Antimicrobial Resistance, an Update from the Ward: Increased Incidence of New Potential Pathogens and Site of Infection-Specific Antibacterial Resistances
Abstract
1. Introduction
2. Results
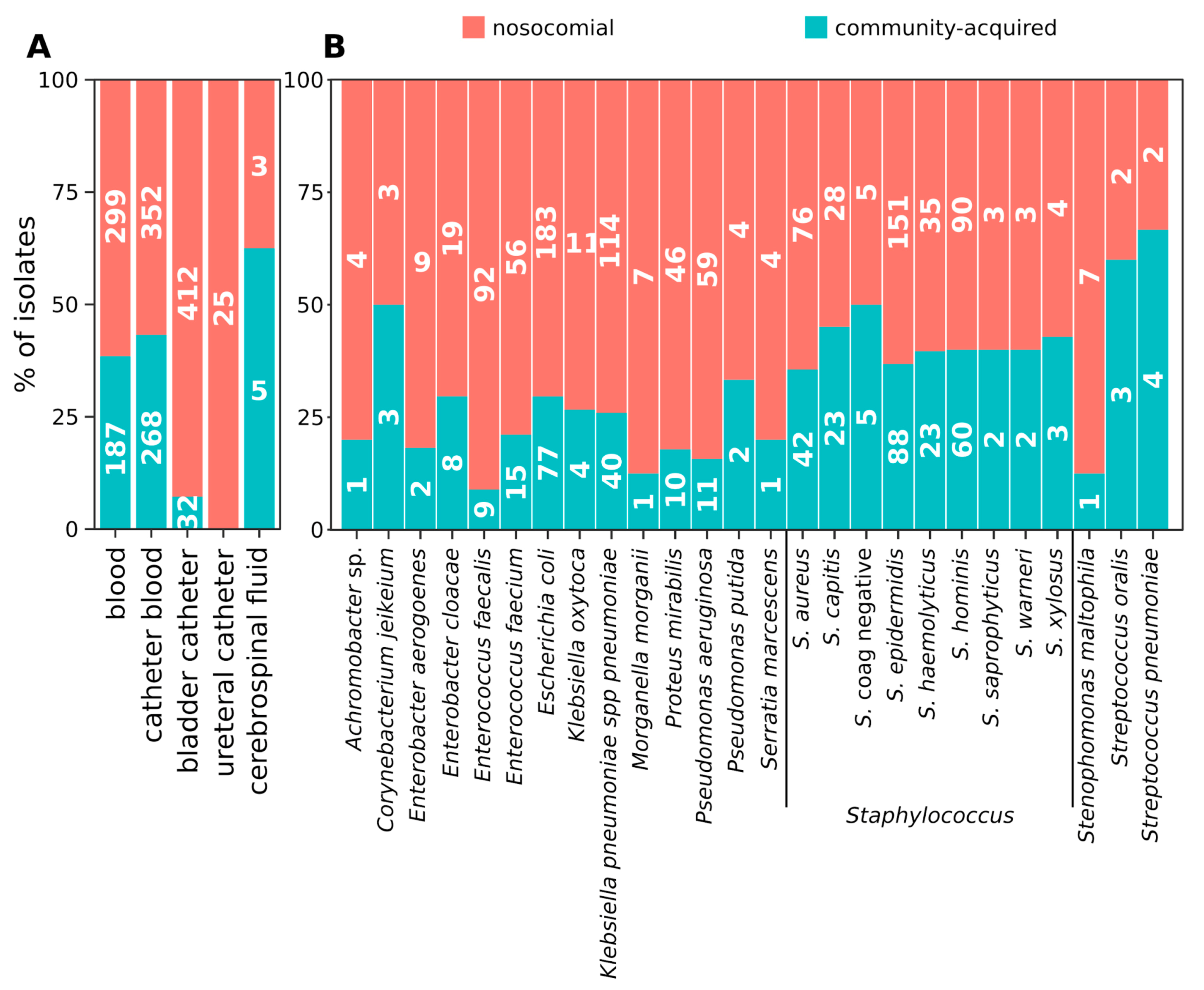
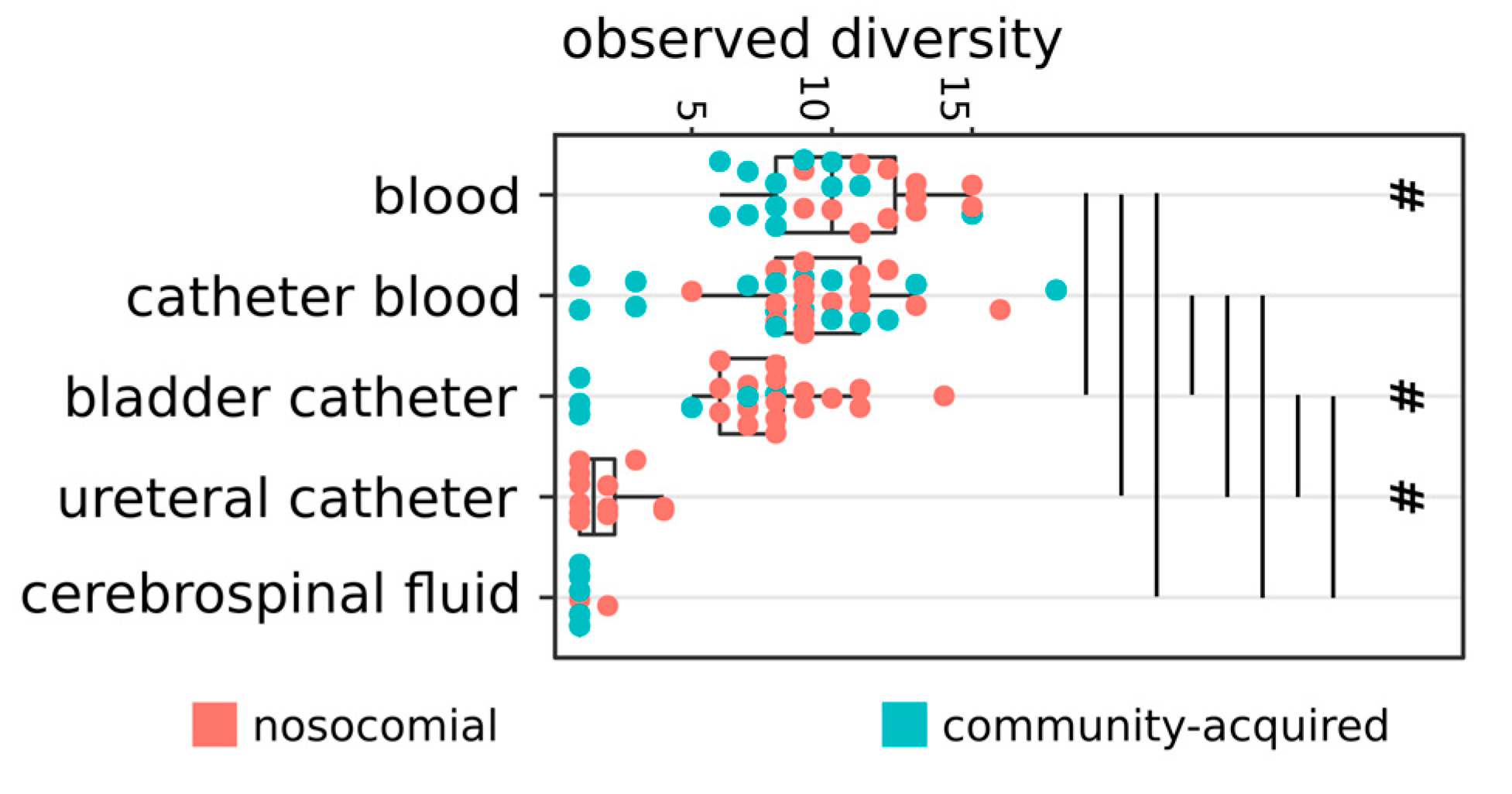
2.1. Distribution of Isolates
2.2. Hospital (HA) and Community-Acquired (CA) Infections
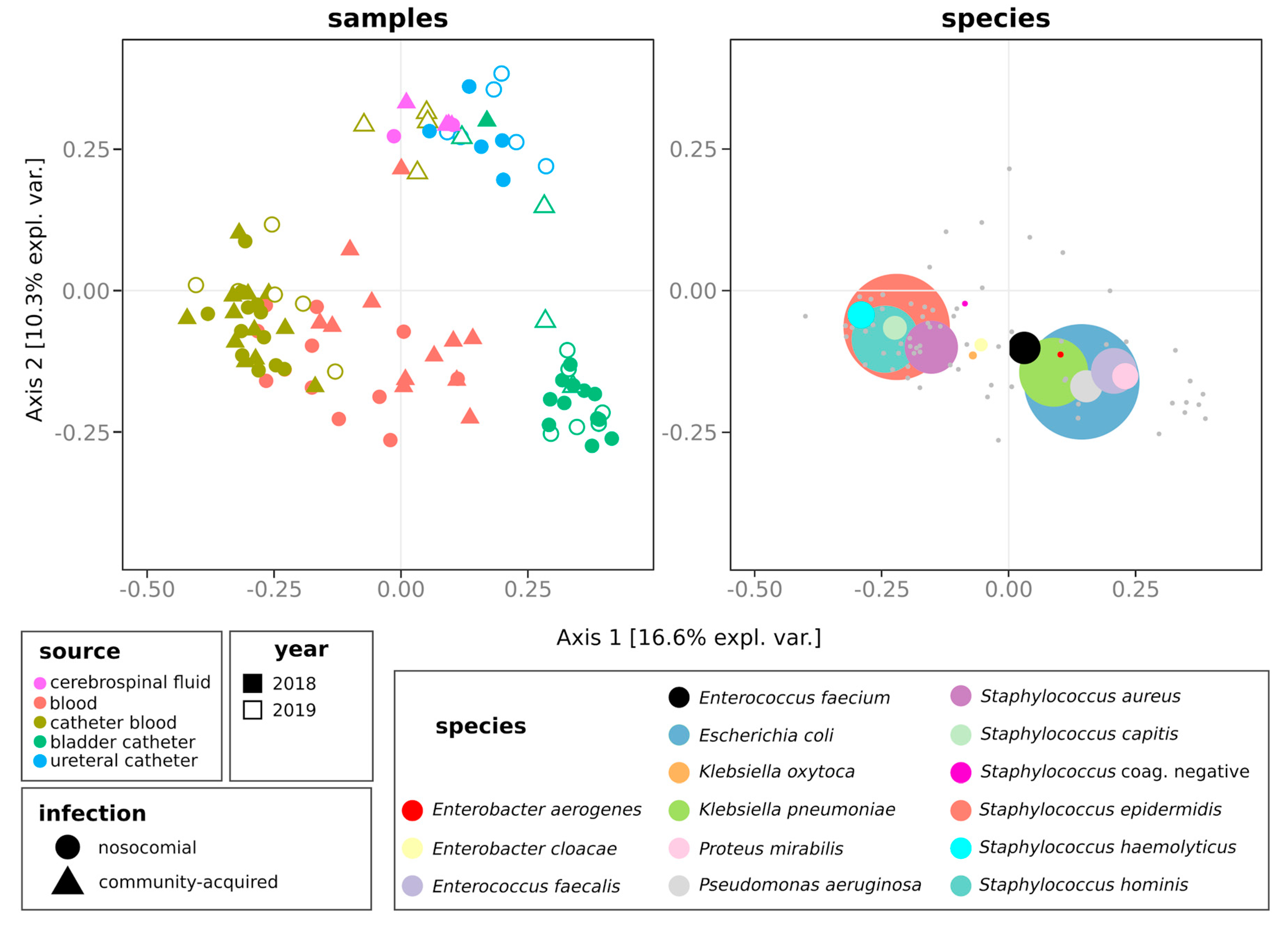
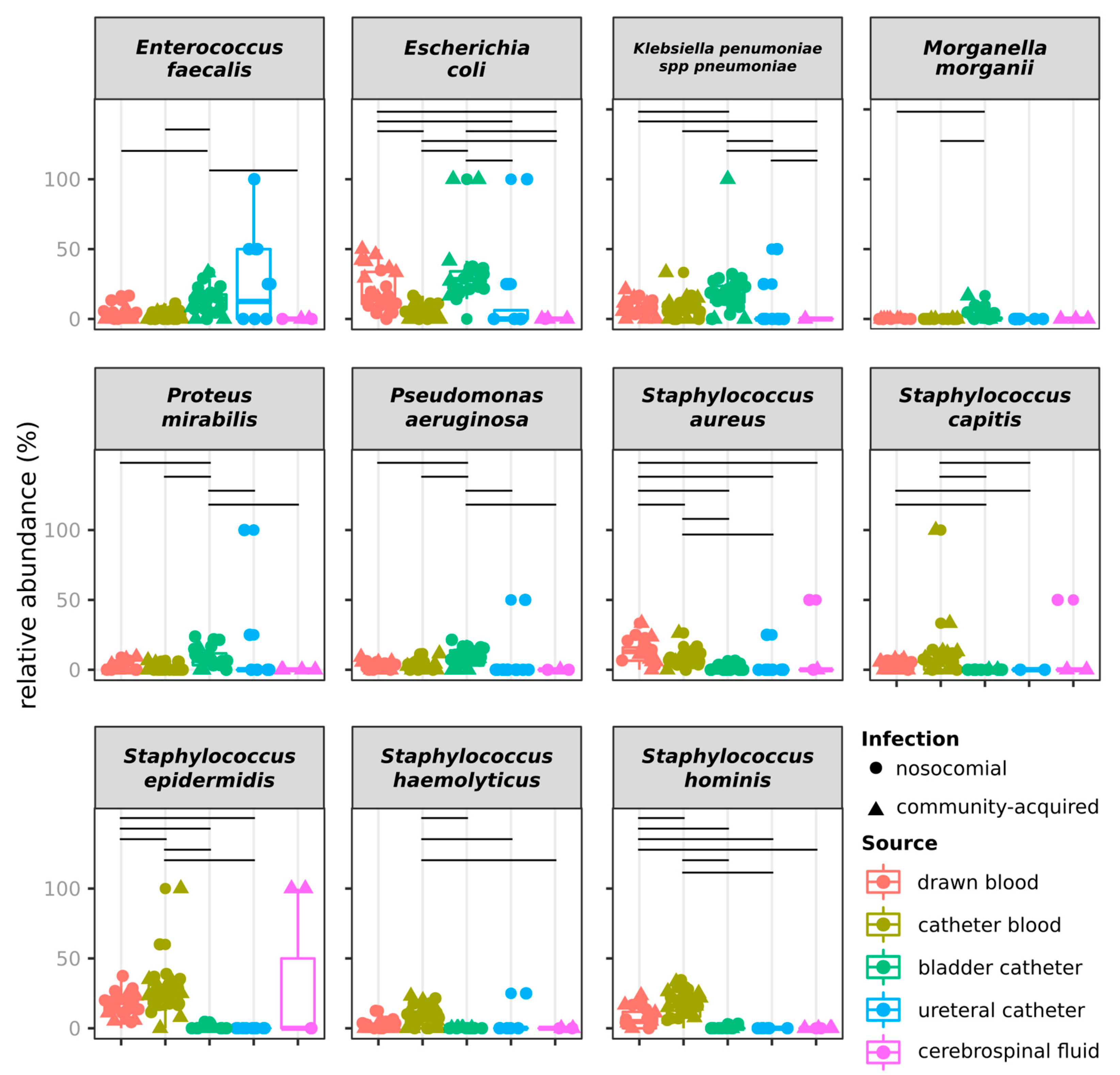
2.3. Distribution of Species
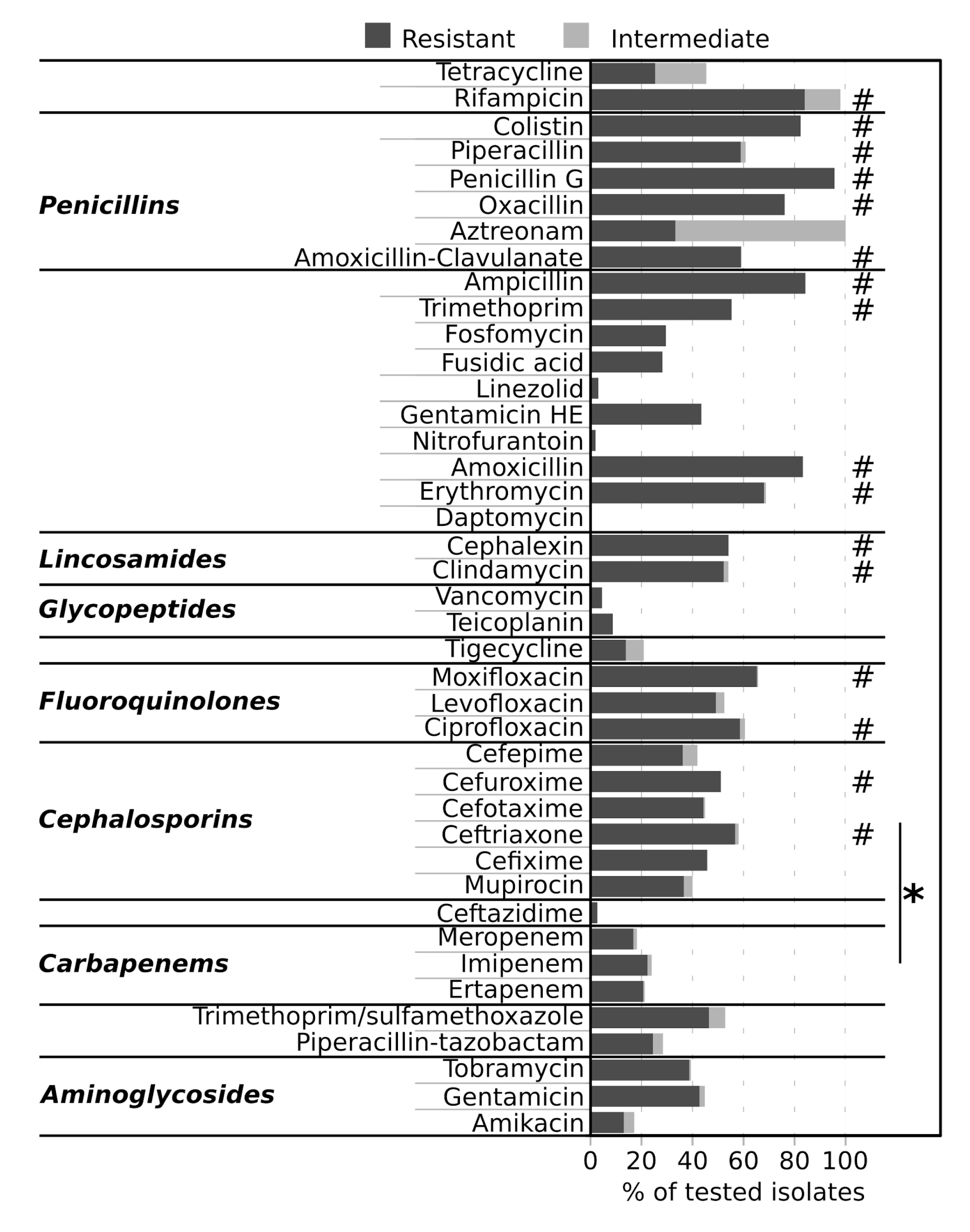
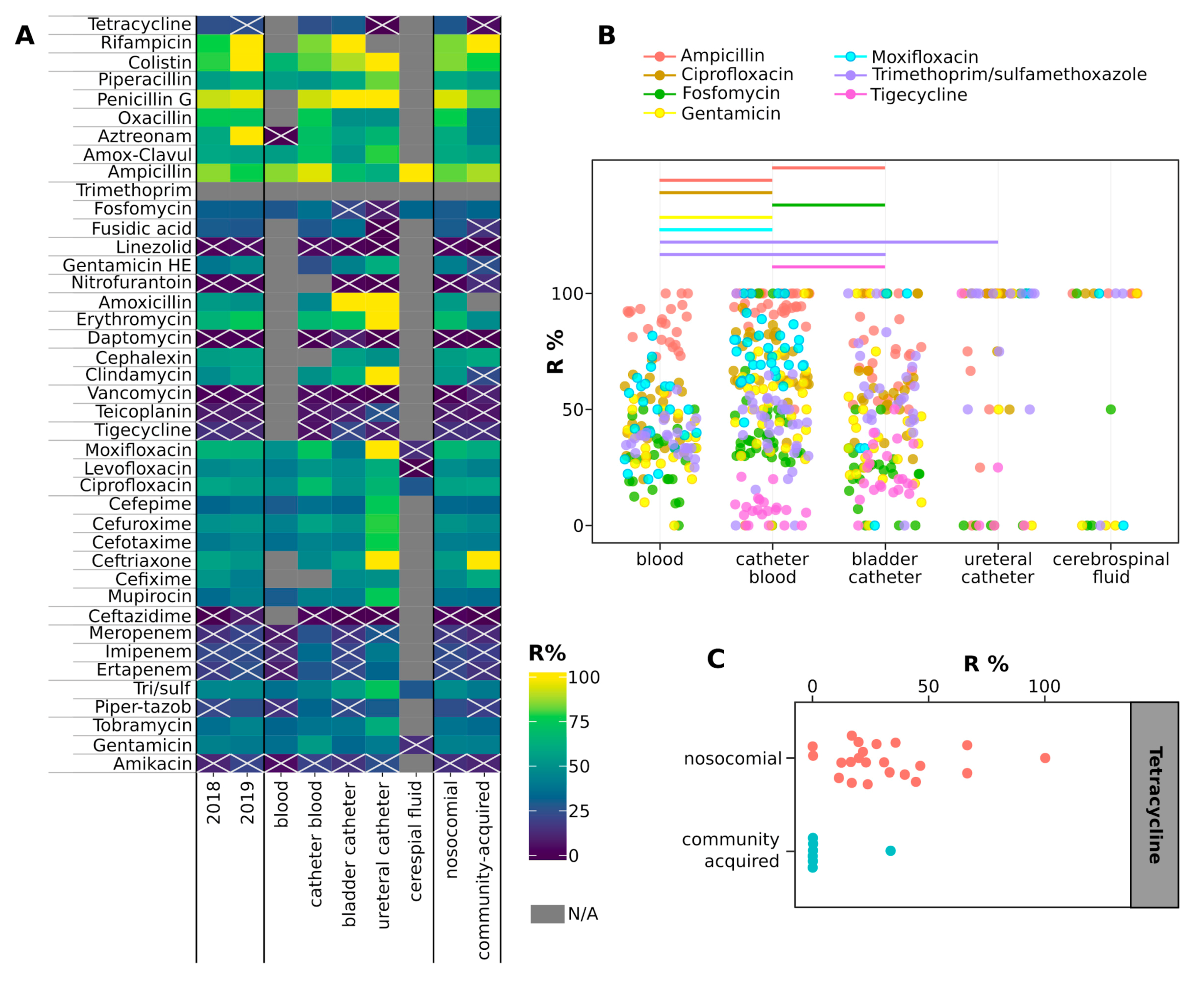
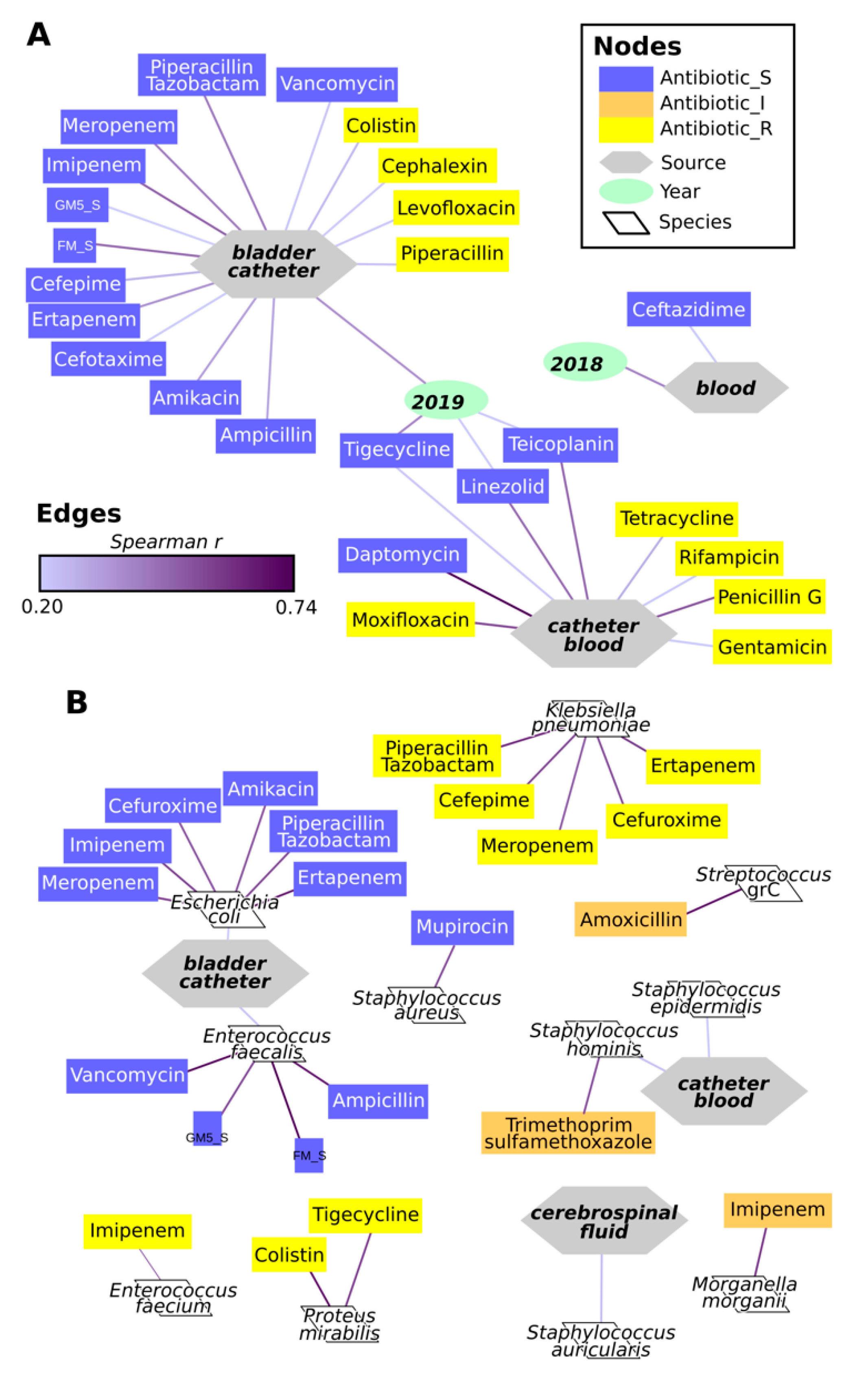
2.4. Antimicrobial Resistance of Isolates
3. Discussion
4. Materials and Methods
4.1. Isolation of Strains
4.2. Antimicrobial Testing
4.3. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- WHO. WHO—Antimicrobial Resistance. Available online: https://www.who.int/health-topics/antimicrobial-resistance (accessed on 7 July 2020).
- WHO. Prioritization of Pathogens to Guide Discovery, Research and Development of New Antibiotics for Drug-Resistant Bacterial Infections Including Tuberculosis. 2017. Available online: https://www.who.int/medicines/areas/rational_use/PPLreport_2017_09_19.pdf?ua=1 (accessed on 7 July 2020).
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. WHO Pathogens Priority List Working Group. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Goossens, H.; Ferech, M.; Stichele, R.V.; Elseviers, M.; ESAC Project Group. Outpatient antibiotic use in Europe and association with resistance: A cross-national database study. Lancet 2005, 365, 579–587. [Google Scholar] [CrossRef]
- Malhotra-Kumar, S.; Lammens, C.; Coenen, S.; van Herck, K.; Goossens, H. Effect of azithromycin and clarithromycin therapy on pharyngeal carriage of macrolide-resistant streptococci in healthy volunteers: A randomised, double-blind, placebo-controlled study. Lancet 2007, 369, 482–490. [Google Scholar] [CrossRef]
- Fridkin, S.K.; Edwards, J.R.; Courval, J.M.; Hill, H.; Tenover, F.C.; Lawton, R.; Gaynes, R.P.; McGowan, J.E., Jr.; Intensive Care Antimicrobial Resistance Epidemiology (ICARE) Project; The National Nosocomial Infections Surveillance (NNIS) System Hospitals. The effect of vancomycin and third-generation cephalosporins on prevalence of vancomycin-resistant enterococci in 126 U.S. adult intensive care units. Ann. Intern. Med. 2001, 135, 175–183. [Google Scholar] [CrossRef] [PubMed]
- Costelloe, C.; Metcalfe, C.; Lovering, A.; Mant, D.; Hay, A.D. Effect of antibiotic prescribing in primary care on antimicrobial resistance in individual patients: Systematic review and meta-analysis. BMJ 2010, 340, c2096. [Google Scholar] [CrossRef]
- OsMed. L’uso dei Farmaci in Italia—Rapporto Nazionale 2018. 2018. Available online: https://www.aifa.gov.it/documents/20142/0/Rapporto_OsMed_2018.pdf/c9eb79f9-b791-2759-4a9e-e56e1348a976 (accessed on 7 July 2020).
- WHO. WHO Report On Surveillance Of Antibiotic Consumption 2016–2018 Early Implementation. 2018. Available online: https://apps.who.int/iris/bitstream/handle/10665/277359/9789241514880-eng.pdf?ua=1 (accessed on 5 July 2020).
- Safdar, N.; Maki, D.G. The commonality of risk factors for nosocomial colonization and infection with antimicrobial-resistant Staphylococcus aureus, enterococcus, gram-negative bacilli, Clostridium difficile, and Candida. Ann. Intern. Med. 2002, 136, 834–844. [Google Scholar] [CrossRef]
- van Duin, D.; Paterson, D.L. Multidrug-resistant bacteria in the community: Trends and lessons learned. Inf. Dis. Clin. N. Am. 2016, 30, 377–390. [Google Scholar] [CrossRef]
- Jeukens, J.; Freschi, L.; Kukavica-Ibrulj, I.; Emond-Rheault, J.G.; Tucker, N.P.; Levesque, R.C. Genomics of antibiotic-resistance prediction in Pseudomonas aeruginosa. Ann. N. Y. Acad. Sci. 2019, 1435, 5–17. [Google Scholar] [CrossRef]
- Nguyen, M.; Long, S.W.; McDermott, P.F.; Olsen, R.J.; Olson, R.; Stevens, R.L.; Tyson, G.H.; Zhao, S.; Davis, J.J. Using machine learning to predict antimicrobial MICs and associated genomic features for nontyphoidal Salmonella. J. Clin. Microbiol. 2019, 57, e01260-18. [Google Scholar] [CrossRef]
- Saha, S.K.; Kong, D.C.M.; Thursky, K.; Mazza, D. A nationwide survey of australian general practitioners on antimicrobial stewardship: Awareness, uptake, collaboration with pharmacists and improvement strategies. Antibiotics 2020, 9, 310. [Google Scholar] [CrossRef]
- di Giuseppe, G.; Mitidieri, M.; Cantore, F.; Pelullo, C.P.; Pavia, M. Community-acquired and healthcare-associated sepsis: Characteristics and in-hospital mortality in Italy. Antibiotics 2020, 9, 263. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Ji, X. Analysis of the microbial species, antimicrobial sensitivity and drug resistance in 2652 patients of nursing hospital. Heliyon 2020, 6, e03965. [Google Scholar] [CrossRef] [PubMed]
- Chanda, W.; Manyepa, M.; Chikwanda, E.; Daka, V.; Chileshe, J.; Tembo, M.; Kasongo, J.; Chipipa, A.; Handema, R.; Mulemena, J.A. Evaluation of antibiotic susceptibility patterns of pathogens isolated from routine laboratory specimens at Ndola Teaching Hospital: A retrospective study. PLoS ONE 2019, 14, e0226676. [Google Scholar] [CrossRef]
- Vandenesch, F.; Célard, M.; Arpin, D.; Bes, M.; Greenland, T.; Etienne, J. Catheter-related bacteremia associated with coagulase-positive Staphylococcus intermedius. J. Clin. Microbiol. 1995, 33, 2508–2510. [Google Scholar] [CrossRef]
- Nemeghaire, S.; Argudín, M.A.; Feßler, A.T.; Hauschild, T.; Schwarz, S.; Butaye, P. The ecological importance of the Staphylococcus sciuri species group as a reservoir for resistance and virulence genes. Vet. Microbiol. 2014, 171, 342–356. [Google Scholar] [CrossRef]
- Swe, T.; Naing, A.T.; Baqui, A.; Khillan, R. Methicillin-resistant Staphylococcus schleiferi subspecies coagulans infection in a patient with hepatocellular carcinoma. J. Investig Med. High. Impact Case Rep. 2016, 4, 232470961667114. [Google Scholar] [CrossRef]
- Geerlings, S.E. Clinical presentations and epidemiology of urinary tract infections. Microbiol. Spectr. 2016, 4. [Google Scholar] [CrossRef]
- Martin, R.M.; Zimmermann, L.L.; Huynh, M.; Polage, C.R. Diagnostic approach to health care- and device-associated central nervous system infections. J. Clin. Microbiol. 2018, 56, e00861-18. [Google Scholar] [CrossRef]
- Pailhoriès, H.; Cassisa, V.; Chenouard, R.; Kempf, M.; Eveillard, M.; Lemarié, C. Staphylococcus saprophyticus: Which beta-lactam? Int. J. Infect. Dis. 2017, 65, 63–66. [Google Scholar] [CrossRef][Green Version]
- Navon-Venezia, S.; Kondratyeva, K.; Carattoli, A. Klebsiella pneumoniae: A major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol. Rev. 2017, 41, 252–275. [Google Scholar] [CrossRef]
- Torres, C.; Alonso, C.A.; Ruiz-Ripa, L.; León-Sampedro, R.; del Campo, R.; Coque, T.M. Antimicrobial resistance in Enterococcus spp. of animal origin. Microbiol. Spectr. 2018, 6. [Google Scholar] [CrossRef]
- World Health Organization; Department of Communicable Disease S and R. Prevention of hospital-Acquired Infections. A Practical Guide, 2nd ed.; Lyon: Geneva, Switzerland, 2002; Available online: https://apps.who.int/iris/bitstream/handle/10665/67350/WHO_CDS_CSR_EPH_2002.12.pdf (accessed on 5 July 2020).
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]
- Westfall, P.H.; Young, S.S. Resampling-Based Multiple Testing: Examples and Methods for p-Value Adjustment; Wiley: New York, NY, USA, 1993. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing 2019; R Foundation for Statistical Computing: Vienna, Austria; Available online: https://www.R-project.org/ (accessed on 27 August 2020).
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stefanini, I.; Boni, M.; Silvaplana, P.; Lovera, P.; Pelassa, S.; De Renzi, G.; Mognetti, B. Antimicrobial Resistance, an Update from the Ward: Increased Incidence of New Potential Pathogens and Site of Infection-Specific Antibacterial Resistances. Antibiotics 2020, 9, 631. https://doi.org/10.3390/antibiotics9090631
Stefanini I, Boni M, Silvaplana P, Lovera P, Pelassa S, De Renzi G, Mognetti B. Antimicrobial Resistance, an Update from the Ward: Increased Incidence of New Potential Pathogens and Site of Infection-Specific Antibacterial Resistances. Antibiotics. 2020; 9(9):631. https://doi.org/10.3390/antibiotics9090631
Chicago/Turabian StyleStefanini, Irene, Martina Boni, Paola Silvaplana, Paola Lovera, Stefania Pelassa, Giuseppe De Renzi, and Barbara Mognetti. 2020. "Antimicrobial Resistance, an Update from the Ward: Increased Incidence of New Potential Pathogens and Site of Infection-Specific Antibacterial Resistances" Antibiotics 9, no. 9: 631. https://doi.org/10.3390/antibiotics9090631
APA StyleStefanini, I., Boni, M., Silvaplana, P., Lovera, P., Pelassa, S., De Renzi, G., & Mognetti, B. (2020). Antimicrobial Resistance, an Update from the Ward: Increased Incidence of New Potential Pathogens and Site of Infection-Specific Antibacterial Resistances. Antibiotics, 9(9), 631. https://doi.org/10.3390/antibiotics9090631






