised by a high level of evolutionary conservation. The recently described IL-1R1 co-receptor TILRR is a transcriptional variant of the FREM1 gene. Here we investigate whether innate co-receptor differences between teleosts and mammals extend to the expression of the TILRR isoform of FREM1. Bioinformatic and phylogenetic approaches were used to analyse the genome sequences of FREM1 from eukaryotic organisms including 37 tetrapods and five teleost fish. The TILRR consensus peptide sequence was present in the FREM1 gene of the tetrapods, but not in fish orthologs of FREM1, and neither FREM1 nor TILRR were present in invertebrates. The TILRR gene appears to have arisen via incorporation of adjacent non-coding DNA with a contiguous exonic sequence after the teleost divergence. Comparing co-receptors in other systems, points to their origin during the same stages of evolution. Our results show that modern teleost fish do not possess the IL-1RI co-receptor TILRR, but that this is maintained in tetrapods as early as amphibians. Further, they are consistent with data showing that co-receptors are recent additions to these regulatory systems and suggest this may underlie differences in innate immune responses between mammals and fish.
Keywords:
TILRR; IL-1RI; co-receptor; FREM1; bioinformatics; evolutionary development; signal transduction; TIR activation; IL-1 1. Introduction
The innate immune system is generally well conserved throughout the animal kingdom with the same characteristic features of regulatory components present in species ranging from insects to mammals [1]. Activation is induced through members of the Toll-like and IL-1 receptor (TIR) family, characterized by the cytoplasmic TIR domain [2]. The high level of conservation of the intracellular domains of both Toll and IL-1R1 and of cytoplasmic regulatory components is consistent with a signaling system that is broadly conserved throughout evolution prior even to the divergence of plants [3] and animals [4]. However, it is increasingly recognized that mechanisms of ligand recognition and co-receptor association, a potent regulator of signal amplification at the level of the receptor complex, are less well conserved [5,6]. In evolutionary terms, such co-receptors appeared relatively late in the development of their respective signaling networks which they control.
Recent studies have revealed that fish, which have been shown to possess certain inflammatory receptors, may lack co-receptors found in more modern organisms, suggesting that signaling mechanisms in earlier species are functionally distinct and less refined. Thus, for example the zebrafish (Danio rerio) possesses two paralogs of TLR4, neither of which is stimulated by LPS, and lacks the co-receptors MD2 and CD14 [7,8]. Similarly, phylogenetic studies of the synteny of the syndecan genes in fish and tetrapods has revealed that while the four mammalian syndecan genes arose due to gene duplication, Syndecan 1 (an FGFR co-receptor) is absent from fish genomes probably as a result of deletion following this duplication event [9].
We recently identified the IL-1R co-receptor, TILRR (Toll-like/IL-1 receptor regulator), a 715 amino acid heparan sulfate glycoprotein encoded within the gene for the extracellular matrix protein FREM1 [10]. FREM1 has a distinct function in embryogenesis and development, and is ubiquitously expressed [11].
TILRR binds the cell membrane through a C-terminal lectin domain, associates with IL-1R1 and increases receptor expression and ligand binding. TILRR association potentiates recruitment of the MyD88 adapter and receptor signal amplification, and enhances activation of NF-κB and inflammatory genes [10]. We earlier confirmed expression and function of TILRR in mouse and human cells [10].
The current studies examine the presence of TILRR throughout evolution and demonstrate that TILRR is a transcriptional variant of FREM1 whose transcriptional start site lies within the intronic sequence of FREM1. TILRR is lacking in early species such as teleosts and invertebrates, being first identifiable in amphibians. These findings highlight that although innate immunity is evolutionarily ancient, refinements to the system have continued to arise until more recently and that important differences exist between model organisms used to study inflammation.
2. Results and Discussion
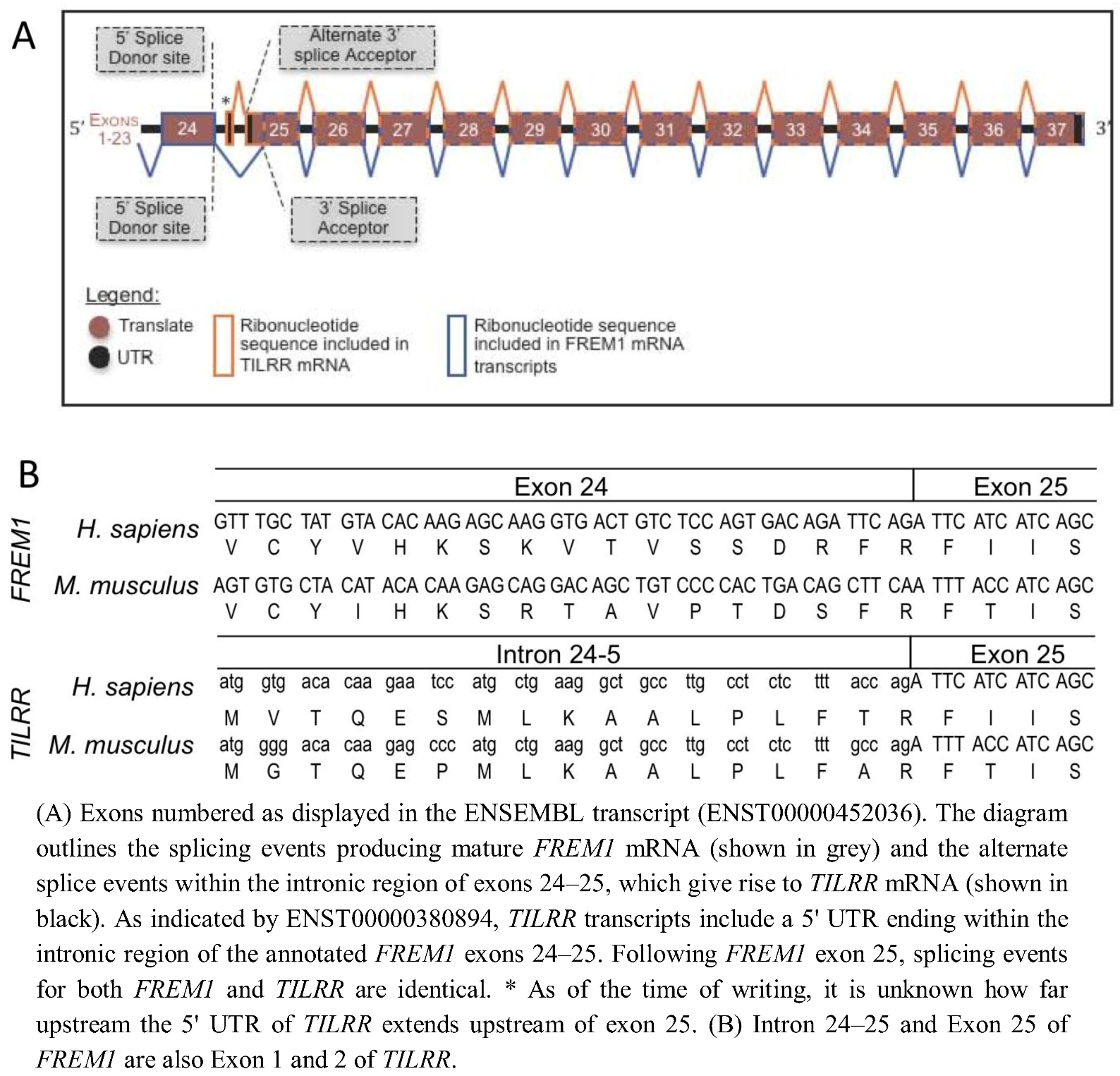
In order to determine the evolutionary development of the IL-1RI co-receptor TILRR, we identified the TILRR isoform of FREM1 within the genomes of multiple organisms and defined the source of the TILRR peptide sequence within the nucleotide sequence of the FREM1 loci. Alignment of the peptide sequences of human TILRR and FREM1 show that they are identical from R17 of TILRR (R1481 of FREM1). This TILRR/FREM1 consensus region is encoded by exons 25 onwards in the human FREM1 gene.
To predict the location within the Human FREM1 locus where TILRR transcripts initiate, we analysed the Ensembl annotation of the 2,179 amino acid encoding Human FREM1 gene (ENST00000452036). Studying the cDNA sequence of this transcript revealed R1481 to be encoded within a residue overlap splice site and due to ligation of the final two nucleotides of exon 24, and the first of exon 25 (Figure 1). A lack of homology between the 16 N-terminal TILRR residues and any other part of FREM1 suggested that no early FREM1 coding exon is ligated to exon 25 to encode TILRR. Therefore, we reasoned that the N-terminus (translational start site) of TILRR must reside within a sequence of FREM1 not incorporated into the processed FREM1 mRNA. We hypothesized that the N-terminus of TILRR could be located by examining the annotation of the FREM1 transcript, prior to the first exon common to both TILRR and FREM1 (exon 25). Translation in all three reading frames of the intronic nucleotide sequence immediately preceding exon 25 produced the unique 16 TILRR N-terminal residues that we previously sequenced using MALDI-TOF [10]: MVTQESMLKAALPLFT followed by R17 and the remaining residues up to Q155. As the N-terminus of TILRR is produced by a series of consecutive nucleotides immediately prior to and in frame with the third nucleotide of the R1481 codon of FREM1, during RNA processing, FREM1 mRNA transcripts arise when exon 24 is ligated to a 3' splice acceptor site prior to the third nucleotide of the R1481 codon, whereas exon 1 of TILRR commences within intron 23–24 and runs into exon 24 without the requirement for such an acceptor splice site (Figure 1B). This initiation of a novel “orphan” gene from a non-coding sequence is a recently described mechanism, which in many cases allows the organism to adapt to novel conditions [12,13,14].
Analysis of the Mouse Frem1 locus (ENSMUSG00000059049) in the same manner as Human FREM1, revealed a similar splicing mechanism: the N-terminal 16 residues of Mouse TILRR (MGTQEPMLKAALPLFA, as we previously showed by peptide sequencing) [10] are encoded by an intronic nucleotide sequence upstream of exon 25 of the Frem1 transcript. This is consistent with the suggestion that TILRR and FREM1 mRNAs arise due to alternative transcriptional initiation of the TILRR mRNA within intron 24–25 of the FREM1 gene.
Since analysis of both the Human FREM1 and Mouse Frem1 loci clearly identified the 5' TILRR coding sequence within the intron preceding exon 25, we reasoned that examining this region in other species would allow determination of whether each organism possesses an ortholog of TILRR.
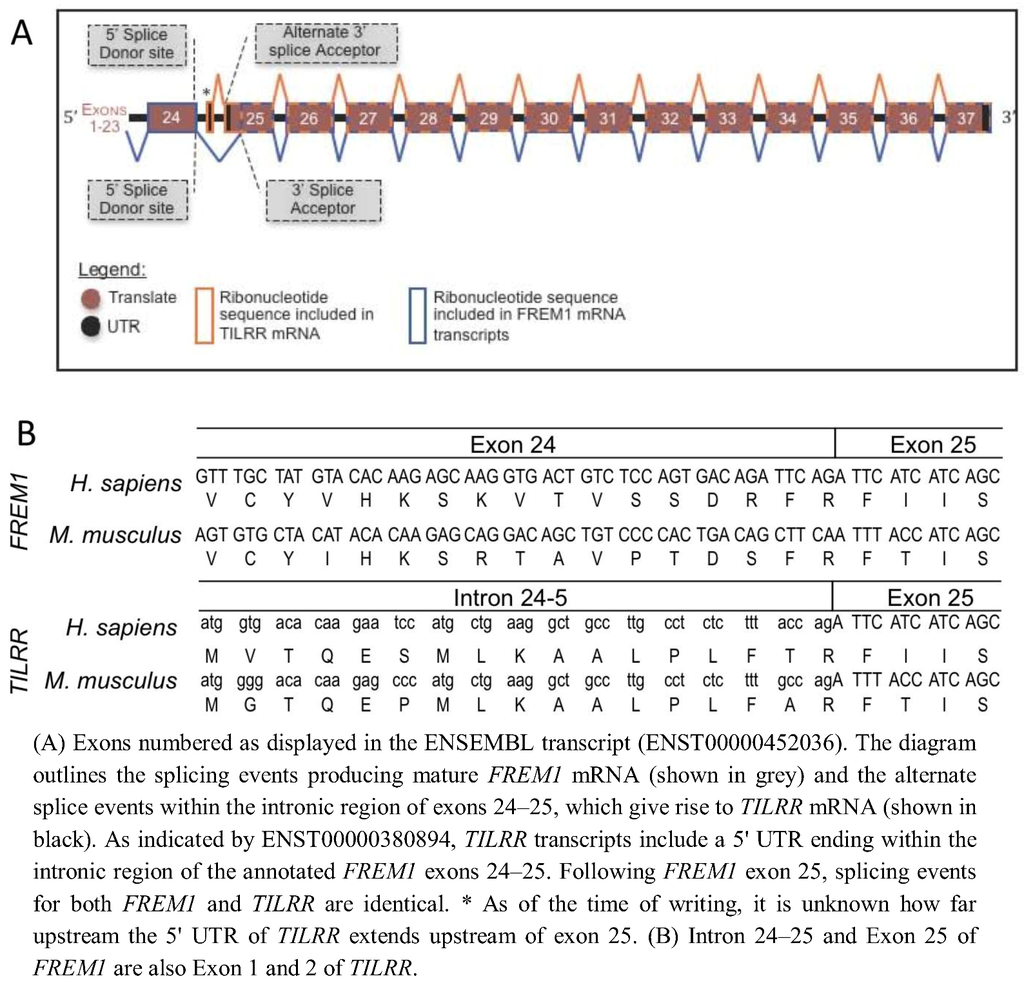
Figure 1.
(A) Schematic diagram of Human and Mouse FREM1 pre-mRNA. (B) DNA sequence and peptide translation of human and mouse FREM1 in region of boundary between exons 24 and 25 (upper panel) and TILRR showing genomic sequence and translated peptide.
Figure 1.
(A) Schematic diagram of Human and Mouse FREM1 pre-mRNA. (B) DNA sequence and peptide translation of human and mouse FREM1 in region of boundary between exons 24 and 25 (upper panel) and TILRR showing genomic sequence and translated peptide.
We therefore extended our investigation to examining the genomes of 37 tetrapod organisms to identify the 5' end of the TILRR coding sequence within each Frem1 ortholog. We used the predicted FREM1 cDNA transcript sequences to identify the region of each locus corresponding to the exon containing alternative 3' splice acceptor sites as in exon 25 of Human FREM1. The preceding nucleotides were translated in frame with the nucleotide sequence of the identified exon and the resulting peptide sequence aligned with the TILRR N-termini. For 33 tetrapod species, 16 consecutive amino acids could be produced in frame with R17, suggesting that a strongly conserved TILRR homolog exists in these organisms (Figure 2).
Although four tetrapod FREM1 loci (C. familiaris, D. ordii, T. syrichta and E. telfairi) could not immediately be translated into the 5' TILRR residues, we found that all four possess a single nucleotide alteration compared to the consensus TILRR peptide sequence, but that with this exception the consensus N-terminal TILRR sequence was preserved, indicating these species are likely to possess the TILRR isoform of FREM1 (Figure 3). Alternatively, in these species TILRR may constitute a pseudogene, which produces a non-functional protein, although it is highly likely that all tetrapod homologues arose from a common ancestor. Future studies are needed to assess the significance of these mutations in signal amplification of the TIR domain.
Since we had identified TILRR orthologues in all tetrapod species studied, we next analysed teleost FREM1 homologs using annotations of all identified teleost FREM1 orthologs in the Ensembl database. All teleost species possessed at least one FREM1 orthologue. However, in these organisms no such FREM1 ortholog contained a conserved 5' coding sequence indicative of a tilrr transcript (Figure 4).
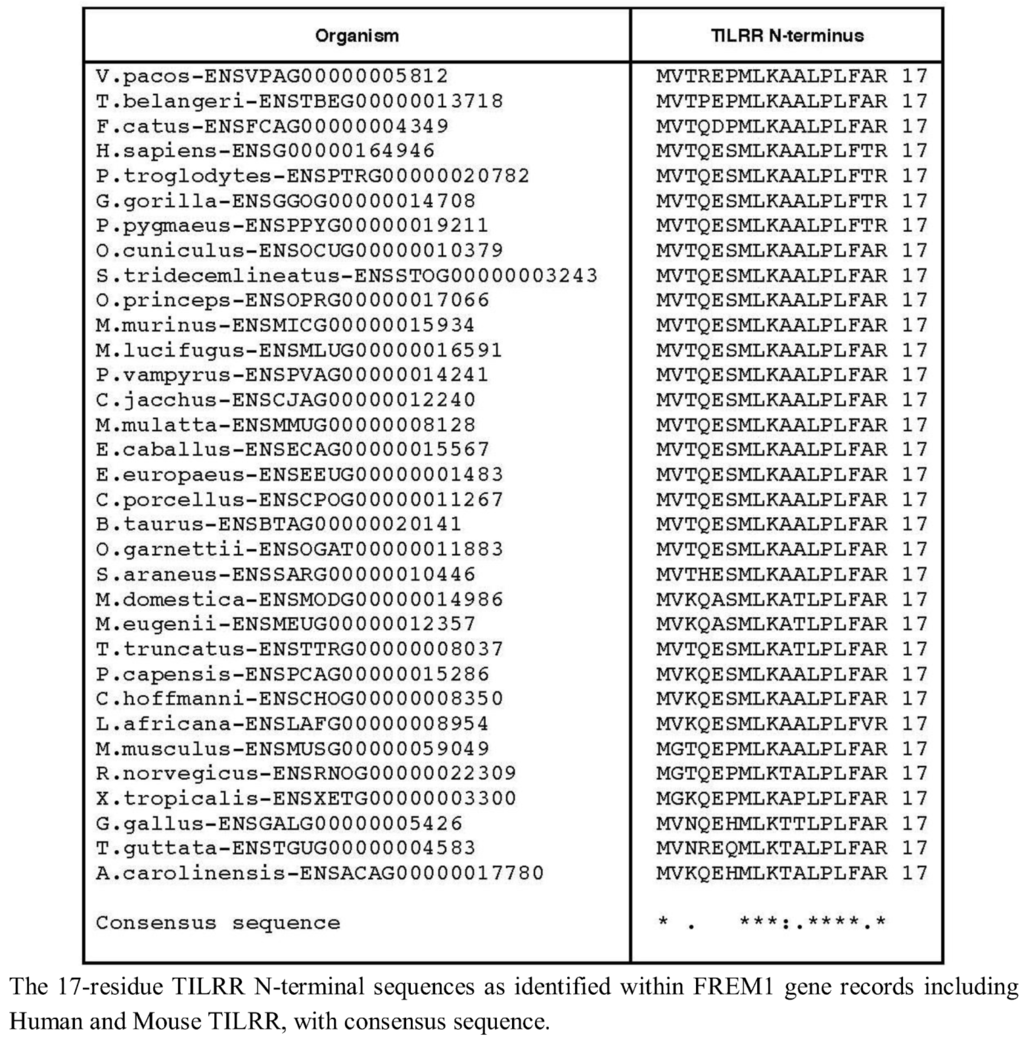
Figure 2.
Comparison of TILRR N-terminal sequence.
Figure 2.
Comparison of TILRR N-terminal sequence.
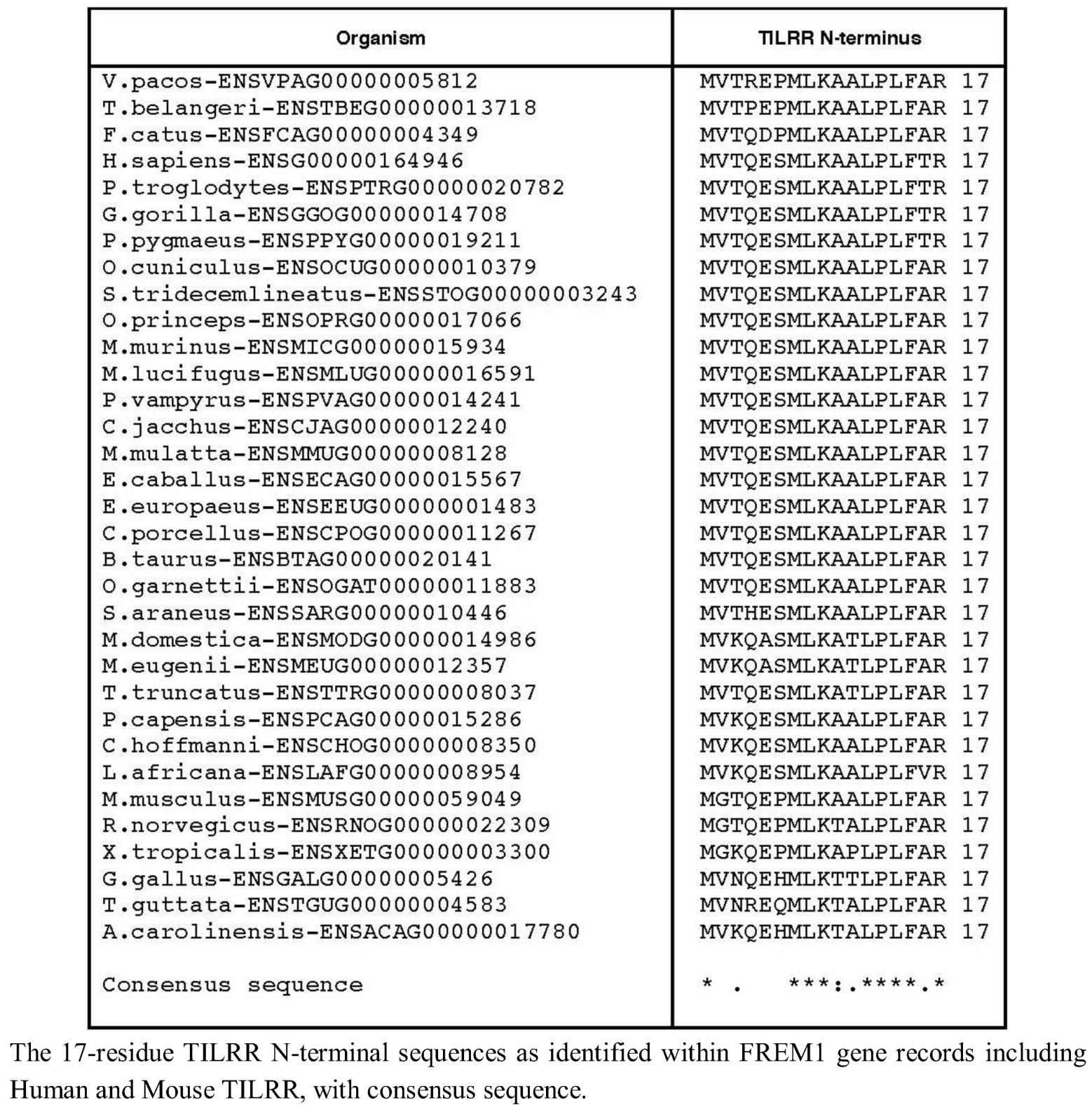

Figure 3.
Identification of single base mutations responsible for non-homologous sequence in C. familiaris, D. ordii, T. syrichta and E. telfairi.
Figure 3.
Identification of single base mutations responsible for non-homologous sequence in C. familiaris, D. ordii, T. syrichta and E. telfairi.
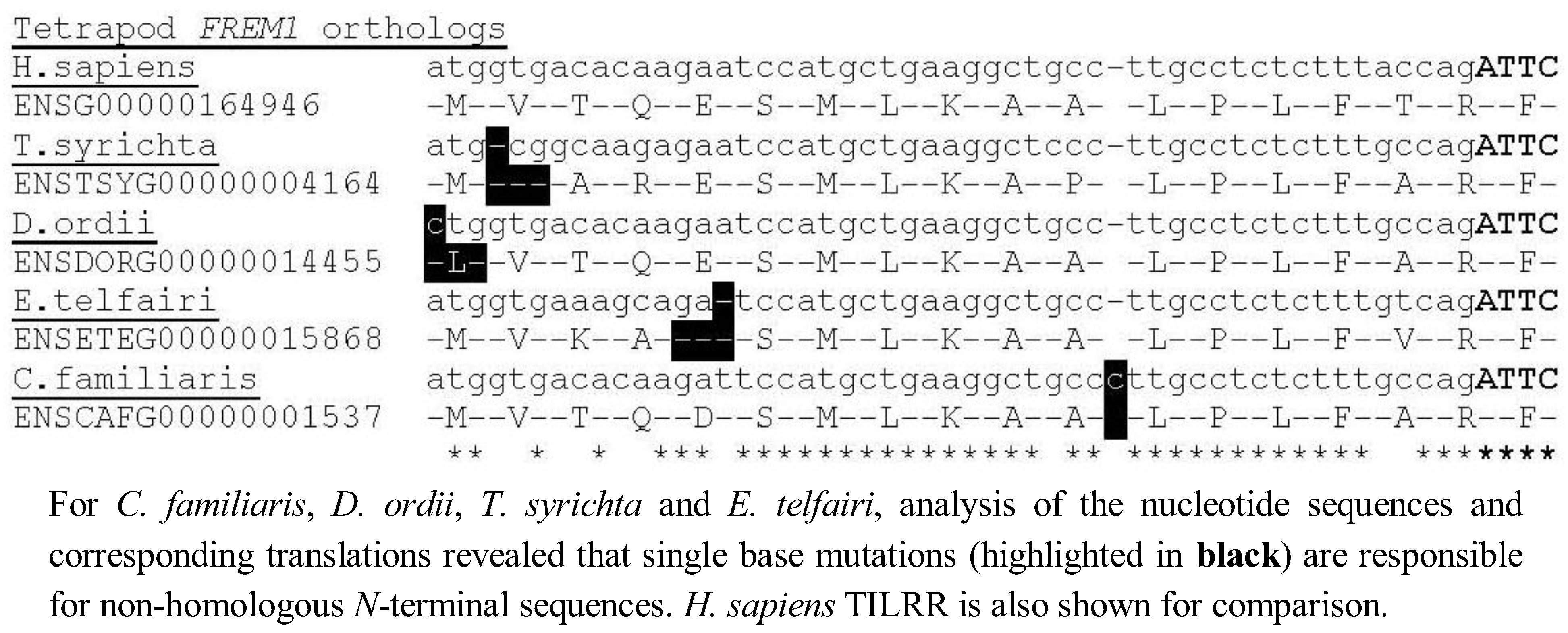

Figure 4.
FREM1 orthologs in fish lack the TILRR N-terminus.
Figure 4.
FREM1 orthologs in fish lack the TILRR N-terminus.

We next examined invertebrate species for orthologues of FREM1/TILRR. Using BLASTP, we were unable to locate orthologues of either the consensus regions shared between Frem1 and Tilrr or the Tilrr 5' sequence in non-vertebrates (Drosophila melanogaster, Ciona intestinalis, Caenorhabditis elegans). We therefore concluded that invertebrates do not possess FREM1, nor a TILRR orthologue. Thus, FREM1 arose after the evolution of vertebrates, but TILRR only becomes detectable after the divergence of the teleosts.
To investigate possible mechanisms for the absence of TILRR in teleosts, we examined the exon/exon boundary sequences of FREM1 in human, mouse, xenopus and four teleosts (Danio rerio [zebrafish], Oryzias latipes [medaka], Takifugu rubripes [fugu] and Gasterosteus aculeatus [stickleback] and the corresponding intron/exon sequences that encode for TILRR in tetrapods but not in teleosts (Figure 5). We found that whereas the exonic and particularly translated sequence of FREM1 were reasonably well conserved even between mammals and telosts, comparison of the intronic region encoding for TILRR in tetrapods reveals marked divergence. Although all introns end with the major spliceosomal AG consensus splice site, the actual intronic sequences diverged markedly between teleosts and mammals. Although N termini often vary greatly in length and sequence between homologues, in zebrafish, medaka and stickleback translation of the ORF of the contiguous intronic sequence preceding the shared frem1 exon leads to a stop codon between the shared exon and the earliest possible methionine start codon (Figure 5). In Fugu there is a methionine in the N-terminal sequence that could represent a TILRR start codon, although there is no equivalent of this in the human sequence (Figure 5). Given the otherwise high conservation of the frem1 gene between teleosts, it seems unlikely that Fugu possess a TILRR homologue when the other three teleosts do not. It seems likely that TILRR arose through a major alteration of intronic sequence, rather than a more subtle perturbation that gave rise to generation of novel intronic transcription binding sites.
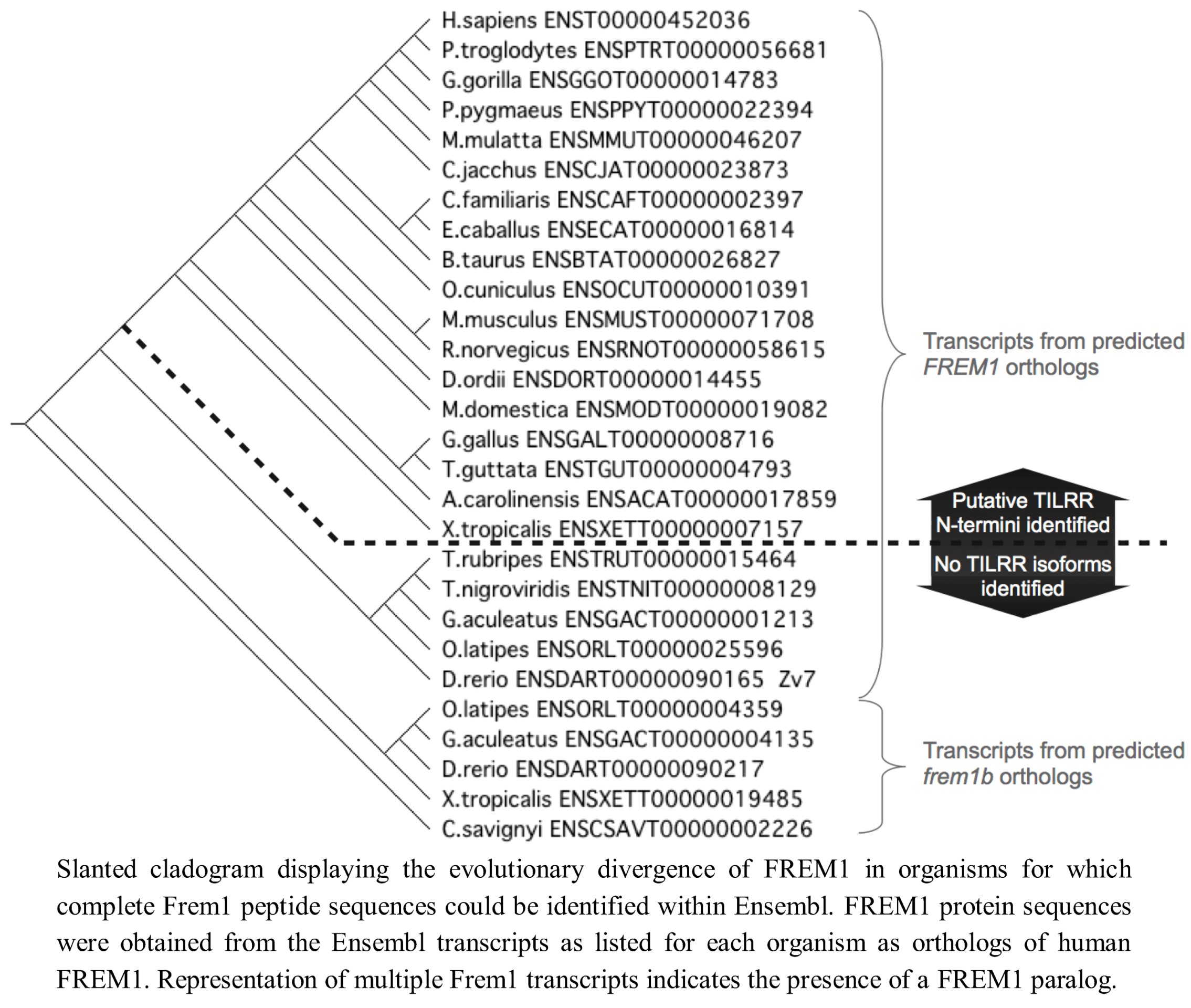
We therefore conclude that the TILRR isoform of FREM1 is present only in tetrapod organisms, presenting two possibilities for the origin of TILRR. This may reflect that it originates from an ancestor common to both teleost and tetrapod organisms that arose after the invertebrates, and was lost in FREM1 paralogs prior to the evolution of modern teleosts. Alternatively, (an explanation we favour) TILRR may have originated following the divergence of a common ancestor into the tetrapod lineage, hence its first detection within amphibian FREM1 (Figure 6).
Either possibility suggests that, in contrast to a majority of IL-1RI complex components, which are present in primitive species such as D. rerio [7,9], TILRR is not involved in IL-1R1 controlled responses to pathogenic invasion in ancestral or modern day teleosts.
The work in this study shows the maturation of the IL-1 receptor complex within the timeframe between the divergent evolution of teleost fish and tetrapod amphibians some 360–450 million years ago [15]. The conservation of TILRR within the genomes of tetrapod organisms likely represents refinement of IL-1 signaling over the course of vertebrate evolution, to allow increased sensitivity of system control through ligand concentration and receptor levels.
The finding that TILRR does not exist in any teleost studied suggests that distinct components of the vertebrate IL-1R1 complex may have evolved at different stages of the evolutionary tree, perhaps reflecting functions related to TILRR controlled environmental sensing and attachment. The lack of TILRR expression in primitive species, such as D. rerio, in addition to the absence of Syndecan 1 and the TLR4 co-receptors [8,9], also supports the notion that inflammatory signaling regulatory mechanisms in mammals are not all synonymous with that of lower vertebrates. Common features of the co-receptors of these systems are related to ligand/receptor interactions and receptor sensitivity to ligand, allowing for increasing variability and specificity over a range of ligand and receptor levels. Similarly, recently identified TILRR mutants demonstrate selective regulation of distinct cellular responses related to inflammation and cell survival, thus contributing refined control and increased specificity [16].
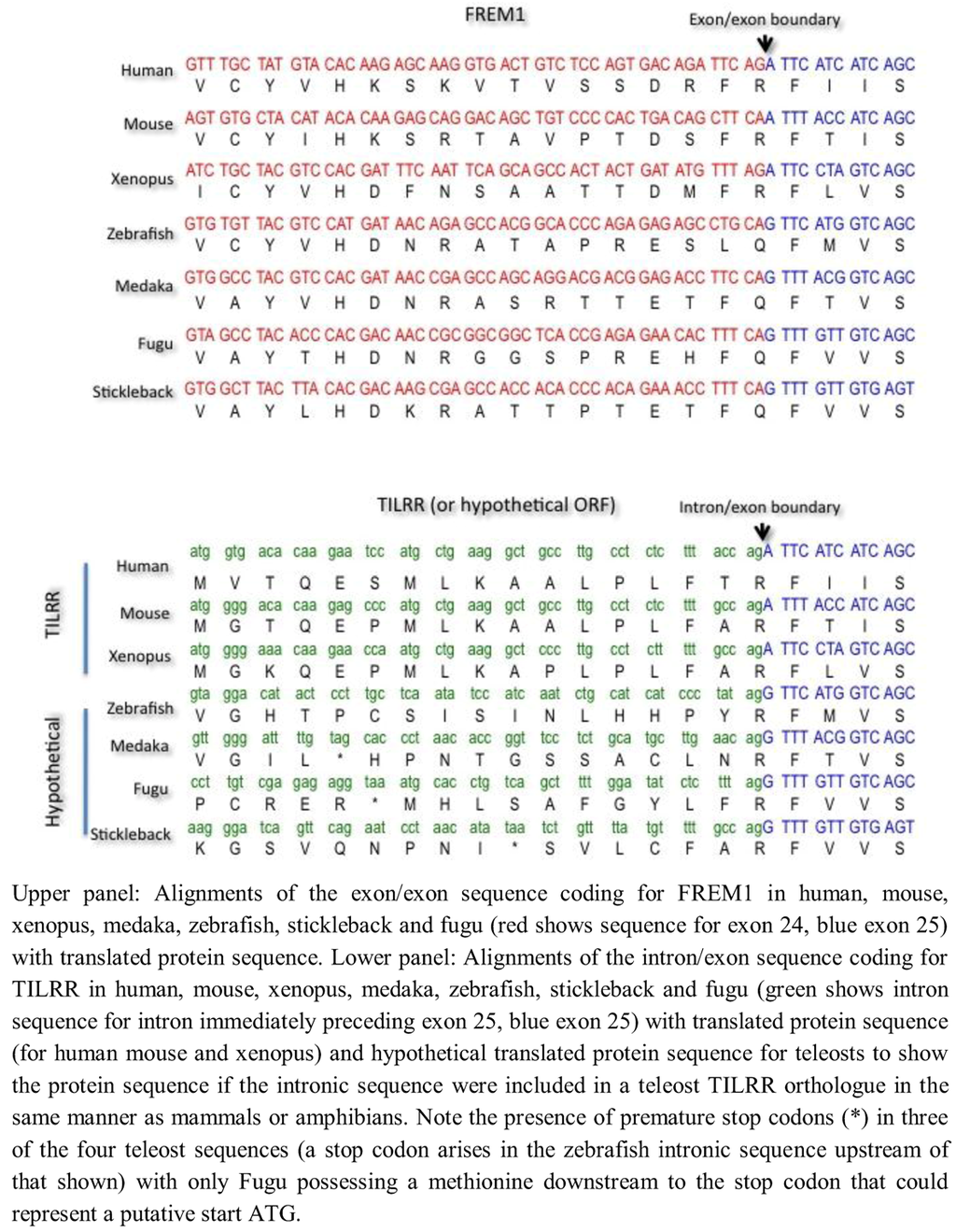
Figure 5.
Comparative alignment of coding sequence of FREM1 and TILRR in mammals, amphibians and teleosts.
Figure 5.
Comparative alignment of coding sequence of FREM1 and TILRR in mammals, amphibians and teleosts.
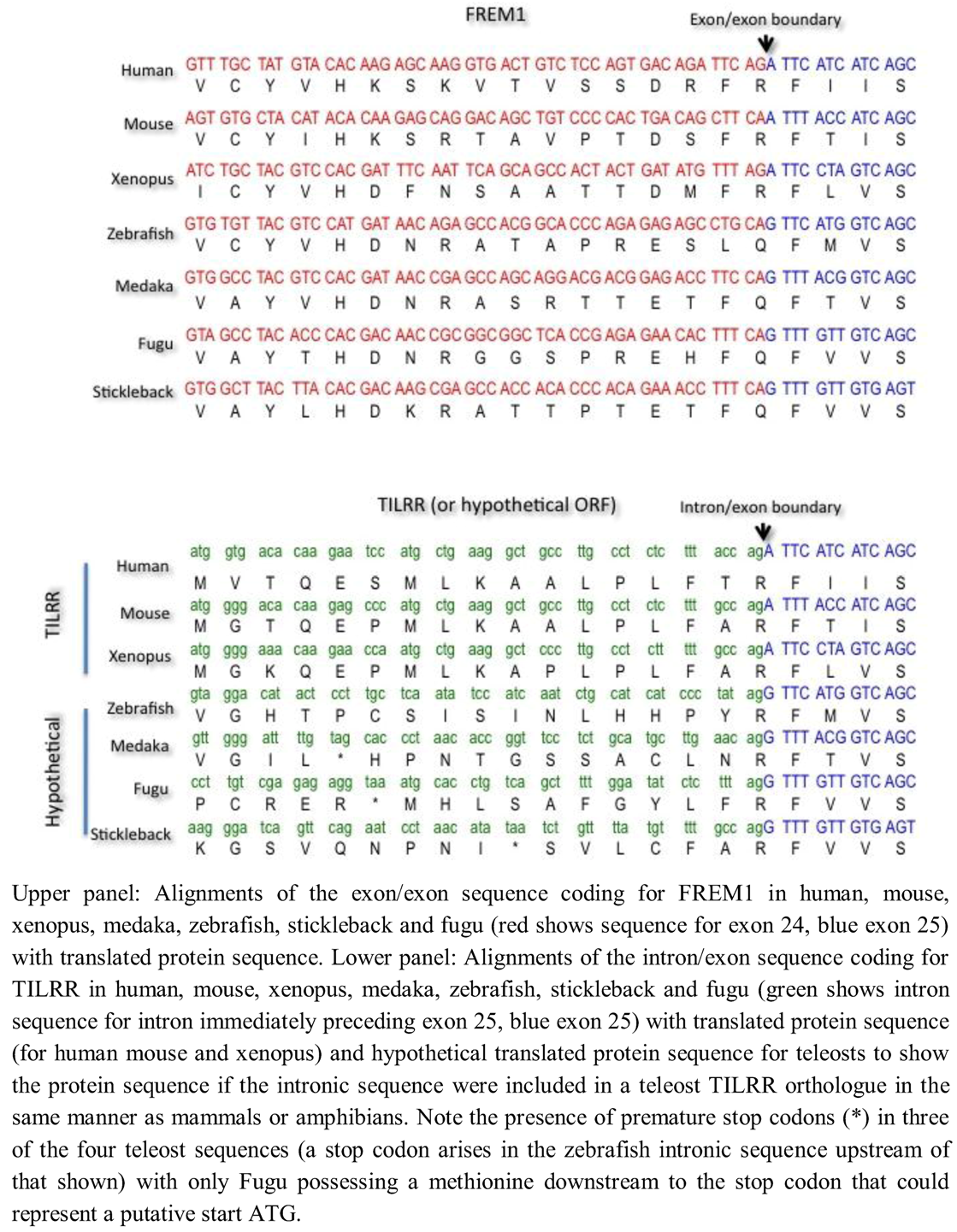
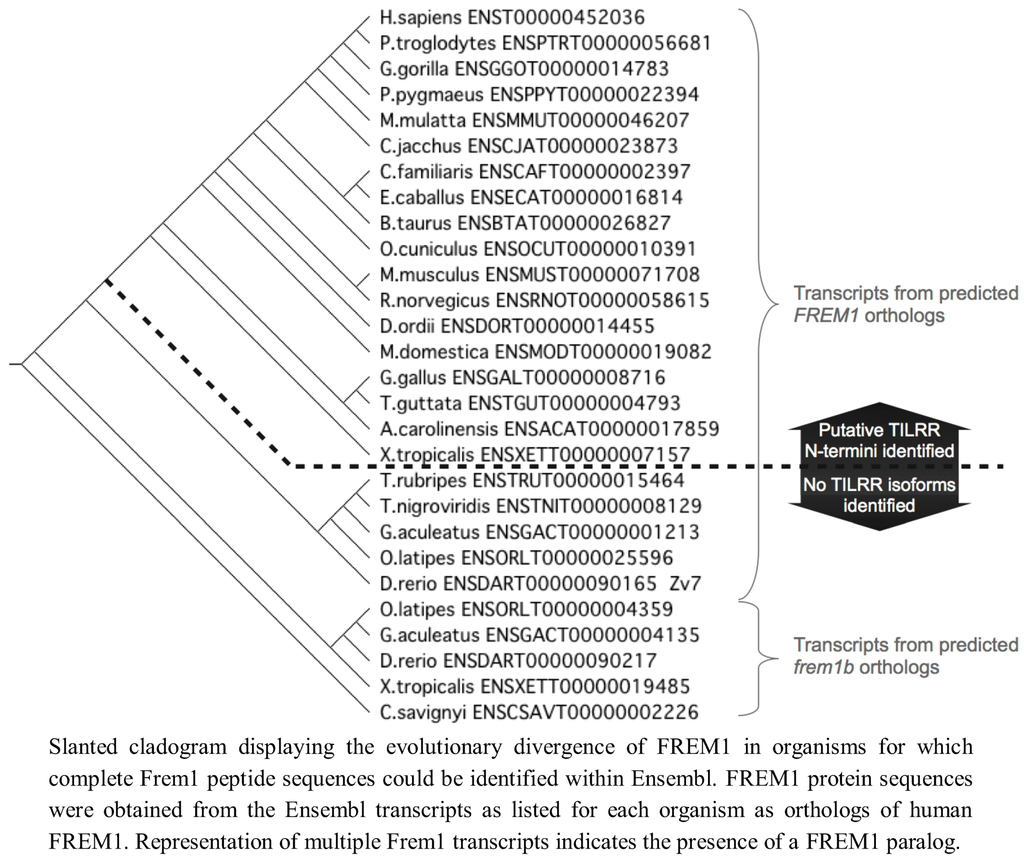
Figure 6.
Evolutionary development of FREM1 and presence of putative TILRR sequence.
Figure 6.
Evolutionary development of FREM1 and presence of putative TILRR sequence.
3. Experimental Section
3.1. Obtaining Predicted Frem1 Peptide Sequences
Genbank [17] was used as a source for TILRR and FREM1 peptide sequences. GeneIDs and NCBI [18] Reference sequences for the peptides used to compare with predicted transcripts are listed below (Table 1).

Table 1.
Accession numbers of FREM1 sequences.
| Organism | GeneID | Protein Name | NCBI Reference sequence |
|---|---|---|---|
| H. sapiens | 158326 | FREM1 isoform 2 (TILRR) | NP_001171175 |
| FREM1isoform 1 precursor | NP_659403 | ||
| M. musculus | 329872 | FREM1 precursor | NP_808531 |
| D. rerio | 100216326 | Frem1a | NP_001177237 |
| 557221 | Frem1b | NP_001131130 |
Ensembl was used to locate predicted FREM1 gene loci, identified as orthologs of the Ensembl annotation of Human FREM1 (ENSG00000164946) or of Zebrafish frem1b (ENSDARG00000062402) [19].
Predicted Ensembl FREM1 ortholog transcripts are shown in Supplementary Figure 1.
Previous studies of D. rerio Frem1 used probes deduced by sequence analysis to analyse expression of the two orthologs, Frem1a and Frem1b, which we aligned to the predicted protein sequences as listed in Ensembl. By this method, we found that D. rerio Frem1a had a greater similarity to the ENSDART00000090165 transcript annotation in the Zv7 genome as compared to the more recent Zv8 genome annotation. Conversely, the frem1b transcript had perfect similarity to ENSDART00000090217 on Zv8. For these reasons we used the Zv7 annotation of ENSDART00000090165 as a basis for the exon structure of frem1a, and the Zv8 annotation of ENSDART00000090217 as a basis for the exon structure of frem1b.
3.2. Sequence Alignments
All peptide and nucleotide sequence alignments were performed using CLUSTALW and CLUSTALX. A slanted cladogram of FREM1 peptide sequences was constructed using the UPGMA algorithm in CLUSTALX displayed using TreeViewX.
4. Conclusions
Our data show that TILRR is a recent addition to the IL-1RI signaling system. These findings, and those of others looking at evolutionary development of regulatory pathways, are consistent with a role for co-receptors in modulating response control in higher organisms. This opens interesting possibilities for investigating development of regulatory intermediates and delineating mechanisms underlying the increased sensitivity and complexity characteristic of maturation of biological systems during evolution.
Supplementary Files
Abbreviations
| IL-1 | interleukin 1 |
| IL-1RI | interleukin 1 receptor type I |
| TILRR | Toll-like and Il-1 receptor regulator |
| NF-κB | nuclear factor kappa B |
| NCBI | National Center for Biotechnology Information |
| FREM-1 | FRAS Related Extracellular Matrix gene 1 |
Acknowledgments
The studies were supported by grants from the Biotechnology and Biological Sciences Research Council (BB/C515798/1; BBS/B/04056), and the British Heart Foundation (PG/2000094; PG/07/094/23742) to EEQ and from the Medical Research Council (centre grant G0700091) and British Heart Foundation (PG/09/087/28051) to TC.
References
- Hoffmann, J.A.; Kafatos, F.C.; Janeway, C.A.; Ezekowitz, R.A. Phylogenetic perspectives in innate immunity. Science 1999, 284, 1313–1318. [Google Scholar] [CrossRef]
- Bowie, A.; O’Neill, L.A.J. The interleukin-1 receptor/toll-like receptor superfamily: Signal generators for pro-inflammatory interleukins and microbial products. J. Leukoc. Biol. 2000, 67, 508–514. [Google Scholar]
- Whitham, S.; Dinesh-Kumar, S.P.; Choi, D.; Hehl, R.; Corr, C.; Baker, B. The product of the tobacco mosaic virus resistance gene n: Similarity to toll and the interleukin-1 receptor. Cell 1994, 78, 1101–1115. [Google Scholar] [CrossRef]
- Belvin, M.P.; Anderson, K.V. A conserved signaling pathway: The drosophila toll-dorsal pathway. Annu. Rev. Cell Dev. Biol. 1996, 12, 393–416. [Google Scholar] [CrossRef]
- Michel, T.; Reichhart, J.M.; Hoffmann, J.A.; Royet, J. Drosophila toll is activated by gram-positive bacteria through a circulating peptidoglycan recognition protein. Nature 2001, 414, 756–759. [Google Scholar] [CrossRef]
- Poltorak, A.; He, X.L.; Smirnova, I.; Liu, M.Y.; van Huffel, C.; Du, X.; Birdwell, D.; Alejos, E.; Silva, M.; Galanos, C.; Freudenberg, M.; Ricciardi-Castagnoli, P.; Layton, B.; Beutler, B. Defective lps signaling in c3h/hej and c57bl/10sccr mice: Mutations in tlr4 gene. Science 1998, 282, 2085–2088. [Google Scholar]
- Iliev, D.B.; Roach, J.C.; Mackenzie, S.; Planas, J.V.; Goetz, F.W. Endotoxin recognition: In fish or not in fish? FEBS Lett. 2005, 579, 6519–6528. [Google Scholar] [CrossRef]
- Sullivan, C.; Charette, J.; Catchen, J.; Lage, C.R.; Giasson, G.; Postlethwait, J.H.; Millard, P.J.; Kim, C.H. The gene history of zebrafish tlr4a and tlr4b is predictive of their divergent functions. J. Immunol. 2009, 183, 5896–5908. [Google Scholar]
- Chakravarti, R.; Adams, J.C. Comparative genomics of the syndecans defines an ancestral genomic context associated with matrilins in vertebrates. BMC Genomics 2006, 7, 83. [Google Scholar] [CrossRef]
- Zhang, X.; Shephard, F.; Kim, H.B.; Palmer, I.R.; McHarg, S.; Fowler, G.J.; O’Neill, L.A.; Kiss-Toth, E.; Qwarnstrom, E.E. Tilrr, a novel il-1ri co-receptor, potentiates myd88 recruitment to control ras-dependent amplification of nf-kappab. J. Biol. Chem. 2010, 285, 7222–7232. [Google Scholar]
- FREM1-FRAS1 Related Extracellular Matrix 1- Genetic Home Reference. Available online: http://ghr.nlm.nih.gov/gene/FREM1/ (accessed on 19 September 2012).
- Levine, M.T.; Jones, C.D.; Kern, A.D.; Lindfors, H.A.; Begun, D.J. Novel genes derived from noncoding DNA in drosophila melanogaster are frequently x-linked and exhibit testis-biased expression. Proc. Natl. Acad. Sci. USA 2006, 103, 9935–9939. [Google Scholar]
- Zhou, Q.; Zhang, G.; Zhang, Y.; Xu, S.; Zhao, R.; Zhan, Z.; Li, X.; Ding, Y.; Yang, S.; Wang, W. On the origin of new genes in drosophila. Genome Res. 2008, 18, 1446–1455. [Google Scholar] [CrossRef]
- Toll-Riera, M.; Bosch, N.; Bellora, N.; Castelo, R.; Armengol, L.; Estivill, X.; Alba, M.M. Origin of primate orphan genes: A comparative genomics approach. Mol. Biol. Evol. 2009, 26, 603–612. [Google Scholar]
- Volff, J.N. Genome evolution and biodiversity in teleost fish. Heredity (Edinb) 2005, 94, 280–294. [Google Scholar] [CrossRef]
- Zhang, X.; Pino, G.M.; Shephard, F.; Kiss-Toth, E.; Qwarnstrom, E.E. Distinct control of myd88-dependent and akt-regulated responses by the il-1ri co-receptor, tilrr. J. Biol. Chem. 2012, 287, 12348–12352. [Google Scholar]
- NCBI Resources Genbank. Available online: http://www.ncbi.nlm.nih.gov/genbank/ (accessed on 19 September 2012).
- NCBI Resources Protein. Available online: http://www.ncbi.nlm.nih.gov/protein/ (accessed on 19 September 2012).
- Ensembl Genome Browser. Available online: http://www.ensembl.org/index.html/ (accessed on 19 September 2012).
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
