Biosynthetic Gene Clusters and Liquid Chromatography Coupled to Mass Spectrometry Analysis of Aryl Polyene Pigments from Chryseobacterium sp. kr6 and Lysobacter sp. A03
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Media
2.2. Genome Mining
2.3. Pigment Extraction
2.4. Spectrophotometry
2.5. LC-DAD-MS Analysis
3. Results and Discussion
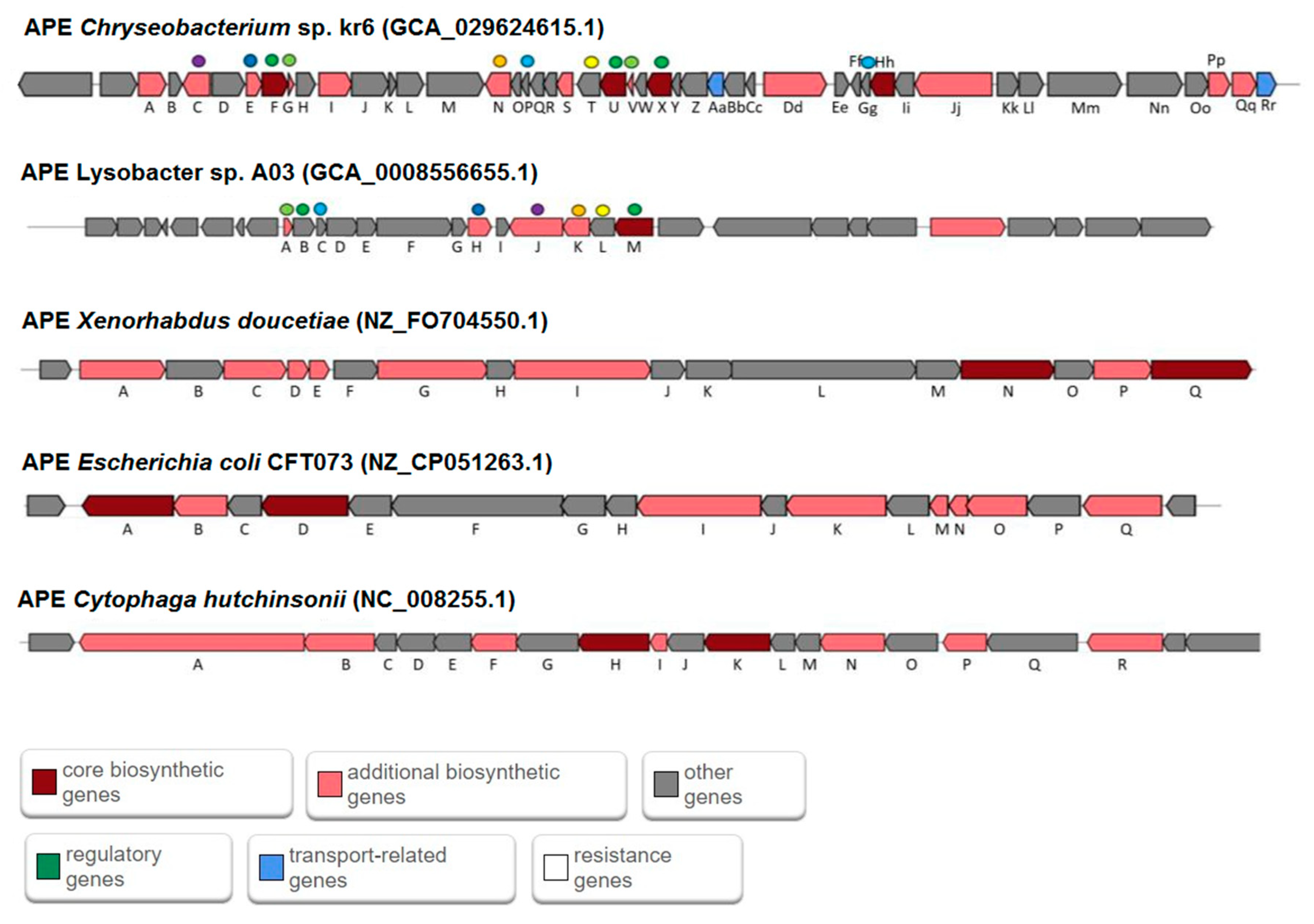
3.1. Identification of Biosynthetic Gene Clusters
3.2. Comparative Analysis of BGCs
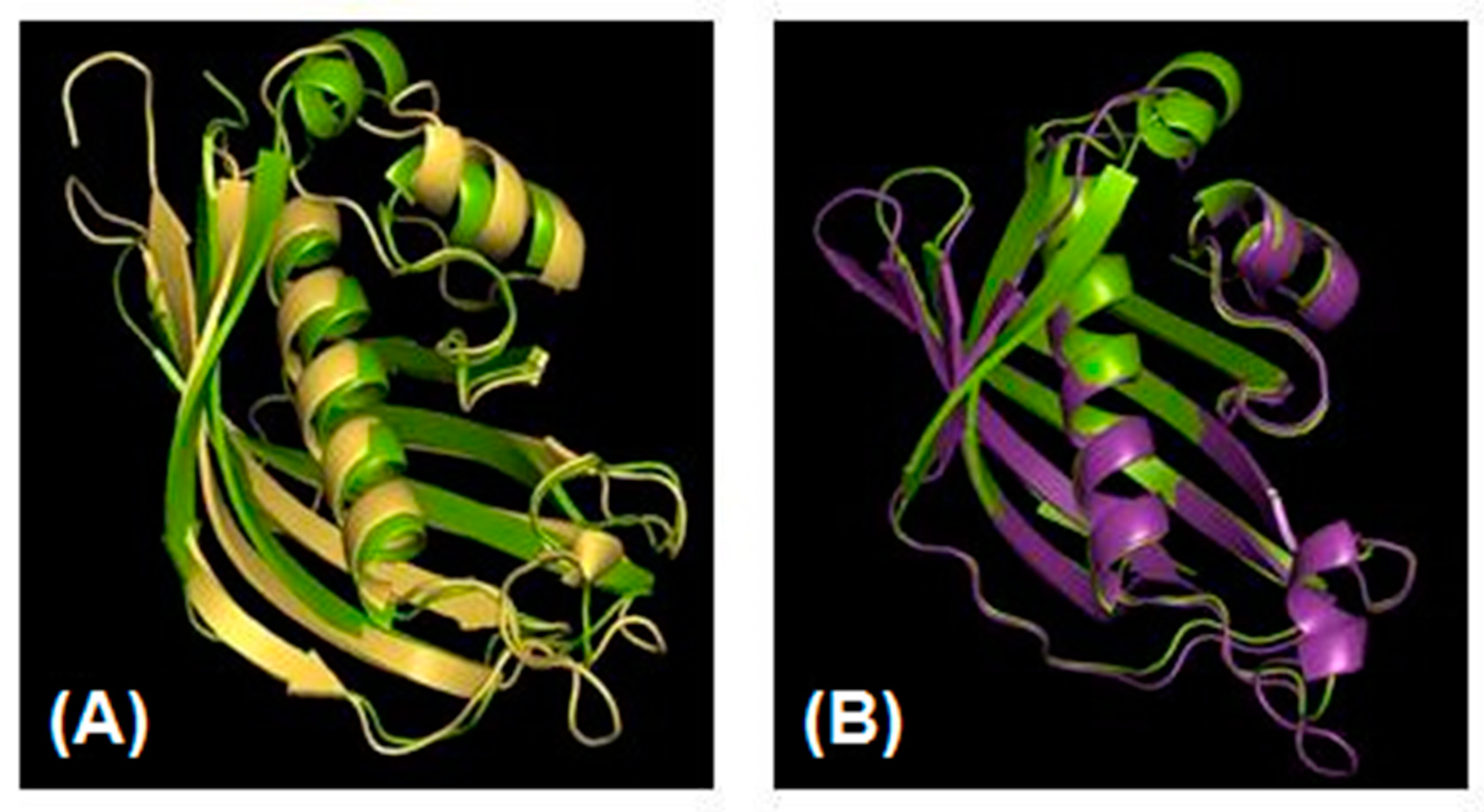
3.3. Analysis of Protein FabA/Z from Strains kr6 and A03
3.4. UV-Vis Spectroscopy and LC-DAD-MS Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Venil, C.K.; Dufossé, L.; Devi, P.R. Bacterial pigments: Sustainable compounds with market potential for pharma and food industry. Front. Sustain. Food Syst. 2020, 4, 100. [Google Scholar] [CrossRef]
- De Mejia, E.G.; Zhang, Q.; Penta, K.; Eroglu, A.; Lila, M.A. The colors of health: Chemistry, bioactivity, and market demand for colorful foods and natural food sources of colorants. Annu. Rev. Food Sci. Technol. 2020, 11, 145–182. [Google Scholar] [CrossRef] [PubMed]
- Sajjad, W.; Din, G.; Rafaq, M.; Iqbal, A.; Khan, S.; Zada., S.; Ali., B.; Kang., S. Pigment production by cold-adapted bacteria and fungi: Colorful tale of cryosphere with wide range applications. Extremophiles 2020, 24, 447–473. [Google Scholar] [CrossRef] [PubMed]
- Velmurugan, P.; Venil, C.K.; Ravi, A.V.; Dufossé, L. Marine bacteria is the cell factory to produce bioactive pigments: A prospective pigment source in the ocean. Front. Sustain. Food Syst. 2020, 4, 589655. [Google Scholar] [CrossRef]
- Wang, Y.; Qian, G.; Li, Y.; Wang, Y.; Wang, Y.; Wright, S.; Li, Y.; Hen, Y.; Liu, F.; Du, L. Biosynthetic mechanism for sunscreens of the biocontrol agent Lysobacter enzymogenes. PLoS ONE 2013, 8, e66633. [Google Scholar] [CrossRef]
- Kumar, S.; Kumar, V.; Ambika, A.A.A.; Nag, D.; Kumar, V.; Darnal, S.; Thakur, V.; Patial, V.; Singh, D. Microbial pigments: Learning from Himalayan perspective to industrial applications. J. Ind. Microbiol. Biotechnol. 2022, 49, kuac017. [Google Scholar] [CrossRef]
- Bai, X.; Zhu, S.; Wang, X.; Zhang, W.; Liu, C.; Lu, X. Identification of a fabZ gene essential for flexirubin synthesis in Cytophaga hutchinsonii. FEMS Microbiol. Lett. 2017, 364, fnx197. [Google Scholar] [CrossRef]
- Sánchez-Muñoz, S.; Mariano-Silva, G.; Leite, M.O.; Mura, F.B.; Verma, M.L.; Da Silva, S.S.; Chandel, A.K. Production of fungal and bacterial pigments and their applications. In Biotechnological Production of Bioactive Compounds; Verma, M.L., Chandel, A.K., Eds.; Elsevier: New York, NY, USA, 2019; pp. 327–361. [Google Scholar]
- Schöner, T.A.; Gassel, S.; Osawa, A.; Tobias, N.J.; Okuno, Y.; Sakakibara, Y.; Shindo, K.; Sandmann, G.; Bode, H.B. Aryl polyenes, a highly abundant class of bacterial natural products, are functionally related to antioxidative carotenoids. ChemBioChem 2016, 17, 247–253. [Google Scholar] [CrossRef]
- Choksi, J.; Vora, J.; Shrivastava, N. Bioactive pigments from isolated bacteria and its antibacterial, antioxidant and sun protective application useful for cosmetic products. Indian J. Microbiol. 2020, 60, 379–382. [Google Scholar] [CrossRef]
- Cao, X.Q.; Wang, J.Y.; Zhou, L.; Chen, B.; Jin, Y.; He, Y.W. Biosynthesis of the yellow xanthomonadin pigments involves an ATP-dependent 3-hydroxybenzoic acid: Acyl carrier protein ligase and an unusual type II polyketide synthase pathway. Mol. Microbiol. 2018, 110, 16–32. [Google Scholar] [CrossRef]
- Johnston, I.; Osborn, L.J.; Markley, R.L.; McManus, E.A.; Kadam, A.; Schultz, K.B.; Nagajothi, N.; Ahern, P.P.; Brown, J.M.; Claesen, J. Identification of essential genes for Escherichia coli aryl polyene biosynthesis and function in biofilm formation. NPJ Biofilms Microbiomes 2021, 7, 56. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.C.; Choi, S.; Jang, A.; Son, K.; Kim, Y. Structural comparison of Acinetobacter baumannii β-ketoacyl-acyl carrier protein reductases in fatty acid and aryl polyene biosynthesis. Sci. Rep. 2021, 11, 7945. [Google Scholar] [CrossRef] [PubMed]
- Grammbitter, G.L.C.; Shi, Y.M.; Shi, Y.N.; Vemulapalli, S.P.B.; Richter, C.; Schwalbe, H.; Alanjary, M.; Schüffler, A.; Witt, M.; Griesinger, C.; et al. The chemical structures of widespread microbial aryl polyene lipids. bioRxiv 2020. [Google Scholar] [CrossRef]
- Jiménez, M.E.P.; Pinilla, C.M.B.; Rodrigues, E.; Brandelli, A. Extraction and partial characterisation of antioxidant pigment produced by Chryseobacterium sp. kr6. Nat. Prod. Res. 2019, 33, 1541–1549. [Google Scholar] [CrossRef]
- Pailliè-Jiménez, M.E.; Stincone, P.; Pereira, J.Q.; Santagapita, P.R.; Rodrigues, E.; Brandelli, A. Isolation and characterization of an antioxidant aryl polyene pigment from Antarctic bacterium Lysobacter sp. A03. Mol. Biotechnol. 2024. ahead of print. [Google Scholar] [CrossRef]
- Pailliè-Jiménez, M.E.; Stincone, P.; Brandelli, A. Natural pigments of microbial origin. Front. Sustain. Food Syst. 2020, 4, 590439. [Google Scholar] [CrossRef]
- Riffel, A.; Brandelli, A. Keratinolytic bacteria isolated from feather waste. Braz. J. Microbiol. 2006, 37, 395–399. [Google Scholar] [CrossRef]
- Pereira, J.Q.; Lopes, F.C.; Petry, M.V.; Medina, L.F.C.; Brandelli, A. Isolation of three novel Antarctic psychrotolerant feather-degrading bacteria and partial purification of keratinolytic enzyme from Lysobacter sp. A03. Int. Biodeterior. Biodegrad. 2014, 88, 1–7. [Google Scholar] [CrossRef]
- Pereira, J.Q.; Ambrosini, A.; Sant’Anna, F.H.; Tadra-Sfeir, M.; Faoro, H.; Pedrosa, F.O.; Souza, E.M.; Brandelli, A.; Passaglia, L.M. Whole-genome shotgun sequence of the keratinolytic bacterium Lysobacter sp. A03, isolated from the Antarctic environment. Genome Announc. 2015, 3, e00246-15. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 12 August 2024).
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef] [PubMed]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongj, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar]
- Blin, K.; Shaw, S.; Augustijn, H.E.; Reitz, Z.L.; Biermann, F.; Alanjary, M.; Fetter, A.; Terlouw, B.R.; Metcalf, W.W.; Helfrich, E.J.; et al. antiSMASH 7.0: New and improved predictions for detection, regulation, chemical structures, and visualization. Nucleic Acids Res. 2023, 51, W46–W50. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Medema, M.H.; Kazempour, D.; Fischbach, M.A.; Breitling, R.; Takano, E.; Weber, T. antiSMASH 2.0—A versatile platform for genome mining of secondary metabolite producers. Nucleic Acids Res. 2013, 41, 204–212. [Google Scholar] [CrossRef]
- He, Y.W.; Cao, X.Q.; Poplawsky, A. Chemical structure, biological roles, biosynthesis and regulation of the yellow xanthomonadin pigments in the phytopathogen Xanthomonas. Mol. Plant-Microbe Interact. 2020, 33, 705–714. [Google Scholar] [CrossRef]
- Kang, D.; Shoaie, S.; Jacquiod, S.; Sørensen, S.J.; Ledesma-Amaro, R. Comparative genomics analysis of Chryseobacterium sp. KMC2 reveals metabolic pathways involved in keratinous utilization and natural product biosynthesis. Microorganisms 2021, 9, 1042. [Google Scholar] [CrossRef]
- Schöner, T.A.; Fuchs, S.W.; Schönau, C.; Bode, H.B. Initiation of the flexirubin biosynthesis in Chitinophaga pinensis. Microb. Biotechnol. 2014, 7, 232–241. [Google Scholar] [CrossRef]
- Sullivan, J.T.; Brown, S.D.; Yocum, R.R.; Ronson, C.W. The bio operon on the acquired symbiosis island of Mesorhizobium sp. strain R7A includes a novel gene involved in pimeloyl-CoA synthesis. Microbiology 2001, 147, 1315–1322. [Google Scholar] [CrossRef][Green Version]
- Sirithanakorn, C.; Cronan, J.E. Biotin, a universal and essential cofactor: Synthesis, ligation and regulation. FEMS Microbiol. Rev. 2021, 45, fuab003. [Google Scholar] [CrossRef]
- Yu, Y.; Ma, J.; Guo, Q.; Ma, J.; Wang, H. A novel 3-oxoacyl-ACP reductase (FabG3) is involved in the xanthomonadin biosynthesis of Xanthomonas campestris pv. campestris. =Mol. Plant Pathol. 2019, 20, 1696–1709. [Google Scholar] [CrossRef]
- Schöner, T.A.; Fuchs, S.W.; Reinhold-Hurek, B.; Bode, H.B. Identification and biosynthesis of a novel xanthomonadin-dialkylresorcinol-hybrid from Azoarcus sp. BH72. PLoS ONE 2014, 9, e90922. [Google Scholar] [CrossRef] [PubMed]
- Jehlička, J.; Osterrothová, K.; Oren, A.; Edwards, H.G.M. Raman spectrometric discrimination of flexirubin pigments from two genera of Bacteroidetes. FEMS Microbiol. Lett. 2013, 348, 97–102. [Google Scholar] [CrossRef] [PubMed]
- McBride, M.J.; Xie, G.; Martens, E.C.; Lapidus, A.; Henrissat, B.; Rhodes, R.G.; Goltsman, E.; Wang, W.; Xu, J.; Hunnicutt, D.W.; et al. Novel features of the polysaccharide-digesting gliding bacterium Flavobacterium johnsoniae as revealed by genome sequence analysis. Appl. Environ. Microbiol. 2009, 75, 6864–6875. [Google Scholar] [CrossRef] [PubMed]
- Brameyer, S.; Kresovic, D.; Bode, H.B.; Heermann, R. Dialkylresorcinols as bacterial signaling molecules. Proc. Natl. Acad. Sci. USA 2015, 112, 572–577. [Google Scholar] [CrossRef]
- Cimermancic, P.; Medema, M.H.; Claesen, J.; Kurita, K.; Brown, L.C.W.; Mavrommatis, K.; Pati, A.; Godfrey, P.A.; Koehrsen, M.; Clardy, J.; et al. Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Cell 2014, 158, 412–421. [Google Scholar] [CrossRef]
- Turrini, P.; Artuso, I.; Tescari, M.; Lugli, G.A.; Frangipani, E.; Ventura, M.; Visca, P. Draft genome sequence and secondary metabolite biosynthetic potential of the Lysobacter niastensis type strain DSM 18481. Microbiol. Resour. Announc. 2021, 10, e01296-20. [Google Scholar] [CrossRef]
- Taniguchi, M.; Lindsey, J.S. Database of absorption and fluorescence spectra of >300 common compounds for use in PhotochemCAD. Photochem. Photobiol. 2018, 94, 290–327. [Google Scholar] [CrossRef]
- PhotochemCAD. Available online: https://www.photochemcad.com/databases/naturally-derived-porphyrins/znii-uroporphyrin-i-in-dmso (accessed on 2 December 2024).
- Andrewes, A.G.; Jenkins, C.L.; Starr, M.P.; Shepherd, J.; Hope, H. Structure of xanthomonadin I, a novel dibrominated aryl-polyene pigment produced by the bacterium Xanthomonas juglandis. Tetrahedron Lett. 1976, 45, 4023–4024. [Google Scholar] [CrossRef]
- Aririatu, L.E.; Kester, A.S. Isolation and characterization of the pigment esters of Xanthomonas juglandis (campestris). Microbiology 1985, 131, 2047–2052. [Google Scholar] [CrossRef][Green Version]
- Madden, K.S.; Laroche, B.; David, S.; Batsanov, A.S.; Thompson, D.; Knowles, J.P.; Whiting, A. Approaches to styrenyl building blocks for the synthesis of polyene xanthomonadin and its analogues. Eur. J. Org. Chem. 2018, 2018, 5312–5322. [Google Scholar] [CrossRef]
- Grammbitter, G.L.C.; Schmallhofer, M.; Karimi, K.; Shi, Y.M.; Schöner, T.A.; Tobias, N.J.; Morgner, N.; Groll, M.; Bode, H.B. An uncommon type II PKS catalyzes biosynthesis of aryl polyene pigments. J. Am. Chem. Soc. 2019, 141, 16615–16623. [Google Scholar] [CrossRef] [PubMed]
- Palazzotto, E.; Weber, T. Omics and multi-omics approaches to study the biosynthesis of secondary metabolites in microorganisms. Curr. Opin. Microbiol. 2018, 45, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Huang, T.W.; Wang, J.Y.; Sun, S.; Chen, G.; Poplawsky, A.; He, Y.W. The rice bacterial pathogen Xanthomonas oryzae pv. oryzae produces 3-hydroxybenzoic acid and 4-hydroxybenzoic acid via XanB2 for use in xanthomonadin, ubiquinone, and exopolysaccharide biosynthesis. Mol. Plant-Microb. Interact. 2013, 26, 1239–1248. [Google Scholar] [CrossRef] [PubMed]
- Starr, M.P.; Jenkins, C.L.; Bussey, L.B.; Andrewes, A.G. Chemotaxonomic significance of the xanthomonadins, novel brominated aryl-polyene pigments produced by bacteria of the genus Xanthomonas. Arch. Microbiol. 1977, 113, 1–9. [Google Scholar] [CrossRef]
- Oren, A. Characterization of pigments of prokaryotes and their use in taxonomy and classification. Meth. Microbiol. 2011, 38, 262–283. [Google Scholar]
- Venil, C.K.; Zakaria, Z.A.; Usha, R.; Ahmad, W.A. Isolation and characterization of flexirubin type pigment Chryseobacterium sp. UTM-3T. Biocatal. Agric. Biotechnol. 2014, 3, 103–107. [Google Scholar] [CrossRef]


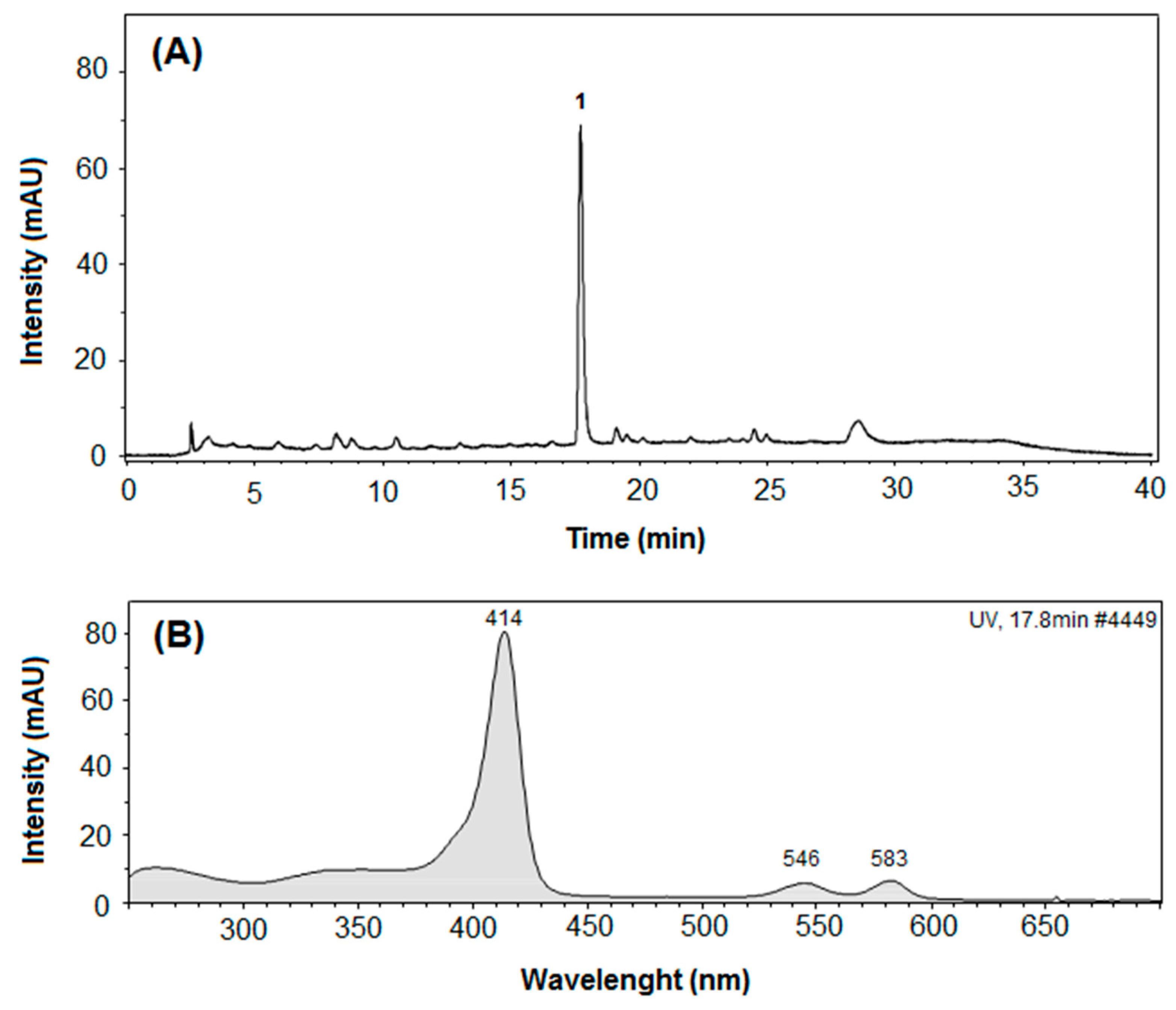

| Peak a | Time (min) b | λmax (nm) | [M − H]− (m/z) | MS/MS (m/z) | [M + Na]+ (m/z) | MS/MS (m/z) |
|---|---|---|---|---|---|---|
| 1 | 17.8 | 414, 548, 583 | 721.5511 | 241, 269, 255 | 745.5109 | 195.0096 |
| 733.5500 | 267, 255, 281, 241 | 757.5127 | n.d. |
| Peak a | Time (min) b | λmax (nm) | [M + H]+ (m/z) | MS/MS (m/z) |
|---|---|---|---|---|
| 1 | 8.0 | 281, 411 | 393.2400 | 149.0291 |
| 2 | 9.8 | 259, 411 | 600.3796 | 310.2496; 210.0609 |
| 3 | 22.2 | 261, 429 | 700.5809 | 133.0926; 177.1192; 561.5103; 299.0695 |
| 4 | 23.2 | 440 | 338.3536 | n.d. |
| 5 | 23.9 | 436 | 334.3175 | n.d. |
| 6 | 24.1 | 448 | 510.4317 | 269.2559 |
| 7 | 24.4 | 440 | 568.4733 | 269.2572 |
| 8 | 24.6 | 448 | 516.4428 | 355.2959 |
| 9 | 25.0 | 448 | 610.2026 | 239.1033; 167.0622; 299.0724; 355.0842 |
| 10 | 25.5 | 448 | 566.4946 | 231.2205 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pailliè-Jiménez, M.E.; Pereira, J.Q.; Rodrigues, E.; Brandelli, A. Biosynthetic Gene Clusters and Liquid Chromatography Coupled to Mass Spectrometry Analysis of Aryl Polyene Pigments from Chryseobacterium sp. kr6 and Lysobacter sp. A03. Colorants 2025, 4, 1. https://doi.org/10.3390/colorants4010001
Pailliè-Jiménez ME, Pereira JQ, Rodrigues E, Brandelli A. Biosynthetic Gene Clusters and Liquid Chromatography Coupled to Mass Spectrometry Analysis of Aryl Polyene Pigments from Chryseobacterium sp. kr6 and Lysobacter sp. A03. Colorants. 2025; 4(1):1. https://doi.org/10.3390/colorants4010001
Chicago/Turabian StylePailliè-Jiménez, Maria Elisa, Jamile Queiroz Pereira, Eliseu Rodrigues, and Adriano Brandelli. 2025. "Biosynthetic Gene Clusters and Liquid Chromatography Coupled to Mass Spectrometry Analysis of Aryl Polyene Pigments from Chryseobacterium sp. kr6 and Lysobacter sp. A03" Colorants 4, no. 1: 1. https://doi.org/10.3390/colorants4010001
APA StylePailliè-Jiménez, M. E., Pereira, J. Q., Rodrigues, E., & Brandelli, A. (2025). Biosynthetic Gene Clusters and Liquid Chromatography Coupled to Mass Spectrometry Analysis of Aryl Polyene Pigments from Chryseobacterium sp. kr6 and Lysobacter sp. A03. Colorants, 4(1), 1. https://doi.org/10.3390/colorants4010001






