Antagonistic Activity against Ascosphaera apis and Functional Properties of Lactobacillus kunkeei Strains
Abstract
1. Introduction
1.1. Ascosphaera apis: The Causative Agent of Chalkbrood Disease
1.2. Chalkbrood Disease Control by Symbiotic Bacteria
2. Results
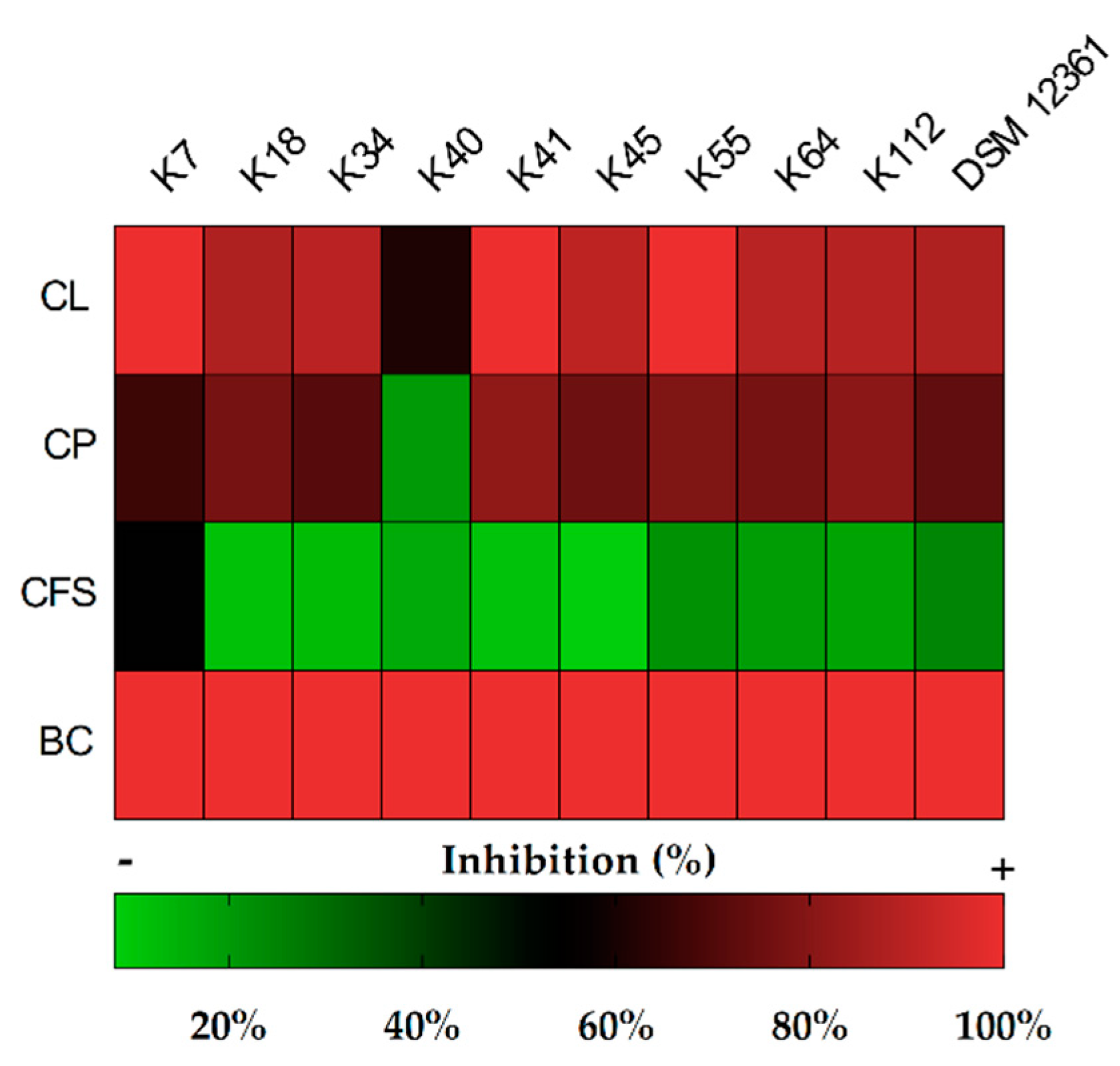
2.1. Screening of Bacteria for Antifungal Activity
2.2. Determination of Inhibitory Activity
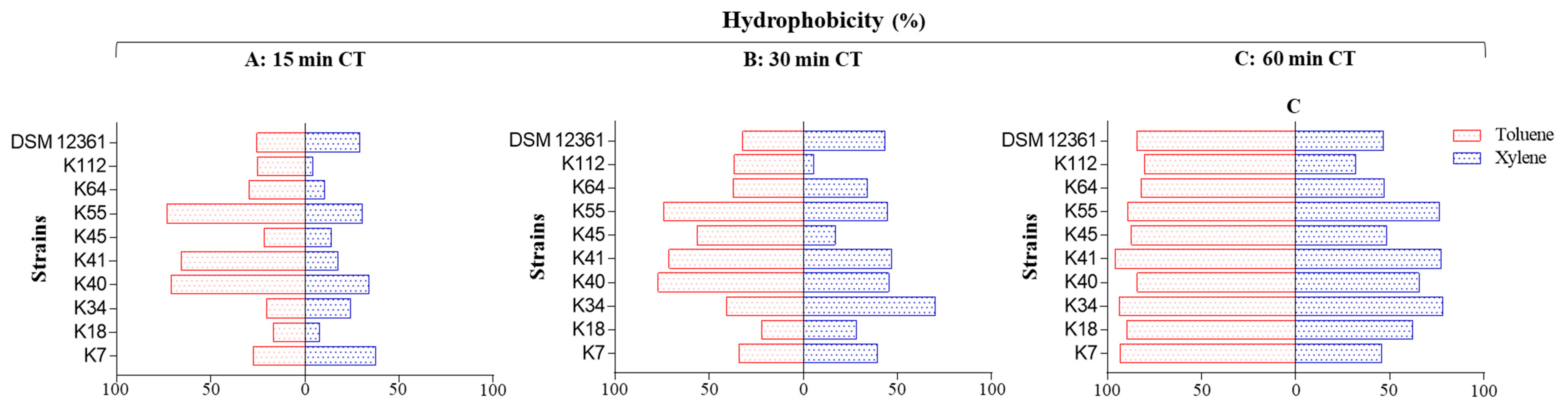
2.3. Hydrophobicity and Auto-Aggregation
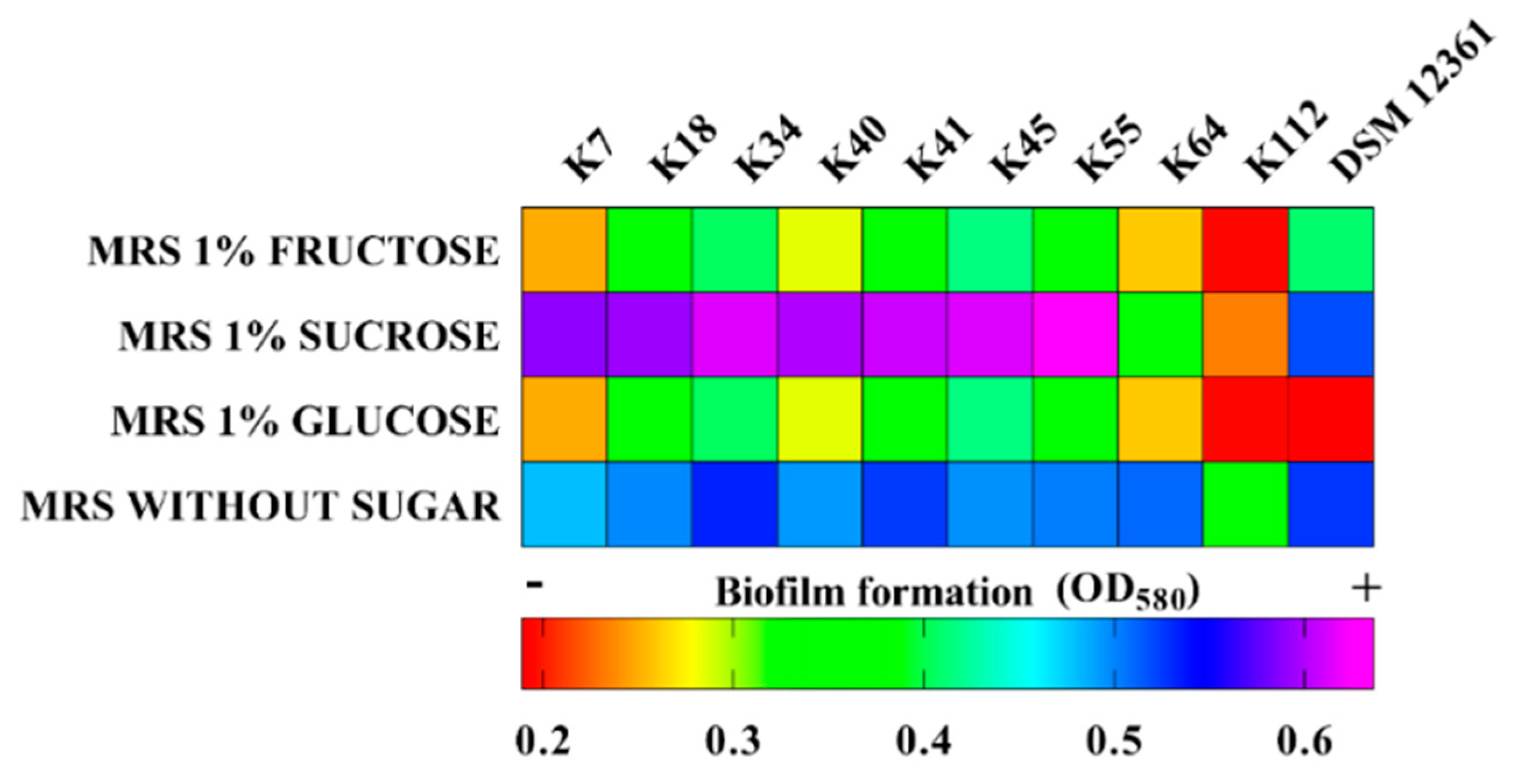
2.4. Biofilm Production
2.5. Bacterial Survival in Sugar Syrup
3. Discussion
3.1. Antifungal Activity
3.2. Cell-Surface Properties
3.3. Survival in Sugar Syrups
3.4. Perspectives
4. Materials and Methods
4.1. Microbial Cultures
4.2. Screening of Bacteria for Antifungal Activity
4.3. Determination of Inhibitory Activity
4.4. Hydrophobicity Assay
4.5. Auto-Aggregation
4.6. Biofilm Production
4.7. Bacterial Survival in Sugar Syrup
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Kwong, W.; Moran, N.A. Gut microbial communities of social bees. Nat. Rev. Microbiol. 2016, 14, 374–384. [Google Scholar] [CrossRef] [PubMed]
- Powell, J.E.; Martinson, V.G.; Urban-Mead, K.; Moran, N.A. Routes of acquisition of the gut microbiota of Apis mellifera. Appl. Environ. Microbiol. 2014, 80, 7378–7387. [Google Scholar] [CrossRef] [PubMed]
- Cox-Foster, D.L.; Conlan, S.; Holmes, E.C.; Palacios, G.; Evans, J.D.; Moran, N.A.; Quan, P.L.; Briese, T.; Hornig, M.; Geiser, D.M.; et al. A metagenomic survey of microbes in honey bee colony collapse disorder. Science 2007, 318, 283–287. [Google Scholar] [CrossRef]
- Hamdi, C.; Balloi, A.; Essanaa, J.; Gonella, E.; Raddadi, N.; Ricci, I.; Boudabous, A.; Borin, S.; Manino, A.; Bandi, C.; et al. Gut microbiome dysbiosis and honeybee health. J. Appl. Entomol. 2011, 135, 524–533. [Google Scholar] [CrossRef]
- Maes, P.W.; Rodrigues, P.A.; Oliver, R.; Mott, B.M.; Anderson, K.E. Diet-related gut bacterial dysbiosis correlates with impaired development, increased mortality and Nosema disease in the honeybee (Apis mellifera). Mol. Ecol. 2016, 25, 5439–5450. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, R.S.; Moran, N.A.; Evans, J.D. Early gut colonizers shape parasite susceptibility and microbiota composition in honey bee workers. Proc. Natl. Acad. Sci. USA 2016, 113, 9345–9350. [Google Scholar] [CrossRef]
- Raymann, K.; Shaffer, Z.; Moran, N.A. Antibiotic exposure perturbs the gut microbiota and elevates mortality in honeybees. PLoS Biol. 2017, 15, e2001861. [Google Scholar] [CrossRef]
- Aronstein, K.A.; Murray, K.D. Chalkbrood disease in honey bees. J. Invertebr. Pathol. 2010, 103, 20–29. [Google Scholar] [CrossRef]
- Evison, S.E. Chalkbrood: Epidemiological perspectives from the host–parasite relationship. Curr. Opin. Insect Sci. 2015, 10, 65–70. [Google Scholar] [CrossRef]
- Garrido-Bailón, E.; Higes, M.; Martínez-Salvador, A.; Antúnez, K.; Botías, C.; Meana, A.; Prieto, L.; Martín-Hernández, R. The prevalence of the honeybee brood pathogens Ascosphaera apis, Paenibacillus larvae and Melissococcus plutonius in Spanish apiaries determined with a new multiplex PCR assay. Microb. Biotechnol. 2013, 6, 31–39. [Google Scholar] [CrossRef]
- Zaghloul, O.A.; Mourad, A.K.; El Kady, M.B.; Nemat, F.M.; Morsy, M.E. Assessment of losses in honey yield due to the chalck-brood disease with reference to the determination of its economic injury levels in Egypt. Commun. Agric. Appl. Biol. Sci. 2005, 70, 703–714. [Google Scholar] [PubMed]
- Reynaldi, F.J.; Lucia, M.; Garcia, M.L.G. Ascosphaera apis, the entomopathogenic fungus affecting larvae of native bees (Xylocopa augusti): First report in South America. Rev. Iberoam. Micol. 2015, 32, 261–264. [Google Scholar] [CrossRef] [PubMed]
- Vojvodic, S.; Boomsma, J.J.; Eilenberg, J.; Jensen, A.B. Virulence of mixed fungal infections in honey bee brood. Front. Zool. 2012, 9, 5. [Google Scholar] [CrossRef]
- Fowler, A.E.; Irwin, R.E.; Adler, L.S. Parasite defense mechanisms in bees: Behavior, immunity, antimicrobials, and symbionts. Emerg. Top. Life Sci. 2019, ETLS20190069. [Google Scholar] [CrossRef]
- Alberoni, D.; Baffoni, L.; Gaggiìa, F.; Ryan, P.M.; Murphy, K.; Ross, P.R.; Santon, C.; Di Gioia, D. Impact of beneficial bacteria supplementation on the gut microbiota, colony development and productivity of Apis mellifera L. Benef. Microbes 2018, 9, 269–278. [Google Scholar] [CrossRef] [PubMed]
- Ptaszyńska, A.A.; Borsuk, G.; Zdybicka-Barabas, A.; Cytryńska, M.; Małek, W. Are commercial probiotics and prebiotics effective in the treatment and prevention of honeybee nosemosis C? Parasitol. Res. 2016, 115, 397–406. [Google Scholar] [CrossRef] [PubMed]
- Pătruică, S.; Mot, D. The effect of using prebiotic and probiotic products on intestinal micro-flora of the honeybee (Apis mellifera carpatica). Bull. Entomol. Res. 2012, 102, 619–623. [Google Scholar] [CrossRef]
- Audisio, M.C. Gram-positive bacteria with probiotic potential for the Apis mellifera L. honey bee: The experience in the Northwest of Argentina. Probiot. Antimicrob. Proteins 2017, 9, 22–31. [Google Scholar] [CrossRef]
- Audisio, M.C.; Benítez-Ahrendts, M.R. Lactobacillus johnsonii CRL1647, isolated from Apis mellifera L. bee-gut, exhibited a beneficial effect on honeybee colonies. Benef. Microbes 2011, 2, 29–34. [Google Scholar] [CrossRef]
- Baffoni, L.; Gaggiìa, F.; Alberoni, D.; Cabbri, R.; Nanetti, A.; Biavati, B.; Di Gioia, D. Effect of dietary supplementation of Bifidobacterium and Lactobacillus strains in Apis mellifera L. against Nosema ceranae. Benef. Microbes 2016, 7, 45–51. [Google Scholar] [CrossRef]
- Vásquez, A.; Forsgren, E.; Fries, I.; Paxton, R.J.; Flaberg, E.; Szekely, L.; Olofsson, T.C. Symbionts as major modulators of insect health: Lactic acid bacteria and honeybees. PLoS ONE 2012, 7, e33188. [Google Scholar] [CrossRef]
- Endo, A.; Irisawa, T.; Futagawa-Endo, Y.; Takano, K.; du Toit, M.; Okada, S.; Dicks, L.M. Characterization and emended description of Lactobacillus kunkeei as a fructophilic lactic acid bacterium. Int. J. Syst. Evol. Microbiol. 2012, 62, 500–504. [Google Scholar] [CrossRef] [PubMed]
- Endo, A.; Salminen, S. Honeybees and beehives are rich sources for fructophilic lactic acid bacteria. Syst. Appl. Microbiol. 2013, 36, 444–448. [Google Scholar] [CrossRef] [PubMed]
- Filannino, P.; Di Cagno, R.; Tlais, A.Z.A.; Cantatore, V.; Gobbetti, M. Fructose-rich niches traced the evolution of lactic acid bacteria toward fructophilic species. Crit. Rev. Microbiol. 2019, 45, 65–81. [Google Scholar] [CrossRef] [PubMed]
- Olofsson, T.C.; Butler, È.; Markowicz, P.; Lindholm, C.; Larsson, L.; Vásquez, A. Lactic acid bacterial symbionts in honeybees–an unknown key to honey’s antimicrobial and therapeutic activities. Int. Wound J. 2016, 13, 668–679. [Google Scholar] [CrossRef]
- Berríos, P.; Fuentes, J.A.; Salas, D.; Carreno, A.; Aldea, P.; Fernandez, F.; Trombert, A.N. Inhibitory effect of biofilm-forming Lactobacillus kunkeei strains against virulent Pseudomonas aeruginosa in vitro and in honeycomb moth (Galleria mellonella) infection model. Benef. Microbes 2018, 9, 257–268. [Google Scholar] [CrossRef]
- Butler, E.; Alsterfjord, M.; Olofsson, T.; Karlsson, C.; Malmström, J.; Vásquez, A. Proteins of novel lactic acid bacteria from Apis mellifera mellifera: An insight into the production of known extra-cellular proteins during microbial stress. BMC Microbiol. 2013, 13, 235. [Google Scholar] [CrossRef]
- Arredondo, D.; Castelli, L.; Porrini, M.P.; Garrido, P.M.; Eguaras, M.J.; Zunino, P.; Antúnez, K. Lactobacillus kunkeei strains decreased the infection by honey bee pathogens Paenibacillus larvae and Nosema ceranae. Benef. Microbes 2018, 9, 279–290. [Google Scholar] [CrossRef]
- Bartel, L.C.; Abrahamovich, E.; Mori, C.; Lopez, A.C.; Alippi, A.M. Bacillus and Brevibacillus strains as potential antagonists of Paenibacillus larvae and Ascosphaera apis. J. Apic. Res. 2019, 58, 117–132. [Google Scholar] [CrossRef]
- Al-Ghamdi, A.; Khan, K.; Ansari, M.; Almasaudi, S.; AL-Kahtani, S. Effect of gut bacterial isolates from Apis mellifera jemenitica on Paenibacillus larvae infected bee larvae. Saudi J. Biol. Sci. 2017, 25, 383–387. [Google Scholar] [CrossRef]
- Forsgren, E.; Olofsson, T.C.; Váasquez, A.; Fries, I. Novel lactic acid bacteria inhibiting Paenibacillus larvae in honey bee larvae. Apidologie 2010, 41, 99–108. [Google Scholar] [CrossRef]
- Janashia, I.; Choiset, Y.; Rabesona, H.; Hwanhlem, N.; Bakuradze, N.; Chanishvili, N.; Haertlé, T. Protection of honeybee Apis mellifera by its endogenous and exogenous lactic flora against bacterial infections. Ann. Agrar. Sci. 2016, 14, 177–181. [Google Scholar] [CrossRef]
- Omar, M.O.M.; Moustafa, A.M.; Ansari, M.J.; Anwar, A.M.; Fahmy, B.F.; Al-Ghamdi, A.; Nuru, A. Antagonistic effect of gut bacteria in the hybrid Carniolan honey bee, Apis Mellifera Carnica, against Ascosphaera apis, the causal organism of chalkbrood disease. J. Apic. Sci. 2014, 58, 17–27. [Google Scholar] [CrossRef]
- Sabaté, D.C.; Carrillo, L.; Audisio, M.C. Inhibition of Paenibacillus larvae and Ascosphaera apis by Bacillus subtilis isolated from honeybee gut and honey samples. Res. Microbiol. 2009, 160, 193–199. [Google Scholar] [CrossRef]
- Tejerina, M.R.; Cabana, M.J.; Carrillo, L.; Benitez-Ahrendts, M.R. Effect of Lactic Bacteria on Ascosphaera apis and A. atra. Asian J. Agric. Food. Sci. 2018, 6, 123–128. [Google Scholar] [CrossRef]
- Kostecka, M.; Niewiadomy, A. Antifungal activity of new series of compound against Ascosphaera apis. Mikol. Lek. 2010, 17, 169–171. [Google Scholar]
- Chorbinski, P. Susceptibility of Ascosphaera apis strains to antifungal preparations. Med. Weter. 2003, 59, 1137–1139. [Google Scholar]
- Sarwar, M. Fungal diseases of honey bees (Hymenoptera: Apidae) that induce considerable losses to colonies and protocol for treatment. Int. J. Zool. Stud. 2016, 1, 8–13. [Google Scholar]
- Frazier, M.; Mullin, C.; Frazier, J.; Ashcraft, S. What have pesticides got to do with it? Am. Bee J. 2008, 148, 521–524. [Google Scholar]
- Ansari, M.J.; Al-Ghamdi, A.; Usmani, S.; Khan, K.A.; Alqarni, A.S.; Kaur, M.; Al-Waili, N. In vitro evaluation of the effects of some plant essential oils on Ascosphaera apis, the causative agent of Chalkbrood disease. Saudi J. Biol. Sci. 2017, 24, 1001–1006. [Google Scholar] [CrossRef]
- Chantawannakul, P.; Puchanichanthranon, T.; Wongsiri, S. Inhibitory effects of some medicinal plant extracts on the growth of Ascosphaera apis. ACTA Hortic. 2005, 678, 183–189. [Google Scholar] [CrossRef]
- Chaimanee, V.; Thongtue, U.; Sornmai, N.; Songsri, S.; Pettis, J.S. Antimicrobial activity of plant extracts against the honeybee pathogens, Paenibacillus larvae and Ascosphaera apis and their topical toxicity to Apis mellifera adults. J. Appl. Microbiol. 2017, 123, 1160–1167. [Google Scholar] [CrossRef] [PubMed]
- Boudegga, H.; Boughalleb, N.; Barbouche, N.; Ben Hamouda, M.H.; Mahjoub, M.E. In vitro inhibitory actions of some essential oils on Ascosphaera apis, a fungus responsible for honey bee chalkbrood. J. Apic. Res. 2010, 49, 236–242. [Google Scholar] [CrossRef]
- Dellacasa, A.D.; Bailac, P.N.; Ponzi, M.I.; Ruffinengo, S.R.; Eguaras, M.J. In vitro activity of essential oils from San Luis-Argentina against Ascosphaera apis. J. Essent. Oil Res. 2003, 15, 282–285. [Google Scholar] [CrossRef]
- Dunn, L.L.; Davidson, P.M.; Critzer, F.J. Antimicrobial efficacy of an array of essential oils against lactic acid bacteria. J. Food Sci. 2016, 81, M438–M444. [Google Scholar] [CrossRef] [PubMed]
- Testa, B.; Lombardi, S.J.; Macciola, E.; Succi, M.; Tremonte, P.; Iorizzo, M. Efficacy of olive leaf extract (Olea europaea L. cv Gentile di Larino) in marinated anchovies (Engraulis encrasicolus, L.) process. Heliyon 2019, 5, e01727. [Google Scholar] [CrossRef]
- García-Ruiz, A.; Bartolomé, B.; Martínez-Rodríguez, A.J.; Pueyo, E.; Martín-Álvarez, P.J.; Moreno-Arribas, M.V. Potential of phenolic compounds for controlling lactic acid bacteria growth in wine. Food Control 2008, 19, 835–841. [Google Scholar] [CrossRef]
- Rivas-Sendra, A.; Landete, J.M.; Alcántara, C.; Zúñiga, M. Response of Lactobacillus casei BL23 to phenolic compounds. J. Appl. Microbiol. 2011, 111, 1473–1481. [Google Scholar] [CrossRef]
- Lombardi, S.J.; Tremonte, P.; Succi, M.; Testa, B.; Pannella, G.; Tipaldi, L.; Sorrentino, E.; Coppola, R.; Iorizzo, M. Effect of phenolic compounds on the growth and L-malic acid metabolism of Oenococcus oeni. J. Life Sci. 2012, 6, 1225. [Google Scholar] [CrossRef]
- Raymann, K.; Moran, N.A. The role of the gut microbiome in health and disease of adult honey bee workers. Curr. Opin. Insect. Sci. 2018, 26, 97–104. [Google Scholar] [CrossRef]
- Wu, M.; Sugimura, Y.; Taylor, D.; Yoshiyama, M. Honeybee gastrointestinal bacteria for novel and sustainable disease control strategies. J. Dev. Sustain. Agric. 2013, 8, 85–90. [Google Scholar] [CrossRef]
- Crowley, S.; Mahony, J.; van Sinderen, D. Current perspectives on antifungal lactic acid bacteria as natural bio-preservatives. Trends Food Sci. Technol. 2013, 33, 93–109. [Google Scholar] [CrossRef]
- Le Lay, C.; Coton, E.; Le Blay, G.; Chobert, J.M.; Haertlé, T.; Choiset, Y.; Van Long, N.N.; Meslet-Cladière, L.; Mounier, J. Identification and quantification of antifungal compounds produced by lactic acid bacteria and propionibacteria. Int. J. Food Microbiol. 2016, 239, 79–85. [Google Scholar] [CrossRef] [PubMed]
- Ngang, J.J.E.; Yadang, G.; Kamdem, S.L.S.; Kouebou, C.P.; Fanche, S.A.Y.; Kougan, D.L.T.; Tsoungui, A.; Etoa, F.X. Antifungal properties of selected lactic acid bacteria and application in the biological control of ochratoxin A producing fungi during cocoa fermentation. Biocontrol Sci. Technol. 2015, 25, 245–259. [Google Scholar] [CrossRef]
- Sorrentino, E.; Tremonte, P.; Succi, M.; Iorizzo, M.; Pannella, G.; Lombardi, S.J.; Sturchio, M.; Coppola, R. Detection of Antilisterial Activity of 3-Phenyllactic Acid Using Listeria innocua as a Model. Front. Microbiol. 2018, 9, 1373. [Google Scholar] [CrossRef] [PubMed]
- Kleerebezem, M.; Hols, P.; Bernard, E.; Rolain, T.; Zhou, M.; Siezen, R.J.; Bron, P.A. The extracellular biology of the lactobacilli. FEMS Microbiol. Rev. 2010, 34, 199–230. [Google Scholar] [CrossRef]
- Kinoshita, H.; Uchida, H.; Kawai, Y.; Kawasaki, T.; Wakahara, N.; Matsuo, H.; Watanabe, M.; Kitazawa, H.; Ohnuma, S.; Miura, K.; et al. Cell surface Lactobacillus plantarum LA 318 glyceraldehyde-3-phosphate dehydrogenase (GAPDH) adheres to human colonic mucin. J. Appl. Microbiol. 2008, 104, 1667–1674. [Google Scholar] [CrossRef]
- Åvall-Jääskeläinen, S.; Palva, A. Lactobacillus surface layers and their applications. FEMS Microbiol. Rev. 2005, 29, 511–529. [Google Scholar] [CrossRef]
- Janashia, I.; Choiset, Y.; Jozefiak, D.; Déniel, F.; Coton, E.; Akbar, A.; Moosavi-Movahedi, A.A.; Chanishvili, N.; Haertlé, T. Beneficial protective role of endogenous lactic acid bacteria against mycotic contamination of honeybee beebread. Probiot. Antimicrob. Proteins 2018, 10, 638–646. [Google Scholar] [CrossRef]
- Daisley, B.A.; Pitek, A.P.; Chmiel, J.A.; Al, F.K.; Chernyshova, A.M.; Faragalla, K.M.; Burton, J.P.; Thompson, G.J.; Reid, G. Novel probiotic approach to counter Paenibacillus larvae infection in honey bees. ISME J. 2020, 14, 476–491. [Google Scholar] [CrossRef]
- Santarmaki, V.; Kourkoutas, Y.; Zoumpopoulou, G.; Mavrogonatou, E.; Kiourtzidis, M.; Chorianopoulos, N.; Tassou, C.; Tsakalidou, E.; Simopoulos, C.; Ypsilantis, P. Survival, intestinal mucosa adhesion, and immunomodulatory potential of Lactobacillus plantarum strains. Curr. Microbiol. 2017, 74, 1061–1067. [Google Scholar] [CrossRef] [PubMed]
- Trunk, T.; Khalil, H.S.; Leo, J.C. Bacterial autoaggregation. AIMS Microbiol. 2018, 4, 140–164. [Google Scholar] [CrossRef] [PubMed]
- Del Re, B.; Sgorbati, B.; Miglioli, M.; Palenzona, D. Adhesion, autoaggregation and hydrophobicity of 13 strains of Bifidobacterium longum. Lett. Appl. Microbiol. 2000, 31, 438–442. [Google Scholar] [CrossRef]
- Pessoa, W.F.B.; Melgaço, A.C.C.; de Almeida, M.E.; Ramos, L.P.; Rezende, R.P.; Romano, C.C. In vitro activity of Lactobacilli with probiotic potential isolated from Cocoa fermentation against Gardnerella vaginalis. BioMed Res. Int. 2017, 2017, 3264194. [Google Scholar] [CrossRef] [PubMed]
- Bouchard, D.S.; Bianca Seridan, B.; Saraoui, T.; Rault, L.; Germon, P.; Gonzalez-Moreno, C.; Nader-Macias, F.M.E.; Baud, D.; François, P.; Chuat, V.; et al. Lactic acid bacteria isolated from bovine mammary microbiota: Potential allies against bovine mastitis. PLoS ONE 2015, 29, e144831. [Google Scholar] [CrossRef]
- Gandomi, H.; Farhangfar, A.; Misaghi, A.; Noori, N. Auto and co-aggregation, hydrophobicity and adhesion properties of Lactobacillus plantarum strains isolated from Siahmazgi traditional cheese. Food Health J. 2019, 2, 1–5. [Google Scholar]
- Collado, M.C.; Meriluoto, J.; Salminen, S. Adhesion and aggregation properties of probiotic and pathogen strains. Eur. Food Res. Technol. 2008, 226, 1065–1073. [Google Scholar] [CrossRef]
- Ekmekci, H.; Aslim, B.; Ozturk, S. Characterization of vaginal lactobacilli coaggregation ability with Escherichia coli. Microbiol. Immunol. 2009, 53, 59–65. [Google Scholar] [CrossRef]
- Marin, M.L.; Benito, Y.; Pin, C.; Fernandez, M.F.; Garcia, M.L.; Selgas, M.D.; Casas, C. Lactic acid bacteria: Hydrophobicity and strength of attachment to meat surfaces. Lett. Appl. Microbiol. 1997, 24, 14–18. [Google Scholar] [CrossRef]
- O’Toole, G.; Kaplan, H.B.; Kolter, R. Biofilm formation as microbial development. Annu. Rev. Microbiol. 2000, 54, 49–79. [Google Scholar] [CrossRef]
- Sakandar, H.A.; Kubow, S.; Sadiq, F.A. Isolation and in-vitro probiotic characterization of fructophilic lactic acid bacteria from Chinese fruits and flowers. LWT Food Sci. Technol. 2019, 104, 70–75. [Google Scholar] [CrossRef]
- Audisio, M.C.; Torres, M.J.; Sabaté, D.C.; Ibarguren, C.; Apella, M.C. Properties of different lactic acid bacteria isolated from Apis mellifera L. bee-gut. Microbiol. Res. 2011, 166, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Van de Guchte, M.; Serror, P.; Chervaux, C.; Smokvina, T.; Ehrlich, S.D.; Maguin, E. Stress responses in lactic acid bacteria. Anton Leeuwenhoek 2002, 82, 187–216. [Google Scholar] [CrossRef]
- Papadimitriou, K.; Alegría, Á.; Bron, P.A.; de Angelis, M.; Gobbetti, M.; Kleerebezem, M.; Lemos, J.A.; Linares, D.M.; Ross, P.; Stanton, C.; et al. Stress physiology of lactic acid bacteria. Microbiol. Mol. Biol. Rev. 2016, 80, 837–890. [Google Scholar] [CrossRef]
- Vasileva, T.; Bivolarski, V.; Michailova, G.; Salim, A.; Rabadjiev, Y.; Ivanova, I.; Iliev, I. Glucansucrases produced by fructophilic lactic acid bacteria Lactobacillus kunkeei H3 and H25 isolated from honeybees. J. Basic Microbiol. 2017, 57, 68–77. [Google Scholar] [CrossRef]
- Meng, X.; Gangoiti, J.; Wang, X.; Grijpstra, P.; Van Leeuwen, S.S.; Pijning, T.; Dijkhuizen, L. Biochemical characterization of a GH70 protein from Lactobacillus kunkeei DSM 12361 with two catalytic domains involving branching sucrase activity. Appl. Microbiol. Biotechnol. 2018, 102, 7935–7950. [Google Scholar] [CrossRef]
- Davey, M.E.; O’toole, G.A. Microbial biofilms: From ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 2000, 64, 847–867. [Google Scholar] [CrossRef]
- Djukic, M.; Poehlein, A.; Strauß, J.; Tann, F.J.; Leimbach, A.; Hoppert, M.; Daniel, R. High quality draft genome of Lactobacillus kunkeei EFB6, isolated from a German European foulbrood outbreak of honeybees. Stand. Genom. Sci. 2015, 10, 16. [Google Scholar] [CrossRef]
- Tamarit, D.; Ellegaard, K.M.; Wikander, J.; Olofsson, T.; Vasquez, A.; Andersson, S.G. Functionally structured genomes in Lactobacillus kunkeei colonizing the honey crop and food products of honeybees and stingless bees. Genome Biol. Evol. 2015, 7, 1455–1473. [Google Scholar] [CrossRef]
- Billiet, A.; Meeus, I.; Cnockaert, M.; Vandamme, P.; Van Oystaeyen, A.; Wäckers, F.; Smagghe, G. Effect of oral administration of lactic acid bacteria on colony performance and gut microbiota in indoor-reared bumblebees (Bombus terrestris). Apidologie 2017, 48, 41–50. [Google Scholar] [CrossRef][Green Version]
- Alberoni, D.; Gaggìa, F.; Baffoni, L.; Di Gioia, D. Beneficial microorganisms for honey bees: Problems and progresses. Appl. Microbiol. Biotechnol. 2016, 100, 9469–9482. [Google Scholar] [CrossRef] [PubMed]
- Crotti, E.; Balloi, A.; Hamdi, C.; Sansonno, L.; Marzorati, M.; Gonella, E.; Favia, G.; Cherif, A.; Bandi, C.; Alma, A.; et al. Microbial symbionts: A resource for the management of insect-related problems. Microb. Biotechnol. 2011, 5, 307–317. [Google Scholar] [CrossRef] [PubMed]
- Maruščáková, I.C.; Schusterová, P.; Bielik, B.; Toporčák, J.; Bíliková, K.; Mudroňová, D. Effect of application of probiotic pollen suspension on immune response and gut microbiota of honey bees (Apis mellifera). Probiot. Antimicrob. Proteins 2020, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Kwong, W.K.; Mancenido, A.L.; Moran, N.A. Immune system stimulation by the native gut microbiota of honey bees. R. Soc. Open Sci. 2017, 4, 170003. [Google Scholar] [CrossRef]
- Di Donato, A.P. Characterization of Lactic Acid Bacteria Isolated from Apis mellifera L. and Hive Products. Master’s Thesis, University of Molise, Campobasso, Italy, 15 February 2016. [Google Scholar]
- Magnusson, J.; Ström, K.; Roos, S.; Sjögren, J.; Schnürer, J. Broad and complex antifungal activity among environmental isolates of lactic acid bacteria. FEMS Microbiol. Lett. 2003, 219, 129–135. [Google Scholar] [CrossRef]
- Cozzolino, A.; Vergalito, F.; Tremonte, P.; Iorizzo, M.; Lombardi, S.J.; Sorrentino, E.; Luongo, D.; Coppola, R.; Di Marco, R.; Succi, M. Preliminary evaluation of the safety and probiotic potential of Akkermansia muciniphila DSM 22959 in Comparison with Lactobacillus rhamnosus GG. Microorganisms 2020, 8, 189. [Google Scholar] [CrossRef] [PubMed]

| Time (Hours) | Auto-Aggregation (%) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| K7 | K18 | K34 | K40 | K41 | K45 | K55 | K64 | K112 | DSM 12361 | |
| 1 | 11.10 ± 0.88Ga | 4.49 ± 0.14Ca | 4.51 ± 0.10Ca | 15.41 ± 0.54Ha | 3.25 ± 0.12Ba | 0.76 ± 0.02Aa | 5.55 ± 0.22Da | 6.76 ± 0.50Ea | 9.48 ± 0.16Fa | 9.32 ± 0.20Fa |
| 2 | 18.40 ± 1.18Fb | 8.62 ± 0.36Cb | 10.94 ± 0.89Db | 17.40 ± 1.08Fb | 7.56 ± 0.32Bb | 2.11 ± 0.10Ab | 10.43 ± 0.27Db | 10.61 ± 0.35Db | 16.10 ± 0.96Fb | 13.66 ± 0.35Eb |
| 5 | 22.71 ± 1.00Fc | 12.41 ± 0.16Cc | 11.31 ± 0.34Bb | 24.94 ± 0.04Hc | 25.20 ± 0.19Ic | 6.23 ± 0.59Ac | 14.82 ± 0.98Dc | 15.27 ± 0.12Dc | 23.42 ± 0.31Gc | 19.70 ± 0.65Ec |
| 24 | 53.42 ± 1.21Bd | 62.23 ± 1.32Dd | 65.62 ± 1.37Ec | 56.52 ± 1.95Cd | 68.10 ± 1.71Fd | 41.81 ± 0.42Ad | 62.30 ± 0.99Dd | 56.12 ± 2.39Cd | 55.80 ± 0.94Cd | 55.42 ± 3.21Cd |
| Time (Hours) | Sugar Syrup Composition | Survival (log CFU/mL) of L. kunkeei Strains | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| K7 | K18 | K34 | K40 | K41 | K45 | K55 | K64 | K112 | DSM 12361 | ||
| T0 | 40% glucose 20% fructose | 8.41 ± 0.02Bc | 8.38 ± 0.04Bc | 8.44 ± 0.06Bc | 8.23 ± 0.03Ac | 8.19 ± 0.04Ac | 8.17 ± 0.02Ac | 8.26 ± 0.05Ac | 8.24 ± 0.01Ac | 8.36 ± 0.02Bc | 8.40 ± 0.04Bc |
| T24 | 7.89 ± 0.01Eb | 8.10 ± 0.01Gb | 7.60 ± 0.07Cb | 7.12 ± 0.05Bb | 7.20 ± 0.04Bb | 7.80 ± 0.06Db | 7.18 ± 0.08Bb | 6.90 ± 0.04Ab | 8.01 ± 0.01Fb | 6.90 ± 0.02Ab | |
| T48 | 6.62 ± 0.03Fa | 7.51 ± 0.02Ha | 4.90 ± 0.02Ba | 5.84 ± 0.05Ca | 6.29 ± 0.01Da | 6.47 ± 0.02Ea | 5.84 ± 0.01Ca | 5.90 ± 0.02Ca | 6.84 ± 0.03Ga | 4.80 ± 0.04Aa | |
| T0 | 40% glucose 30% fructose | 8.66 ± 0.04Ec | 8.50 ± 0.03Dc | 8.43 ± 0.05Dc | 8.06 ± 0.08Ac | 8.46 ± 0.08Dc | 8.43 ± 0.02Dc | 8.25 ± 0.04Bc | 8.23 ± 0.05Bc | 8.30 ± 0.02Cc | 8.45 ± 0.06Dc |
| T24 | 6.23 ± 0.02Db | 7.12 ± 0.05Gb | 6.24 ± 0.04Db | 6.90 ± 0.02Fb | 6.84 ± 0.01Eb | 6.11 ± 0.05Cb | 6.12 ± 0.02Cb | 6.24 ± 0.06Db | 5.97 ± 0.02Bb | 4.97 ± 0.07Ab | |
| T48 | 4.54 ± 0.04Da | 5.75 ± 0.05Ha | 4.61 ± 0.01Ea | 4.01 ± 0.02Ba | 4.98 ± 0.03Ga | 4.73 ± 0.03Fa | 5.59 ± 0.01Ia | 4.94 ± 0.04Ga | 4.20 ± 0.02Ca | 2.91 ± 0.01Aa | |
| T0 | 50% sucrose | 8.20 ± 0.04Bc | 8.10 ± 0.01Ac | 8.50 ± 0.03Fc | 8.40 ± 0.06Dc | 8.49 ± 0.07Ec | 8.23 ± 0.01Bc | 8.10 ± 0.04Ac | 8.53 ± 0.07Fc | 8.50 ± 0.06Fc | 8.30 ± 0.04Cc |
| T24 | 7.20 ± 0.04Bb | 8.20 ± 0.03Gb | 7.90 ± 0.02Fb | 7.79 ± 0.06Eb | 8.50 ± 0.01Hb | 7.60 ± 0.02Db | 7.10 ± 0.01Ab | 7.60 ± 0.05Db | 7.50 ± 0.07Cb | 7.20 ± 0.06Bb | |
| T48 | 7.00 ± 0.05Da | 7.94 ± 0.01Fa | 5.94 ± 0.02Aa | 6.85 ± 0.01Ca | 7.30 ± 0.05Ea | 6.89 ± 0.02Ca | 6.85 ± 0.01Ca | 6.90 ± 0.05Ca | 6.90 ± 0.06Ca | 6.50 ± 0.06Ba | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iorizzo, M.; Lombardi, S.J.; Ganassi, S.; Testa, B.; Ianiro, M.; Letizia, F.; Succi, M.; Tremonte, P.; Vergalito, F.; Cozzolino, A.; et al. Antagonistic Activity against Ascosphaera apis and Functional Properties of Lactobacillus kunkeei Strains. Antibiotics 2020, 9, 262. https://doi.org/10.3390/antibiotics9050262
Iorizzo M, Lombardi SJ, Ganassi S, Testa B, Ianiro M, Letizia F, Succi M, Tremonte P, Vergalito F, Cozzolino A, et al. Antagonistic Activity against Ascosphaera apis and Functional Properties of Lactobacillus kunkeei Strains. Antibiotics. 2020; 9(5):262. https://doi.org/10.3390/antibiotics9050262
Chicago/Turabian StyleIorizzo, Massimo, Silvia Jane Lombardi, Sonia Ganassi, Bruno Testa, Mario Ianiro, Francesco Letizia, Mariantonietta Succi, Patrizio Tremonte, Franca Vergalito, Autilia Cozzolino, and et al. 2020. "Antagonistic Activity against Ascosphaera apis and Functional Properties of Lactobacillus kunkeei Strains" Antibiotics 9, no. 5: 262. https://doi.org/10.3390/antibiotics9050262
APA StyleIorizzo, M., Lombardi, S. J., Ganassi, S., Testa, B., Ianiro, M., Letizia, F., Succi, M., Tremonte, P., Vergalito, F., Cozzolino, A., Sorrentino, E., Coppola, R., Petrarca, S., Mancini, M., & De Cristofaro, A. (2020). Antagonistic Activity against Ascosphaera apis and Functional Properties of Lactobacillus kunkeei Strains. Antibiotics, 9(5), 262. https://doi.org/10.3390/antibiotics9050262









