Phages for Biofilm Removal
Abstract
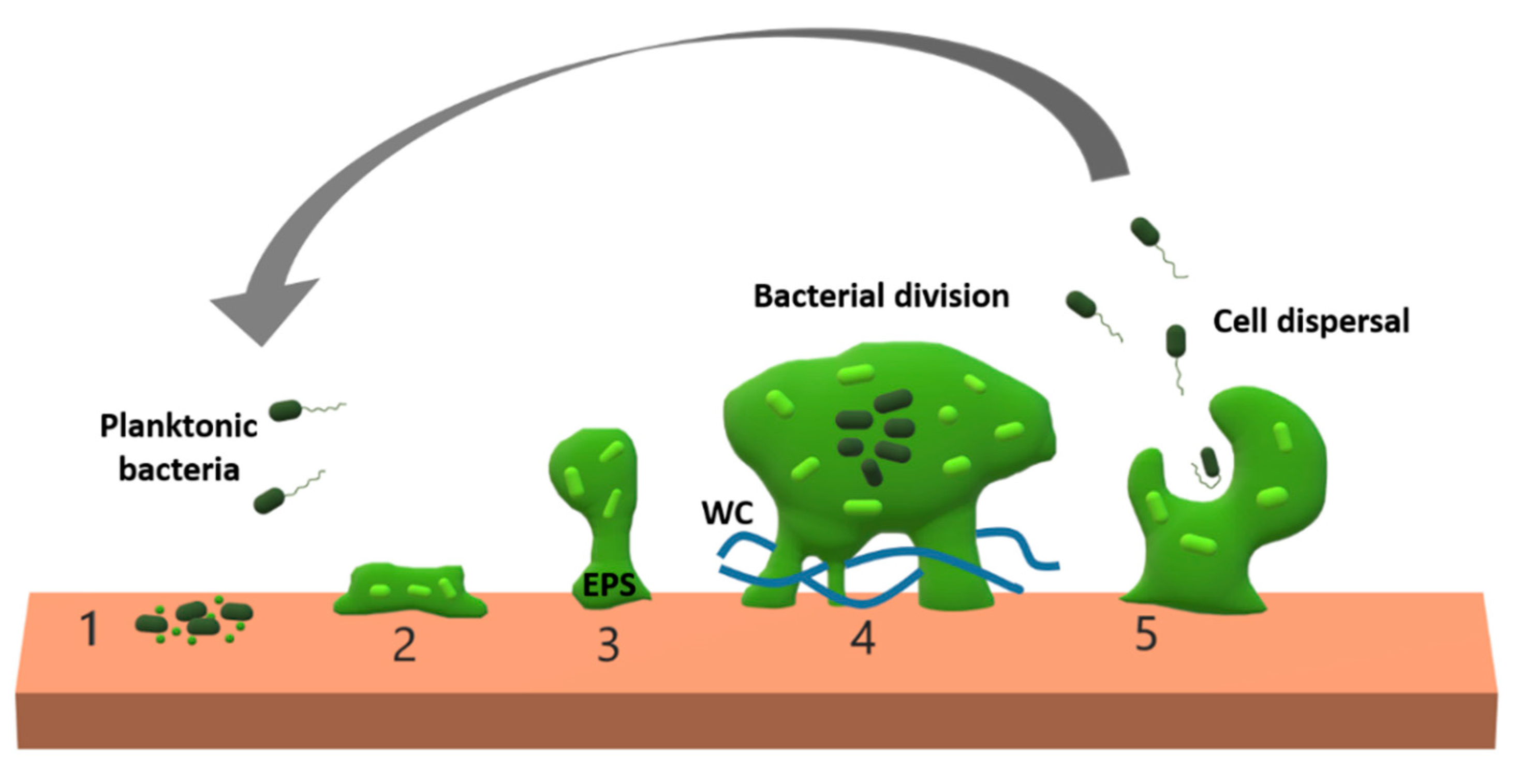
1. Introduction
2. Biofilm Features Affecting Phage Penetration, Diffusion, and Propagation
2.1. Anti-Phage Role of Biofilm Matrices
2.2. Physiological Heterogeneity Inside Biofilms
2.3. Species and Strain Composition of the Biofilm
3. Bacteria–Phage Co-Evolution Within Biofilms
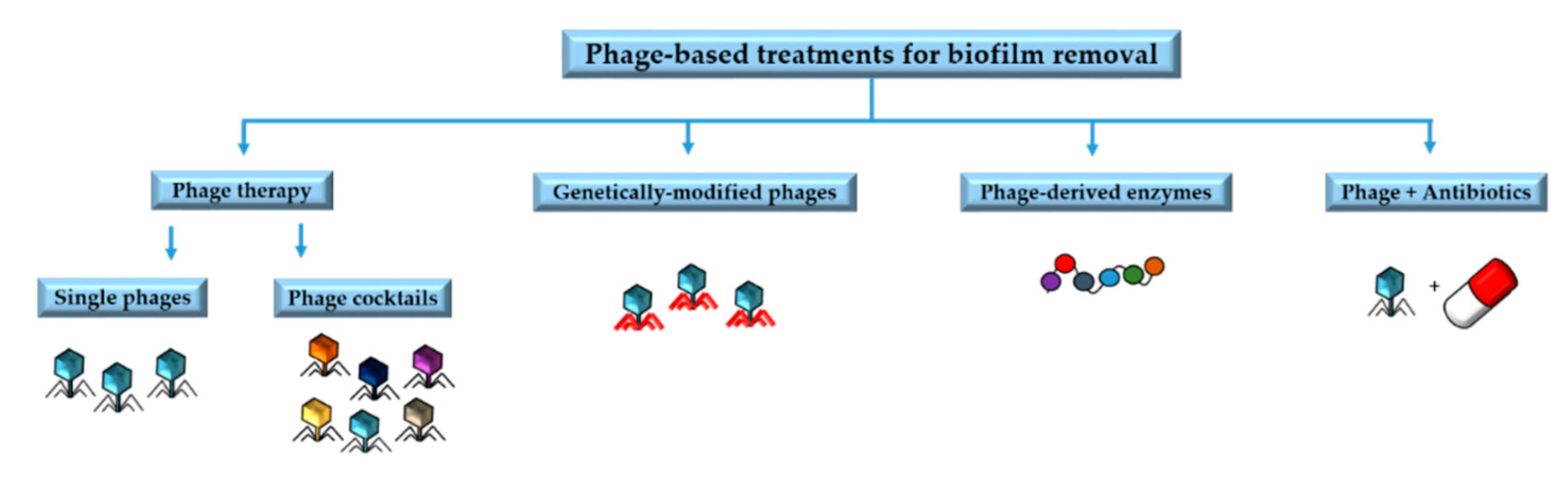
4. Phage Applications against Biofilms
4.1. Phage Therapy
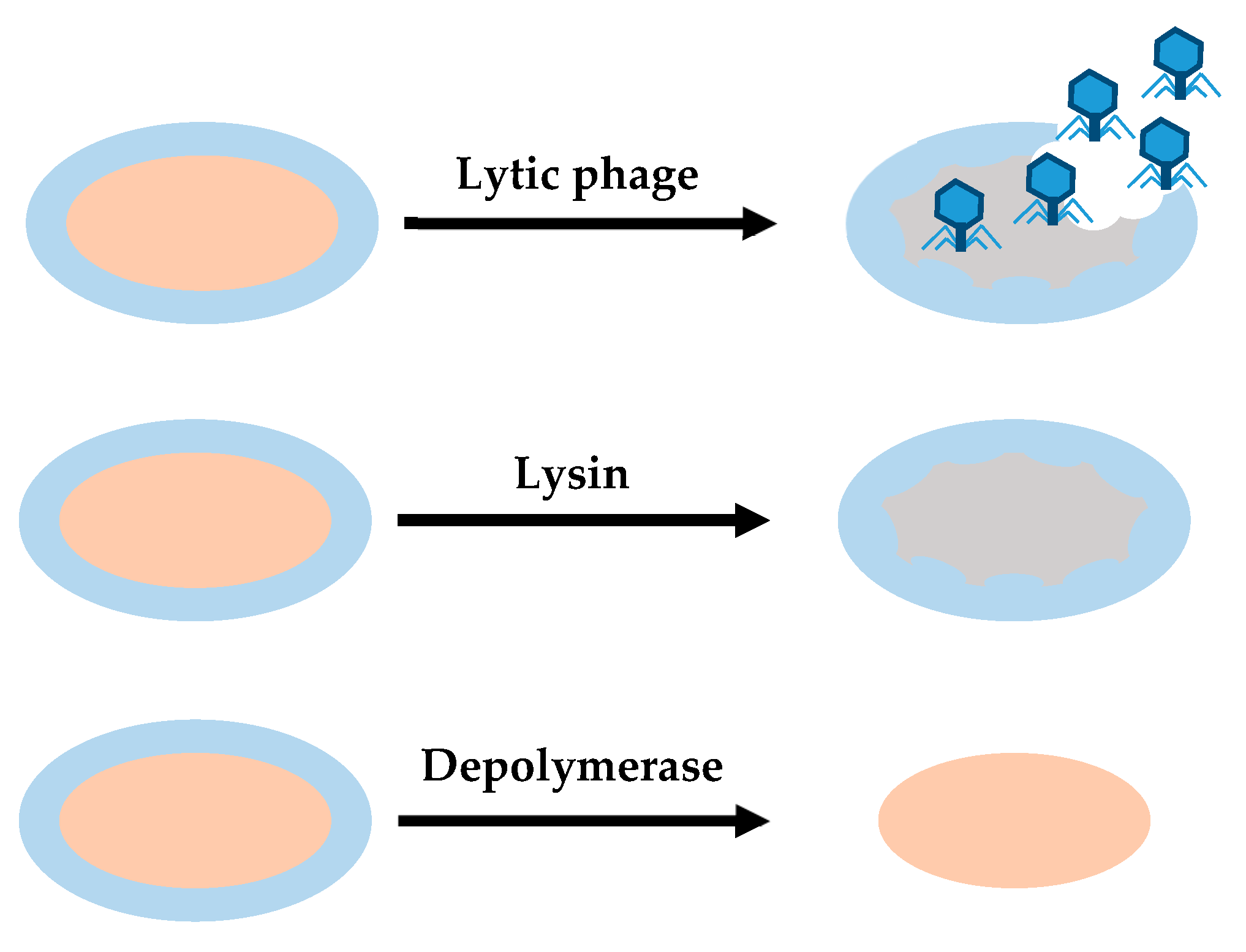
4.2. Phage-Derived Enzymes
4.2.1. Lysins
4.2.2. Depolymerases
4.3. Genetically Modified Phages
4.4. Phages in Combination with Antibiotics
5. Problematic Biofilms in Anthropogenic Spaces
5.1. Infectious Diseases Caused by Biofilms
5.2. Biofilms in Medical Environments and Medical Devices
5.3. Biofilms in Industrial Sectors
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Nazar, C.J. Biofilms bacterianos. Rev. Otorrinolaringol. Cir. Cabeza Cuello 2007, 67, 161–172. [Google Scholar] [CrossRef]
- Jamal, M.; Ahmad, W.; Andleeb, S.; Jalil, F.; Imran, M.; Nawaz, M.A.; Hussain, T.; Ali, M.; Rafiq, M.; Kamil, M.A. Bacterial biofilm and associated infections. J. Chin. Med. Assoc. 2018, 81, 7–11. [Google Scholar] [CrossRef]
- González, S.; Fernández, L.; Campelo, A.B.; Gutiérrez, D.; Martínez, B.; Rodríguez, A.; García, P. The behavior of Staphylococcus aureus dual-species biofilms treated with bacteriophage PhiIPLA-RODI depends on the accompanying microorganism. Appl. Environ. Microbiol. 2017, 83, e02821-16. [Google Scholar] [CrossRef]
- Donlan, R.M. Biofilms: Microbial life on surfaces. Emerg. Infect. Dis. 2002, 8, 881–890. [Google Scholar] [CrossRef]
- Potera, C. Forging a link between biofilms and disease. Science 1999, 283, 1837–1839. [Google Scholar] [CrossRef]
- Flemming, H.-C.; Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. [Google Scholar] [CrossRef]
- Stoodley, P.; Sauer, K.; Davies, D.G.; Costerton, J.W. Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 2002, 56, 187–209. [Google Scholar] [CrossRef]
- Lasa, I.; Pozo, J.L.; Del Penadés, J.R.; Leiva, J. Biofilms bacterianos e infección. An. Sist. Sanit. Navar. 2005, 28, 163–175. [Google Scholar] [CrossRef]
- Lappin-Scott, H.; Burton, S.; Stoodley, P. Revealing a world of biofilms—The pioneering research of Bill Costerton. Nat. Rev. Microbiol. 2014, 12, 781–787. [Google Scholar] [CrossRef]
- Solano, C.; Echeverz, M.; Lasa, I. Biofilm dispersion and quorum sensing. Curr. Opin. Microbiol. 2014, 18, 96–104. [Google Scholar] [CrossRef]
- Musk, D., Jr.; Hergenrother, P. Chemical countermeasures for the control of bacterial biofilms: Effective compounds and promising targets. Curr. Med. Chem. 2006, 13, 2163–2177. [Google Scholar] [CrossRef]
- Fernández, L.; Rodríguez, A.; García, P. Phage or foe: An insight into the impact of viral predation on microbial communities. ISME J. 2018, 12, 1171–1179. [Google Scholar] [CrossRef]
- Rossmann, F.S.; Racek, T.; Wobser, D.; Puchalka, J.; Rabener, E.M.; Reiger, M.; Hendrickx, A.P.A.; Diederich, A.-K.; Jung, K.; Klein, C.; et al. Phage-mediated dispersal of biofilm and distribution of bacterial virulence genes is induced by quorum sensing. PLoS Pathog. 2015, 11, e1004653. [Google Scholar] [CrossRef]
- Li, Y.H.; Tian, X. Quorum sensing and bacterial social interactions in biofilms. Sensors 2012, 12, 2519–2538. [Google Scholar] [CrossRef]
- Parsek, M.R.; Greenberg, E.P. Sociomicrobiology: The connections between quorum sensing and biofilms. Trends Microbiol. 2005, 13, 27–33. [Google Scholar] [CrossRef]
- Mah, T.-F. Biofilm-specific antibiotic resistance. Future Microbiol. 2012, 7, 1061–1072. [Google Scholar] [CrossRef]
- Stewart, P.S. Antimicrobial tolerance in biofilms. Microbiol. Spectr. 2015, 3, 269–285. [Google Scholar] [CrossRef]
- Hall, C.W.; Mah, T.-F. Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol. Rev. 2017, 41, 276–301. [Google Scholar] [CrossRef]
- Davies, D. Understanding biofilm resistance to antibacterial agents. Nat. Rev. Drug Discov. 2003, 2, 114–122. [Google Scholar] [CrossRef]
- Sønderholm, M.; Bjarnsholt, T.; Alhede, M.; Kolpen, M.; Jensen, P.Ø.; Kühl, M.; Kragh, K.N. The consequences of being in an infectious biofilm: Microenvironmental conditions governing antibiotic tolerance. Int. J. Mol. Sci. 2017, 18, 2688. [Google Scholar] [CrossRef]
- Fauconnier, A. Regulating phage therapy. EMBO Rep. 2017, 18, 198–200. [Google Scholar] [CrossRef]
- Moelling, K.; Broecker, F.; Willy, C. A wake-up call: We need phage therapy now. Viruses 2018, 10, 688. [Google Scholar] [CrossRef]
- Penadés, J.R.; Chen, J.; Quiles-Puchalt, N.; Carpena, N.; Novick, R.P. Bacteriophage-mediated spread of bacterial virulence genes. Curr. Opin. Microbiol. 2015, 23, 171–178. [Google Scholar] [CrossRef]
- Abedon, S.T.; Kuhl, S.J.; Blasdel, B.G.; Kutter, E.M. Phage treatment of human infections. Bacteriophage 2011, 1, 66–85. [Google Scholar]
- González, S.; Fernández, L.; Gutiérrez, D.; Campelo, A.B.; Rodríguez, A.; García, P. Analysis of different parameters affecting diffusion, propagation and survival of Staphylophages in bacterial biofilms. Front. Microbiol. 2018, 9, 2348. [Google Scholar] [CrossRef]
- Lebeaux, D.; Ghigo, J.-M.; Beloin, C. Biofilm-related infections: Bridging the gap between clinical management and fundamental aspects of recalcitrance toward antibiotics. Microbiol. Mol. Biol. Rev. 2014, 78, 510–543. [Google Scholar] [CrossRef]
- Simmons, M.; Drescher, K.; Nadell, C.D.; Bucci, V. Phage mobility is a core determinant of phage–bacteria Coexistence in Biofilms. ISME J. 2018, 12, 531–543. [Google Scholar] [CrossRef]
- Hanlon, G.W.; Denyer, S.P.; Olliff, C.J.; Ibrahim, L.J. Reduction in exopolysaccharide viscosity as an aid to bacteriophage penetration through Pseudomonas Aeruginosa biofilms. Appl. Environ. Microbiol. 2001, 67, 2746–2753. [Google Scholar] [CrossRef]
- Abedon, S.T. Phage “delay” towards enhancing bacterial escape from biofilms: A more comprehensive way of viewing resistance to bacteriophages. AIMS Microbiol. 2017, 3, 186–226. [Google Scholar] [CrossRef]
- Romaní, A.M.; Fund, K.; Artigas, J.; Schwartz, T.; Sabater, S.; Obst, U. Relevance of polymeric matrix enzymes during biofilm formation. Microb. Ecol. 2008, 56, 427–436. [Google Scholar] [CrossRef]
- Abedon, S.T. Bacteriophage exploitation of bacterial biofilms: Phage preference for less mature targets? FEMS Microbiol. Lett. 2016, 363, 246. [Google Scholar] [CrossRef]
- Briandet, R.; Lacroix-Gueu, P.; Renault, M.; Lecart, S.; Meylheuc, T.; Bidnenko, E.; Steenkeste, K.; Bellon-Fontaine, M.-N.; Fontaine-Aupart, M.-P. Fluorescence correlation spectroscopy to study diffusion and reaction of bacteriophages inside biofilms. Appl. Environ. Microbiol. 2008, 74, 2135–2143. [Google Scholar] [CrossRef]
- Vilas Boas, D.; Almeida, C.; Sillankorva, S.; Nicolau, A.; Azeredo, J.; Azevedo, N.F. Discrimination of bacteriophage infected cells using locked nucleic acid fluorescent in situ hybridization (LNA-FISH). Biofouling 2016, 32, 179–190. [Google Scholar] [CrossRef] [PubMed]
- Pires, D.; Melo, L.; Vilas Boas, D.; Sillankorva, S.; Azeredo, J. Phage therapy as an alternative or complementary strategy to prevent and control biofilm-related infections. Curr. Opin. Microbiol. 2017, 39, 48–56. [Google Scholar] [CrossRef] [PubMed]
- Chibeu, A.; Lingohr, E.J.; Masson, L.; Manges, A.; Harel, J.; Ackermann, H.-W.; Kropinski, A.M.; Boerlin, P. Bacteriophages with the ability to degrade uropathogenic Escherichia coli biofilms. Viruses 2012, 4, 471–487. [Google Scholar] [CrossRef] [PubMed]
- Cornelissen, A.; Ceyssens, P.-J.; T’Syen, J.; Van Praet, H.; Noben, J.-P.; Shaburova, O.V.; Krylov, V.N.; Volckaert, G.; Lavigne, R. The T7-related Pseudomonas putida Phage Φ15 displays virion-associated biofilm degradation properties. PLoS ONE 2011, 6, e18597. [Google Scholar] [CrossRef]
- Kay, M.K.; Erwin, T.C.; McLean, R.J.C.; Aron, G.M. Bacteriophage ecology in Escherichia coli and Pseudomonas aeruginosa mixed-biofilm communities. Appl. Environ. Microbiol. 2011, 77, 821–829. [Google Scholar] [CrossRef]
- Sillankorva, S.; Neubauer, P.; Azeredo, J. Phage control of dual species biofilms of Pseudomonas fluorescens and Staphylococcus lentus. Biofouling 2010, 26, 567–575. [Google Scholar] [CrossRef]
- Harcombe, W.R.; Bull, J.J. Impact of phages on two-species bacterial communities. Appl. Environ. Microbiol. 2005, 71, 5254–5259. [Google Scholar] [CrossRef]
- Örmälä, A.M.; Jalasvuori, M. Phage therapy. Bacteriophage 2013, 3, e24219. [Google Scholar] [CrossRef]
- Motlagh, A.M.; Bhattacharjee, A.S.; Goel, R. Biofilm control with natural and genetically-modified phages. World J. Microbiol. Biotechnol. 2016, 32, 67. [Google Scholar] [CrossRef] [PubMed]
- Nordström, K.; Forsgren, A. Effect of protein A on adsorption of bacteriophages to Staphylococcus aureus. J. Virol. 1974, 14, 198–202. [Google Scholar] [CrossRef]
- Pedruzzi, I.; Rosenbusch, J.P.; Locher, K.P. Inactivation in vitro of the Escherichia coli outer membrane protein FhuA by a phage T5-encoded lipoprotein. FEMS Microbiol. Lett. 1998, 168, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Riede, I.; Eschbach, M.L. Evidence that TraT interacts with OmpA of Escherichia coli. FEBS Lett. 1986, 205, 241–245. [Google Scholar] [CrossRef]
- Labrie, S.J.; Samson, J.E.; Moineau, S. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 2010, 8, 317–327. [Google Scholar] [CrossRef] [PubMed]
- Sorek, R.; Kunin, V.; Hugenholtz, P. CRISPR—A widespread system that provides acquired resistance against phages in bacteria and archaea. Nat. Rev. Microbiol. 2008, 6, 181–186. [Google Scholar] [CrossRef]
- Høyland-Kroghsbo, N.M.; Maerkedahl, R.B.; Svenningsen, S.L. A quorum-sensing-induced bacteriophage defense mechanism. MBio 2013, 4, e00362. [Google Scholar] [CrossRef]
- Qin, X.; Sun, Q.; Yang, B.; Pan, X.; He, Y.; Yang, H. Quorum sensing influences phage infection efficiency via affecting cell population and physiological state. J. Basic Microbiol. 2017, 57, 162–170. [Google Scholar] [CrossRef]
- Scanlan, P.D.; Buckling, A.; Hall, A.R. Experimental evolution and bacterial resistance: (co)evolutionary costs and trade-offs as opportunities in phage therapy research. Bacteriophage 2015, 5, e1050153. [Google Scholar] [CrossRef]
- Domingo-Calap, P.; Delgado-Martínez, J. Bacteriophages: Protagonists of a post-antibiotic era. Antibiotics 2018, 7, 66. [Google Scholar] [CrossRef]
- Tinoco, J.M.; Buttaro, B.; Zhang, H.; Liss, N.; Sassone, L.; Stevens, R. Effect of a genetically engineered bacteriophage on Enterococcus faecalis biofilms. Arch. Oral Biol. 2016, 71, 80–86. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Miyanaga, K.; Tanji, Y. Diffusion properties of bacteriophages through agarose gel membrane. Biotechnol. Prog. 2010, 26, 1213–1221. [Google Scholar] [CrossRef] [PubMed]
- Waters, E.M.; Neill, D.R.; Kaman, B.; Sahota, J.S.; Clokie, M.R.J.; Winstanley, C.; Kadioglu, A. Phage therapy is highly effective against chronic lung infections with Pseudomonas aeruginosa. Thorax 2017, 72, 666–667. [Google Scholar] [CrossRef] [PubMed]
- Szafrański, S.P.; Winkel, A.; Stiesch, M. The use of bacteriophages to biocontrol oral biofilms. J. Biotechnol. 2017, 250, 29–44. [Google Scholar] [CrossRef] [PubMed]
- Chan, B.K.; Abedon, S.T.; Loc-Carrillo, C. Phage cocktails and the future of phage therapy. Future Microbiol. 2013, 8, 769–783. [Google Scholar] [CrossRef]
- Chan, B.K.; Abedon, S.T. Phage therapy pharmacology phage cocktails. Adv. Appl. Microbiol. 2012, 78, 1–23. [Google Scholar] [PubMed]
- Schmerer, M.; Molineux, I.J.; Bull, J.J. Synergy as a rationale for phage therapy using phage cocktails. PeerJ 2014, 2, e590. [Google Scholar] [CrossRef]
- Maszewska, A.; Zygmunt, M.; Grzejdziak, I.; Różalski, A. Use of polyvalent bacteriophages to combat biofilm of Proteus mirabilis causing catheter-associated urinary tract infections. J. Appl. Microbiol. 2018, 125, 1253–1265. [Google Scholar] [CrossRef]
- Alves, D.R.; Pérez-Esteban, P.; Kot, W.; Bean, J.E.; Arnot, T.; Hansen, L.H.; Enright, M.C.; Jenkins, A.T.A. A novel bacteriophage cocktail reduces and disperses Pseudomonas aeruginosa biofilms under static and flow conditions. Microb. Biotechnol. 2015, 9, 61–74. [Google Scholar] [CrossRef]
- Forti, F.; Roach, D.R.; Cafora, M.; Pasini, M.E.; Horner, D.S.; Fiscarelli, E.V.; Rossitto, M.; Cariani, L.; Briani, F.; Debarbieux, L.; et al. Design of a broad-range bacteriophage cocktail that reduces Pseudomonas aeruginosa biofilms and treats acute infections in two animal models. Antimicrob. Agents Chemother. 2018, 62, e02573-17. [Google Scholar] [CrossRef]
- Khalifa, L.; Gelman, D.; Shlezinger, M.; Dessal, A.L.; Coppenhagen-Glazer, S.; Beyth, N.; Hazan, R. Defeating antibiotic- and phage-resistant Enterococcus faecalis using a phage cocktail in vitro and in a clot model. Front. Microbiol. 2018, 9, 326. [Google Scholar] [CrossRef] [PubMed]
- Tkhilaishvili, T.; Wang, L.; Tavanti, A.; Trampuz, A.; Di Luca, M. Antibacterial efficacy of two commercially available bacteriophage formulations, staphylococcal bacteriophage and PYO bacteriophage, against methicillin-resistant Staphylococcus aureus: Prevention and eradication of biofilm formation and control of a systemic infection of Galleria mellonella larvae. Front. Microbiol. 2020, 11, 110. [Google Scholar]
- Fish, R.; Kutter, E.; Wheat, G.; Blasdel, B.; Kutateladze, M.; Kuhl, S. Bacteriophage treatment of intransigent diabetic toe ulcers: A case series. J. Wound Care. 2016, 25, 27–33. [Google Scholar] [CrossRef]
- Markoishvili, K.; Tsitlanadze, G.; Katsarava, R.; Morris, J.G.; Sulakvelidze, A. A novel sustained-release matrix based on biodegradable poly (ester amide)s and impregnated with bacteriophages and an antibiotic shows promise in management of infected venous stasis ulcers and other poorly healing wounds. Int. J. Dermatol. 2002, 41, 453–458. [Google Scholar] [CrossRef] [PubMed]
- PhageGuard Listeria. Available online: https://phageguard.com/es/solucion-listeria/ (accessed on 14 May 2020).
- Intralytix, Inc. Available online: http://www.intralytix.com/index.php?page=prod&id=1 (accessed on 14 May 2020).
- Intralytix, Inc. Available online: http://www.intralytix.com/index.php?page=prod&id=2 (accessed on 14 May 2020).
- Iacumin, L.; Manzano, M.; Comi, G. Phage Inactivation of Listeria monocytogenes on San Daniele Dry-Cured Ham and Elimination of Biofilms from Equipment and Working Environments. Microorganisms 2016, 4, 4. [Google Scholar] [CrossRef]
- Maciejewska, B.; Olszak, T.; Drulis-Kawa, Z. Applications of bacteriophages versus phage enzymes to combat and cure bacterial infections: An ambitious and also a realistic application? Appl. Microbiol. Biotechnol. 2018, 102, 2563–2581. [Google Scholar] [CrossRef]
- Vázquez, R.; García, E.; García, P. Phage Lysins for fighting bacterial respiratory infections: A new generation of antimicrobials. Front. Immunol. 2018, 9, 2252. [Google Scholar] [CrossRef]
- Gilmer, D.B.; Schmitz, J.E.; Euler, C.W.; Fischetti, V.A. Novel bacteriophage lysin with broad lytic Activity protects against mixed infection by Streptococcus pyogenes and Methicillin-Resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2013, 57, 2743–2750. [Google Scholar] [CrossRef]
- Sharma, U.; Vipra, A.; Channabasappa, S. Phage-derived lysins as potential agents for eradicating biofilms and persisters. Drug Discov. Today 2018, 23, 848–856. [Google Scholar] [CrossRef]
- Nobrega, F.L.; Costa, A.R.; Kluskens, L.D.; Azeredo, J. Revisiting phage therapy: New applications for old resources. Trends Microbiol. 2015, 23, 185–191. [Google Scholar] [CrossRef]
- Latka, A.; Maciejewska, B.; Majkowska-Skrobek, G.; Briers, Y.; Drulis-Kawa, Z. Bacteriophage-encoded virion-associated enzymes to overcome the carbohydrate barriers during the infection process. Appl. Microbiol. Biotechnol. 2017, 101, 3103–3119. [Google Scholar] [CrossRef] [PubMed]
- Nelson, D.; Loomis, L.; Fischetti, V.A. Prevention and elimination of upper respiratory colonization of mice by group A Streptococci by using a bacteriophage lytic enzyme. PNAS 2001, 98, 4107–4112. [Google Scholar] [CrossRef] [PubMed]
- Hermoso, J.A.; García, J.L.; García, P. Taking aim on bacterial pathogens: From phage therapy to enzybiotics. Curr. Opin. Microbiol. 2007, 10, 461–472. [Google Scholar] [CrossRef] [PubMed]
- Briers, Y.; Walmagh, M.; Grymonprez, B.; Biebl, M.; Pirnay, J.-P.; Defraine, V.; Michiels, J.; Cenens, W.; Aertsen, A.; Miller, S.; et al. Art-175 is a highly efficient antibacterial against multidrug-resistant strains and persisters of Pseudomonas aeruginosa. Antimicrob. Agents. Chemother 2014, 58, 3774–3784. [Google Scholar] [CrossRef]
- López, R.; García, E. Recent trends on the molecular biology of Pneumococcal capsules, lytic enzymes, and bacteriophage. FEMS Microbiol. Rev. 2004, 28, 553–580. [Google Scholar] [CrossRef]
- Vázquez, R.; Domenech, M.; Iglesias-Bexiga, M.; Menéndez, M.; García, P. Csl2, a novel chimeric bacteriophage lysin to fight infections caused by Streptococcus suis, an emerging zoonotic pathogen. Sci. Rep. 2017, 7, 16506. [Google Scholar] [CrossRef]
- Pires, D.P.; Oliveira, H.; Melo, L.D.R.; Sillankorva, S.; Azeredo, J. Bacteriophage-encoded depolymerases: Their diversity and biotechnological applications. Appl. Microbiol. Biotechnol. 2016, 100, 2141–2151. [Google Scholar] [CrossRef]
- Gutiérrez, D.; Briers, Y.; Rodríguez-Rubio, L.; Martínez, B.; Rodríguez, A.; Lavigne, R.; García, P. Role of the pre-neck appendage protein (Dpo7) from phage VB_SepiS-PhiIPLA7 as an anti-biofilm agent in Staphylococcal species. Front. Microbiol. 2015, 6, 1315. [Google Scholar] [CrossRef]
- Guo, Z.; Huang, J.; Yan, G.; Lei, L.; Wang, S.; Yu, L.; Zhou, L.; Gao, A.; Feng, X.; Han, W.; et al. Identification and characterization of Dpo42, a novel depolymerase derived from the Escherichia coli phage VB_EcoM_ECOO78. Front. Microbiol. 2017, 8, 1460. [Google Scholar] [CrossRef]
- Olsen, N.M.C.; Thiran, E.; Hasler, T.; Vanzieleghem, T.; Belibasakis, G.N.; Mahillon, J.; Loessner, M.J.; Schmelcher, M. Synergistic removal of static and dynamic Staphylococcus aureus biofilms by combined treatment with a bacteriophage endolysin and a polysaccharide depolymerase. Viruses 2018, 10, 438. [Google Scholar] [CrossRef]
- Donlan, R.M. Preventing biofilms of clinically relevant organisms using bacteriophage. Trends Microbiol. 2009, 17, 66–72. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.K.; Collins, J.J. Dispersing biofilms with engineered enzymatic bacteriophage. Proc. Natl. Acad. Sci. USA 2007, 104, 11197–11202. [Google Scholar] [CrossRef] [PubMed]
- Lemon, D.J.; Kay, M.K.; Titus, J.K.; Ford, A.A.; Chen, W.; Hamlin, N.J.; Hwang, Y.Y. Construction of a genetically modified T7Select phage system to express the antimicrobial peptide 1018. J. Microbiol. 2019, 57, 532–538. [Google Scholar] [CrossRef] [PubMed]
- Yosef, I.; Manor, M.; Kiro, R.; Qimron, U. Temperate and lytic bacteriophages programmed to sensitize and kill antibiotic-resistant bacteria. Proc. Natl. Acad. Sci. USA 2015, 112, 7267–7272. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Borg, R.E.; Dow, L.P.; Pruitt, B.L.; Chen, I.A. Controlled phage therapy by photothermal ablation of specific bacterial species using gold nanorods targeted by chimeric phages. Proc. Natl. Acad. Sci. USA 2020, 117, 1951–1961. [Google Scholar] [CrossRef]
- Torres-Barceló, C.; Hochberg, M.E. Evolutionary rationale for phages as complements of antibiotics. Trends Microbiol. 2016, 24, 249–256. [Google Scholar] [CrossRef]
- Tkhilaishvili, T.; Lombardi, L.; Klatt, A.-B.; Trampuz, A.; Di Luca, M. Bacteriophage Sb-1 enhances antibiotic activity against biofilm, degrades exopolysaccharide matrix and targets persisters of Staphylococcus aureus. Int. J. Antimicrob. Agents 2018, 52, 842–853. [Google Scholar] [CrossRef]
- Coulter, L.B.; McLean, R.J.C.; Rohde, R.E.; Aron, G.M. Effect of bacteriophage infection in combination with tobramycin on the emergence of resistance in Escherichia coli and Pseudomonas aeruginosa biofilms. Viruses 2014, 6, 3778–3786. [Google Scholar] [CrossRef]
- Chai, Z.; Wang, J.; Tao, S.; Mou, H. Application of bacteriophage-borne enzyme combined with chlorine dioxide on controlling bacterial biofilm. LWT Food Sci. Technol. 2014, 59, 1159–1165. [Google Scholar] [CrossRef]
- Pena-Miller, R.; Laehnemann, D.; Jansen, G.; Fuentes-Hernandez, A.; Rosenstiel, P.; Schulenburg, H.; Beardmore, R. When the most potent combination of antibiotics selects for the greatest bacterial load: The smile-frown transition. PLoS Biol. 2013, 11, e1001540. [Google Scholar] [CrossRef]
- Abedon, S.T. Phage-antibiotic combination treatments: Antagonistic impacts of antibiotics on the pharmacodynamics of phage therapy? Antibiotics 2019, 8, 182. [Google Scholar] [CrossRef] [PubMed]
- Tagliaferri, T.L.; Jansen, M.; Horz, H.-P. Fighting pathogenic bacteria on two fronts: Phages and antibiotics as combined strategy. Front. Cell. Infect. Microbiol. 2019, 9, 22. [Google Scholar] [CrossRef] [PubMed]
- Matsui, T.; Yoshikawa, G.; Mihara, T.; Chatchawankanphanich, O.; Kawasaki, T.; Nakano, M.; Fujie, M.; Ogata, H.; Yamada, T. Replications of two closely related groups of Jumbo phages show different level of dependence on host-encoded RNA polymerase. Front. Microbiol. 2017, 8, 1010. [Google Scholar] [CrossRef] [PubMed]
- Lehman, S.M.; Donlan, R.M. Bacteriophage-mediated control of a two-species biofilm formed by microorganisms causing catheter-associated urinary tract infections in an in vitro urinary catheter model. Antimicrob. Agents Chemother. 2015, 59, 1127–1137. [Google Scholar] [CrossRef]
- Lewis, K. Riddle of biofilm resistance. Antimicrob. Agents Chemother. 2001, 45, 999–1007. [Google Scholar] [CrossRef]
- Pinto, G.; Silva, M.D.; Peddey, M.; Sillankorva, S.; Azeredo, J. The role of bacteriophages in periodontal health and disease. Future Microbiol. 2016, 11, 1359–1369. [Google Scholar] [CrossRef]
- Delisle, A.L. Bacteriophage-Encoded Enzymes for the Treatment and Prevention of Dental Caries and Periodontal Diseases. US6635238B2. Available online: https://patents.google.com/patent/US6635238B2/en (accessed on 14 May 2020).
- Blasi, F.; Page, C.; Rossolini, G.M.; Pallecchi, L.; Matera, M.G.; Rogliani, P.; Cazzola, M. The effect of N-acetylcysteine on biofilms: Implications for the treatment of respiratory tract infections. Respir. Med. 2016, 117, 190–197. [Google Scholar] [CrossRef]
- Percival, S.L. Importance of biofilm formation in surgical infection: Biofilm formation in surgical infection. Br. J. Surg. 2017, 104, e85–e94. [Google Scholar] [CrossRef]
- Phagoburn: Evaluation of Phage Therapy for the Treatment of Burn Wound Infections. Available online: www.phagoburn.eu (accessed on 14 May 2020).
- Upadhyay, M.P.; Srinivasan, M.; Whitcher, J.P. Diagnosing and managing microbial keratitis. Community Eye Health 2015, 28, 3–6. [Google Scholar]
- Syed, M.A.; Manzoor, U.; Shah, I.; Bukhari, S.H.A. Antibacterial effects of tungsten nanoparticles on the Escherichia coli strains isolated from catheterized urinary tract infection (UTI) cases and Staphylococcus aureus. New Microbiol. 2010, 33, 329–335. [Google Scholar] [PubMed]
- Niveditha, S.; Pramodhini, S.; Umadevi, S.; Kumar, S.; Stephen, S. The isolation and the biofilm formation of uropathogens in the patients with catheter associated urinary tract infections (UTIs). J. Clin. Diagn. Res. 2012, 6, 1478–1482. [Google Scholar] [CrossRef] [PubMed]
- Bagshaw, S.M.; Laupland, K.B. Epidemiology of intensive care unit-acquired urinary tract infections. Curr. Opin. Infect. Dis. 2006, 19, 67–71. [Google Scholar] [CrossRef] [PubMed]
- Sabir, N.; Ikram, A.; Zaman, G.; Satti, L.; Gardezi, A.; Ahmed, A.; Ahmed, P. Bacterial biofilm-based catheter-associated urinary tract infections: Causative pathogens and antibiotic resistance. Am. J. Infect. Control. 2017, 45, 1101–1105. [Google Scholar] [CrossRef] [PubMed]
- Macleod, S.M.; Stickler, D.J. Species interactions in mixed-community crystalline biofilms on urinary catheters. J. Med. Microbiol. 2007, 56, 1549–1557. [Google Scholar] [CrossRef] [PubMed]
- François, B.; Luyt, C.-E.; Stover, C.; Brubaker, J.; Chastre, J.; Jafri, H. New strategies targeting virulence factors of Staphylococcus aureus and Pseudomonas aeruginosa. Semin. Respir. Crit. Care. Med. 2017, 38, 346–358. [Google Scholar] [CrossRef] [PubMed]
- Bouchart, F.; Vidal, O.; Lacroix, J.M.; Spriet, C.; Chamary, S.; Brutel, A.; Hornez, J.C. 3D printed bioceramic for phage therapy against bone nosocomial infections. Mater. Sci. Eng. Mater. Biol. Appl. 2020, 111, 10840. [Google Scholar] [CrossRef] [PubMed]
- Bridier, A.; Sanchez-Vizuete, P.; Guilbaud, M.; Piard, J.-C.; Naïtali, M.; Briandet, R. Biofilm-associated persistence of food-borne pathogens. Food Microbiol. 2015, 45, 167–178. [Google Scholar] [CrossRef]
- Giaouris, E.; Heir, E.; Hébraud, M.; Chorianopoulos, N.; Langsrud, S.; Møretrø, T.; Habimana, O.; Desvaux, M.; Renier, S.; Nychas, G.-J. Attachment and biofilm formation by foodborne bacteria in meat processing environments: Causes, implications, role of bacterial interactions and control by alternative novel methods. Meat Sci. 2014, 97, 298–309. [Google Scholar] [CrossRef]
- Camargo, A.C.; Woodward, J.J.; Call, D.R.; Nero, L.A. Listeria monocytogenes in food-processing facilities, food contamination, and human listeriosis: The brazilian scenario. Foodborne Pathog. Dis. 2017, 14, 623–636. [Google Scholar] [CrossRef] [PubMed]
- Charlebois, A.; Jacques, M.; Boulianne, M.; Archambault, M. Tolerance of Clostridium perfringens biofilms to disinfectants commonly used in the food industry. Food Microbiol. 2017, 62, 32–38. [Google Scholar] [CrossRef]
- Milho, C.; Silva, M.D.; Sillankorva, S.; Harper, D. Biofilm Applications of Bacteriophages. In Bacteriophages; Springer: Cham, Switzerland, 2019; pp. 1–35. [Google Scholar]
- Di Pippo, F.; Di Gregorio, L.; Congestri, R.; TanDOI, V.; Rossetti, S. Biofilm growth and control in cooling water industrial systems. FEMS Microbiol. Ecol. 2018, 94, 1–13. [Google Scholar] [CrossRef]
- Di Gregorio, L.; TanDOI, V.; Congestri, R.; Rossetti, S.; Di Pippo, F. Unravelling the core microbiome of biofilms in cooling tower systems. Biofouling 2017, 33, 793–806. [Google Scholar] [CrossRef]
- Liu, S.; Gunawan, C.; Barraud, N.; Rice, S.A.; Harry, E.J.; Amal, R. Understanding, monitoring, and controlling biofilm growth in drinking water distribution systems. Environ. Sci. Technol. 2016, 50, 8954–8976. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ferriol-González, C.; Domingo-Calap, P. Phages for Biofilm Removal. Antibiotics 2020, 9, 268. https://doi.org/10.3390/antibiotics9050268
Ferriol-González C, Domingo-Calap P. Phages for Biofilm Removal. Antibiotics. 2020; 9(5):268. https://doi.org/10.3390/antibiotics9050268
Chicago/Turabian StyleFerriol-González, Celia, and Pilar Domingo-Calap. 2020. "Phages for Biofilm Removal" Antibiotics 9, no. 5: 268. https://doi.org/10.3390/antibiotics9050268
APA StyleFerriol-González, C., & Domingo-Calap, P. (2020). Phages for Biofilm Removal. Antibiotics, 9(5), 268. https://doi.org/10.3390/antibiotics9050268




