Detection of Quorum-Sensing Molecules for Pathogenic Molecules Using Cell-Based and Cell-Free Biosensors
Abstract
1. Introduction
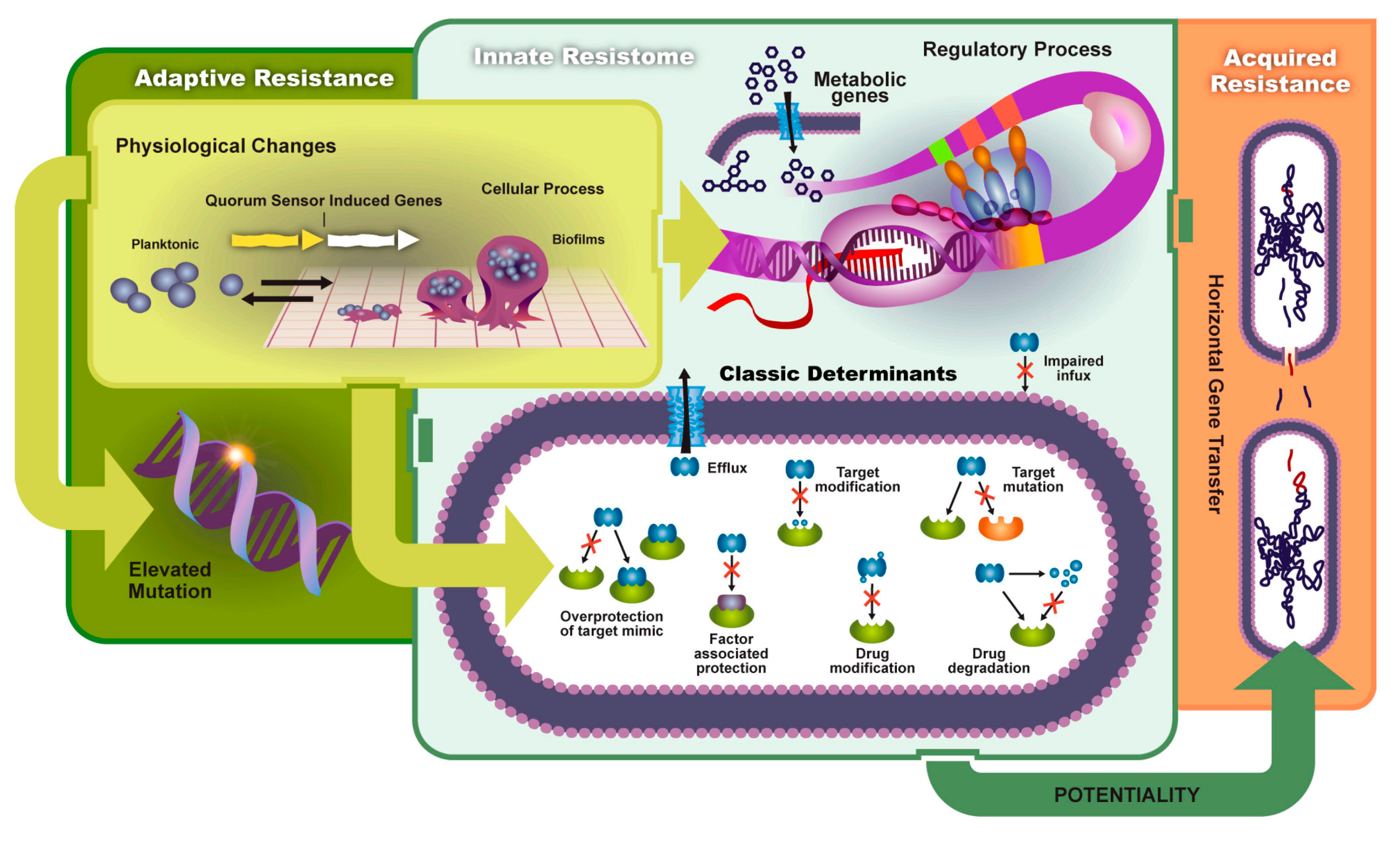
2. Development of Antibiotic Resistance in ESKAPE Bacteria
3. QS in Gram-Positive Pathogens
4. QS in Gram-Negative Pathogens
5. State-of-the-Art Biosensing for Quorum-Sensing Molecules
6. Biosensing Developments
6.1. Gram-Positive
6.2. Gram-Negative
7. Outlook and Considerations
Author Contributions
Funding
Conflicts of Interest
References
- Centers for Disease Control and Preventrion. About Antibiotic Resistance. 2019. Available online: https://www.cdc.gov/drugresistance/about.html (accessed on 12 January 2020).
- Ng, W.; Bassler, B.L. Bacterial Quorum-Sensing Network Architectures. Annu. Rev. Genet. 2009, 43, 197–222. [Google Scholar] [CrossRef] [PubMed]
- Dickschat, J.S. Quorum sensing and bacterial biofilms. Nat. Prod. Rep. 2010, 27, 343–369. [Google Scholar] [CrossRef] [PubMed]
- Rutherford, S.T.; Bassler, B.L. Bacterial quorum sensing: Its role in virulence and possibilities for its control. Cold Spring Harb. Perspect. Med. 2012, 2, a012427. [Google Scholar] [CrossRef] [PubMed]
- Bassler, B.L. How bacteria talk to each other: Regulation of gene expression by quorum sensing. Curr. Opin. Microbiol. 1999, 2, 582–587. [Google Scholar] [CrossRef]
- Gould, T.A.; Herman, J.; Krank, J.; Murphy, R.C.; Churchill, M.E.A. Specificity of acyl-homoserine lactone synthases examined by mass spectrometry. J. Bacteriol. 2006, 188, 773–783. [Google Scholar] [CrossRef]
- Bhardwaj, A.K.; Vinothkumar, K. Quorum Sensing vs Quorum Quenching: A Battle with No End in Sight; Springer: New Delhi, India, 2015; ISBN 9788132219811. [Google Scholar]
- Van Boeckel, T.P.; Gandra, S.; Ashok, A.; Caudron, Q.; Grenfell, B.T.; Levin, S.A.; Laxminarayan, R. Global antibiotic consumption 2000 to 2010: An analysis of national pharmaceutical sales data. Lancet Infect. Dis. 2014, 14, 742–750. [Google Scholar] [CrossRef]
- Bartlett, J.G.; Gilbert, D.N.; Spellberg, B. Seven ways to preserve the Miracle of antibiotics. Clin. Infect. Dis. 2013, 56, 1445–1450. [Google Scholar] [CrossRef]
- Interagency Coordination Group on Antimicrobial Resistance. No Time to Wait: Securing the Future from Drug-Resistant Infections; WHO: Geneva, Switzerland, 2019. [Google Scholar]
- Talbot, G.H.; Jezek, A.; Murray, B.E.; Jones, R.N.; Ebright, R.H.; Nau, G.J.; Rodvold, K.A.; Newland, J.G.; Boucher, H.W. The infectious diseases society of America’s 10 × ’20 initiative (10 new systemic antibacterial agents US food and drug administration approved by 2020): Is 20 × ’20 a possibility? Clin. Infect. Dis. 2019, 69, 1–11. [Google Scholar] [CrossRef]
- Pantosti, A.; Sanchini, A.; Monaco, M. Mechanisms of antibiotic resistance in Staphylococcus aureus. Future Microbiol. 2007, 2, 323–334. [Google Scholar] [CrossRef]
- Reygaert, W.C. An overview of the antimicrobial resistance mechanisms of bacteria. AIMS Microbiol. 2018, 4, 482–501. [Google Scholar] [CrossRef]
- Flemming, H.C.; Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. [Google Scholar] [CrossRef] [PubMed]
- Mohammad Reza, S. Bacterial Biofilm and its Clinical Implications. Ann. Microbiol. Res. 2018, 2, 2. [Google Scholar] [CrossRef]
- Schroeder, M.; Brooks, B.D.; Brooks, A.E. The complex relationship between virulence and antibiotic resistance. Genes 2017, 8, 39. [Google Scholar] [CrossRef] [PubMed]
- Ali, L.; Goraya, M.U.; Arafat, Y.; Ajmal, M.; Chen, J.L.; Yu, D. Molecular mechanism of quorum-sensing in Enterococcus faecalis: Its role in virulence and therapeutic approaches. Int. J. Mol. Sci. 2017, 18, 960. [Google Scholar] [CrossRef] [PubMed]
- Tong, S.Y.C.; Davis, J.S.; Eichenberger, E.; Holland, T.L.; Fowler, V.G. Staphylococcus aureus infections: Epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 2015, 28, 603–661. [Google Scholar] [CrossRef]
- Le, K.Y.; Otto, M. Quorum-sensing regulation in staphylococci—An overview. Front. Microbiol. 2015, 6, 1174. [Google Scholar] [CrossRef]
- Yarwood, J.M.; Schlievert, P.M. Quorum sensing in Staphylococcus infections. J. Clin. Investig. 2003, 112, 1620–1625. [Google Scholar] [CrossRef]
- Gomes-Fernandes, M.; Laabei, M.; Pagan, N.; Hidalgo, J.; Molinos, S.; Villar Hernandez, R.; Domínguez-Villanueva, D.; Jenkins, A.T.A.; Lacoma, A.; Prat, C. Accessory gene regulator (Agr) functionality in Staphylococcus aureus derived from lower respiratory tract infections. PLoS ONE 2017, 12, e0175552. [Google Scholar] [CrossRef]
- Novick, R.P. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 2003, 48, 1429–1449. [Google Scholar] [CrossRef]
- Pinkston, K.L.; Gao, P.; Diaz-Garcia, D.; Sillanpää, J.; Nallapareddy, S.R.; Murray, B.E.; Harvey, B.R. The Fsr quorum-sensing system of Enterococcus faecalis modulates surface display of the collagen-binding MSCRAMM Ace through regulation of gelE. J. Bacteriol. 2011, 193, 4317–4325. [Google Scholar] [CrossRef]
- Dundar, H.; Brede, D.A.; La Rosa, S.L.; El-Gendy, A.O.; Diep, D.B.; Nes, I.F. The fsr quorum-sensing system and cognate gelatinase orchestrate the expression and processing of proprotein EF_1097 into the mature antimicrobial peptide enterocin O16. J. Bacteriol. 2015, 197, 2112–2121. [Google Scholar] [CrossRef]
- Shankar, J.; Walker, R.G.; Ward, D.; Horsburgh, M.J. The Enterococcus faecalis exoproteome: Identification and temporal regulation by fsr. PLoS ONE 2012, 7, e33450. [Google Scholar] [CrossRef]
- Thurlow, L.R.; Thomas, V.C.; Narayanan, S.; Olson, S.; Fleming, S.D.; Hancock, L.E. Gelatinase contributes to the pathogenesis of endocarditis caused by Enterococcus faecalis. Infect. Immun. 2010, 78, 4936–4943. [Google Scholar] [CrossRef] [PubMed]
- Thomas, V.C.; Thurlow, L.R.; Boyle, D.; Hancock, L.E. Regulation of autolysis-dependent extracellular DNA release by Enterococcus faecalis extracellular proteases influences biofilm development. J. Bacteriol. 2008, 190, 5690–5698. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Preventrion. Pneumococcal Disease. 2019. Available online: https://www.cdc.gov/mmwr/preview/mmwrhtml/00047135.htm (accessed on 12 January 2020).
- Galante, J.; Ho, A.; Tingey, S.; Charalambous, B. Quorum Sensing and Biofilms in the Pathogen, Streptococcus pneumoniae. Curr. Pharm. Des. 2014, 21, 25–30. [Google Scholar] [CrossRef] [PubMed]
- Yadav, M.K.; Vidal, J.E.; Go, Y.Y.; Kim, S.H.; Chae, S.W.; Song, J.J. The LuxS/AI-2 quorum-sensing system of Streptococcus pneumoniae Is required to cause disease, and to regulate virulence- and metabolism-related genes in a rat model of middle ear infection. Front. Cell. Infect. Microbiol. 2018, 8, 138. [Google Scholar] [CrossRef]
- Lin, J.; Cheng, J.; Wang, Y.; Shen, X. The Pseudomonas quinolone signal (PQS): Not just for quorum sensing anymore. Front. Cell. Infect. Microbiol. 2018, 8, 230. [Google Scholar] [CrossRef]
- Oziat, J.; Gougis, M.; Malliaras, G.G.; Mailley, P. Electrochemical Characterizations of four Main Redox–metabolites of Pseudomonas aeruginosa. Electroanalysis 2017, 29, 1332–1340. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. Healthcare-Associated Infections. 2019. Available online: https://www.cdc.gov/hai/organisms/pseudomonas.html (accessed on 12 January 2020).
- Bielecki, P.; Glik, J.; Kawecki, M.; Martins Dos Santos, V.A.P. Towards understanding Pseudomonas aeruginosa burn wound infections by profiling gene expression. Biotechnol. Lett. 2008, 30, 777–790. [Google Scholar] [CrossRef]
- Lee, J.; Zhang, L. The hierarchy quorum sensing network in Pseudomonas aeruginosa. Protein Cell 2014, 6, 26–41. [Google Scholar] [CrossRef]
- Higgins, S.; Heeb, S.; Rampioni, G.; Fletcher, M.P.; Williams, P.; Cámara, M. Differential regulation of the phenazine biosynthetic operons by quorum sensing in Pseudomonas aeruginosa PAO1-N. Front. Cell. Infect. Microbiol. 2018, 8, 252. [Google Scholar] [CrossRef] [PubMed]
- Saipriya, K.; Swathi, C.H.; Ratnakar, K.S.; Sritharan, V. Quorum-sensing system in Acinetobacter baumannii: A potential target for new drug development. J. Appl. Microbiol. 2019, 128, 15–27. [Google Scholar] [CrossRef] [PubMed]
- Niu, C.; Clemmer, K.M.; Bonomo, R.A.; Rather, P.N. Isolation and characterization of an autoinducer synthase from Acinetobacter baumannii. J. Bacteriol. 2008, 190, 3386–3392. [Google Scholar] [CrossRef] [PubMed]
- De Araujo, C.; Balestrino, D.; Roth, L.; Charbonnel, N.; Forestier, C. Quorum sensing affects biofilm formation through lipopolysaccharide synthesis in Klebsiella pneumoniae. Res. Microbiol. 2010, 161, 595–603. [Google Scholar] [CrossRef]
- Balestrino, D.; Haagensen, J.A.J.; Rich, C.; Forestier, C. Characterization of type 2 quorum sensing in Klebsiella pneumoniae and relationship with biofilm formation. J. Bacteriol. 2005, 187, 2870–2880. [Google Scholar] [CrossRef]
- Yin, W.F.; Purmal, K.; Chin, S.; Chan, X.Y.; Koh, C.L.; Sam, C.K.; Chan, K.G. N-Acyl homoserine lactone production by Klebsiella pneumonia isolated from human tongue surface. Sensors 2012, 12, 3472–3483. [Google Scholar] [CrossRef]
- Pereira, C.S.; Thompson, J.A.; Xavier, K.B. AI-2-mediated signalling in bacteria. FEMS Microbiol. Rev. 2013, 37, 156–181. [Google Scholar] [CrossRef]
- Xu, L.; Li, H.; Vuong, C.; Vadyvaloo, V.; Wang, J.; Yao, Y.; Otto, M.; Gao, Q. Role of the luxS quorum-sensing system in biofilm formation and virulence of Staphylococcus epidermidis. Infect. Immun. 2006, 74, 488–496. [Google Scholar] [CrossRef]
- Eugene Sanders, W.E.; Sanders, C.C. Enterobacter spp.: Pathogens poised to flourish at the turn of the century. Clin. Microbiol. Rev. 1997, 10, 220–241. [Google Scholar] [CrossRef]
- Zhou, J.; Lao, Y.M.; Ma, Z.P.; Cai, Z.H. Genome sequence of Enterobacter sp. ST3, a quorum sensing bacterium associated with marine dinoflagellate. Genom. Data 2016, 7, 195–199. [Google Scholar] [CrossRef][Green Version]
- Yin, W.F.; Purmal, K.; Chin, S.; Chan, X.Y.; Chan, K.G. Long chain N-acyl homoserine lactone production by Enterobacter sp. isolated from human tongue surfaces. Sensors 2012, 12, 14307–14314. [Google Scholar] [CrossRef] [PubMed]
- Surette, M.G.; Miller, M.B.; Bassler, B.L. Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: A new family of genes responsible for autoinducer production. Proc. Natl. Acad. Sci. USA 1999, 96, 1639–1644. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Xue, T.; Shang, F.; Sun, H.; Sun, B. Staphylococcus aureus AI-2 quorum sensing associates with the KdpDE two-component system to regulate capsular polysaccharide synthesis and virulence. Infect. Immun. 2010, 78, 3506–3515. [Google Scholar] [CrossRef]
- Hegde, M.; Englert, D.L.; Schrock, S.; Cohn, W.B.; Vogt, C.; Wood, T.K.; Manson, M.D.; Jayaraman, A. Chemotaxis to the quorum-sensing signal AI-2 requires the Tsr chemoreceptor and the periplasmic LsrB AI-2-binding protein. J. Bacteriol. 2011, 193, 768–773. [Google Scholar] [CrossRef] [PubMed]
- Pitt, J.J. Principles and Applications of Liquid Chromatography-Mass Spectrometry in Clinical Biochemistry. Clin. Biochem Rev. 2009, 30, 19–34. [Google Scholar] [PubMed]
- Purohit, A.A.; Johansen, J.A.; Hansen, H.; Leiros, H.K.S.; Kashulin, A.; Karlsen, C.; Smalås, A.; Haugen, P.; Willassen, N.P. Presence of acyl-homoserine lactones in 57 members of the Vibrionaceae family. J. Appl. Microbiol. 2013, 115, 835–847. [Google Scholar] [CrossRef] [PubMed]
- Pratima, N.A. Liquid Chromatography-Mass Spectrometry and Its Applications: A Brief Review. Arch. Org. Inorg. Chem. Sci. 2018, 1, 26–34. [Google Scholar] [CrossRef]
- Crawford Scientific Mass Spectrometry Fundamental LC-MS Introduction. 2004, pp. 1–24. Available online: www.chromacademy.com (accessed on 12 January 2020).
- Yang, Y.; Zhou, M.; Hardwidge, P.R.; Cui, H.; Zhu, G. Isolation and characterization of N-acyl homoserine lactone-producing bacteria from cattle rumen and swine intestines. Front. Cell. Infect. Microbiol. 2018, 8, 155. [Google Scholar] [CrossRef]
- Gless, B.H.; Bojer, M.S.; Peng, P.; Baldry, M.; Ingmer, H.; Olsen, C.A. Identification of autoinducing thiodepsipeptides from staphylococci enabled by native chemical ligation. Nat. Chem. 2019, 11, 463–469. [Google Scholar] [CrossRef]
- Junio, H.A.; Todd, D.A.; Ettefagh, K.A.; Ehrmann, B.M.; Kavanaugh, J.S.; Horswill, A.R.; Cech, N.B. Quantitative analysis of autoinducing peptide I (AIP-I) from Staphylococcus aureus cultures using ultrahigh performance liquid chromatography-high resolving power mass spectrometry. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2013, 930, 7–12. [Google Scholar] [CrossRef]
- Kalkum, M.; Lyon, G.J.; Chait, B.T. Detection of secreted peptides by using hypothesis-driven multistage mass spectrometry. Proc. Natl. Acad. Sci. USA 2003, 100, 2795–2800. [Google Scholar] [CrossRef] [PubMed]
- Miller, C.; Stiglich, M.; Livingstone, M.; Gilmore, J. Impedance-Based Biosensing of Pseudomonas putida via Solution Blow Spun PLA: MWCNT Composite Nanofibers. Micromachines 2019, 10, 876. [Google Scholar] [CrossRef] [PubMed]
- Bellitti, P.; Bona, M.; Fontana, S.; Sardini, E.; Serpelloni, M. Study toward the integration of a system for bacterial growth monitoring in an automated specimen processing platform. Lect. Notes Electr. Eng. 2019, 539, 445–454. [Google Scholar]
- Cristina, P.; Javier, M.; Sandra, C.; van Grieken, R. Implications of Electrical Impedance-Based Microbiological Technology in Pork Meat Processing Industry for the Rapid Detection and Quantification of Salmonella spp. J. Food Sci. Eng. 2017, 7, 1–16. [Google Scholar] [CrossRef][Green Version]
- Lei, K.F. Review on impedance detection of cellular responses in micro/nano environment. Micromachines 2014, 5, 1–12. [Google Scholar] [CrossRef]
- Farooq, U.; Wajid Ullah, M.; Yang, Q.; Wang, S. Applications of Phage-Based Biosensors in the Diagnosis of Infectious Diseases, Food Safety, and Environmental Monitoring. Biosens. Environ. Monit. 2019, 1–18. [Google Scholar] [CrossRef]
- Singh, A.; Poshtiban, S.; Evoy, S. Recent advances in bacteriophage based biosensors for food-borne pathogen detection. Sensors 2013, 13, 1763–1786. [Google Scholar] [CrossRef]
- Sibbald, R.G.; Orsted, H.L.; Schultz, G.; Keast, D.H. Preparing the Wound Bed 2003: Focus on Infection and Inflammation. Ostomy/Wound Manag. 2003, 49, 23–51. [Google Scholar]
- Stojadinovic, A.; Carlson, J.W.; Schultz, G.S.; Davis, T.A.; Elster, E.A. Topical advances in wound care. Gynecol. Oncol. 2008, 111, S70–S80. [Google Scholar] [CrossRef]
- Bjarnsholt, T.; Kirketerp-Møller, K.; Jensen, P.Ø.; Madsen, K.G.; Phipps, R.; Krogfelt, K.; Høiby, N.; Givskov, M. Why chronic wounds will not heal: A novel hypothesis. Wound Repair Regen. 2008, 16, 2–10. [Google Scholar] [CrossRef]
- Yussof, S.J.M.; Omar, E.; Pai, D.R.; Sood, S. Cellular events and biomarkers of wound healing. Indian J. Plast. Surg. 2012, 45, 220–228. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Wang, J.; Boyd, B.J. Peptide-based biosensors. Talanta 2015, 136, 114–127. [Google Scholar] [CrossRef] [PubMed]
- Verbeke, F.; De Craemer, S.; Debunne, N.; Janssens, Y.; Wynendaele, E.; Van de Wiele, C.; De Spiegeleer, B. Peptides as quorum sensing molecules: Measurement techniques and obtained levels in vitro and in vivo. Front. Neurosci. 2017, 11, 183. [Google Scholar] [CrossRef] [PubMed]
- Syvitski, R.T.; Tian, X.L.; Sampara, K.; Salman, A.; Lee, S.F.; Jakeman, D.L.; Li, Y.H. Structure-activity analysis of quorum-sensing signaling peptides from Streptococcus mutans. J. Bacteriol. 2007, 189, 1441–1450. [Google Scholar] [CrossRef]
- Tian, X.; Syvitski, R.T.; Liu, T.; Livingstone, N.; Jakeman, D.L.; Li, Y.H. A method for structure-activity analysis of quorum-sensing signaling peptides from naturally transformable streptococci. Biol. Proced. Online 2009, 11, 207–226. [Google Scholar] [CrossRef]
- La Rosa, S.L.; Solheim, M.; Diep, D.B.; Nes, I.F.; Brede, D.A. Bioluminescence based biosensors for quantitative detection of enterococcal peptide—Pheromone activity reveal inter-strain telesensing in vivo during polymicrobial systemic infection. Sci. Rep. 2015, 5, 8339. [Google Scholar] [CrossRef]
- Lubkowicz, D.; Ho, C.L.; Hwang, I.Y.; Yew, W.S.; Lee, Y.S.; Chang, M.W. Reprogramming Probiotic Lactobacillus reuteri as a Biosensor for Staphylococcus aureus Derived AIP-I Detection. ACS Synth. Biol. 2018, 7, 1229–1237. [Google Scholar] [CrossRef]
- Malone, C.L.; Boles, B.R.; Lauderdale, K.J.; Thoendel, M.; Kavanaugh, J.S.; Horswill, A.R. Fluorescent reporters for Staphylococcus aureus. J. Microbiol. Methods 2009, 77, 251–260. [Google Scholar] [CrossRef]
- Yarwood, J.M.; Bartels, D.J.; Volper, E.M.; Greenberg, E.P. Quorum Sensing in Staphylococcus aureus Biofilms. J. Bacteriol. 2004, 186, 1838–1850. [Google Scholar] [CrossRef]
- Franke, G.C.; Dobinsky, S.; Mack, D.; Wang, C.J.; Sobottka, I.; Christner, M.; Knobloch, J.K.M.; Horstkotte, M.A.; Aepfelbacher, M.; Rohde, H. Expression and functional characterization of gfpmut3.1 and its unstable variants in Staphylococcus epidermidis. J. Microbiol. Methods 2007, 71, 123–132. [Google Scholar] [CrossRef]
- Subramoni, S.; Nathoo, N.; Klimov, E.; Yuan, Z.C. Agrobacterium tumefaciens responses to plant-derived signaling molecules. Front. Plant Sci. 2014, 5, 322. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.Q.; Su, S.; Farrand, S.K. In situ activation of the quorum-sensing transcription factor TraR by cognate and noncognate acyl-homoserine lactone ligands: Kinetics and consequences. J. Bacteriol. 2003, 185, 5665–5672. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Beaber, J.W.; Moré, M.I.; Fuqua, C.; Eberhard, A.; Winans, S.C. Analogs of the autoinducer 3-oxooctanoyl-homoserine lactone strongly inhibit activity of the TraR protein of Agrobacterium tumefaciens. J. Bacteriol. 1998, 180, 5398–5405. [Google Scholar] [CrossRef] [PubMed]
- Cha, C.; Gao, P.; Chen, Y.C.; Shaw, P.D.; Farrand, S.K. Production of acyl-homoserine lactone quorum-sensing signals by gram-negative plant-associated bacteria. Mol. Plant-Microbe Interact. 1998, 11, 1119–1129. [Google Scholar] [CrossRef]
- Farrand, S.K.; Qin, Y.; Oger, P. Quorum-sensing system of Agrobacterium plasmids: Analysis and utility. Methods Enzymol. 2002, 358, 452–484. [Google Scholar]
- Steindler, L.; Venturi, V. Detection of quorum-sensing N-acyl homoserine lactone signal molecules by bacterial biosensors. FEMS Microbiol. Lett. 2007, 266, 1–9. [Google Scholar] [CrossRef]
- Nievas, F.; Bogino, P.; Sorroche, F.; Giordano, W. Detection, characterization, and biological effect of quorum-sensing signaling molecules in Peanut-nodulating bradyrhizobia. Sensors 2012, 12, 2851–2873. [Google Scholar] [CrossRef]
- Chambers, C.E.; Visser, M.B.; Schwab, U.; Sokol, P.A. Identification of N-acylhomoserine lactones in mucopurulent respiratory secretions from cystic fibrosis patients. FEMS Microbiol. Lett. 2005, 244, 297–304. [Google Scholar] [CrossRef]
- Zhu, J.; Chai, Y.; Zhong, Z.; Li, S.; Winans, S.C. Agrobacterium Bioassay Strain for Ultrasensitive Detection of N-acylhomoserine lactone-type quorum-sensing molecules: Detection of autoinducers in Mesorhizobium huakuii. Appl. Environ. Microbiol. 2003, 69, 6949–6953. [Google Scholar] [CrossRef]
- Massai, F.; Imperi, F.; Quattrucci, S.; Zennaro, E.; Visca, P.; Leoni, L. A multitask biosensor for micro-volumetric detection of N-3-oxo-dodecanoyl-homoserine lactone quorum sensing signal. Biosens. Bioelectron. 2011, 26, 3444–3449. [Google Scholar] [CrossRef]
- Swift, S.; Karlyshev, A.V.; Fish, L.; Durant, E.L.; Winson, M.K.; Chhabra, S.R.; Williams, P.; Macintyre, S.; Stewart, G.S.A.B. Quorum sensing in Aeromonas hydrophila and Aeromonas salmonicida: Identification of the Luxri homologs AhyRi and AsaRi and their cognate N-acylhomoserine lactone signal molecules. J. Bacteriol. 1997, 179, 5271–5281. [Google Scholar] [CrossRef] [PubMed]
- Pearson, J.P.; Gray, K.M.; Passador, L.; Tucker, K.D.; Eberhard, A.; Iglewski, B.H.; Greenberg, E.P. Structure of the autoinducer required for expression of Pseudomonas aeruginosa virulence genes. Proc. Natl. Acad. Sci. USA 1994, 91, 197–201. [Google Scholar] [CrossRef] [PubMed]
- Winson, M.K.; Swift, S.; Fish, L.; Throup, J.P.; Jørgensen, F.; Chhabra, S.R.; Bycroft, B.W.; Williams, P.; Stewart, G.S.A.B. Construction and analysis of luxCDABE-based plasmid sensors for investigating N-acyl homoserine lactone-mediated quorum sensing. FEMS Microbiol. Lett. 1998, 163, 185–192. [Google Scholar] [CrossRef] [PubMed]
- Rai, N.; Anand, R.; Ramkumar, K.; Sreenivasan, V.; Dabholkar, S.; Venkatesh, K.V.; Thattai, M. Prediction by promoter logic in bacterial quorum sensing. PLoS Comput. Biol. 2012, 8, e1002361. [Google Scholar] [CrossRef]
- Deng, X.; Zhuang, G.; Ma, A.; Yu, Q.; Zhuang, X. Construction of a dual fluorescence whole-cell biosensor to detect N-acyl homoserine lactones. J. Environ. Sci. (China) 2014, 26, 415–422. [Google Scholar] [CrossRef]
- Andersen, J.B.O.; Heydorn, A.; Hentzer, M.; Eberl, L.E.O.; Geisenberger, O. gfp-Based N-Acyl Homoserine-Lactone Sensor Systems for Detection of Bacterial Communication. Microbiology 2001, 67, 575–585. [Google Scholar] [CrossRef]
- Struss, A.; Pasini, P.; Ensor, C.M.; Raut, N.; Daunert, S. Paper strip whole cell biosensors: A portable test for the semiquantitative detection of bacterial quorum signaling molecules. Anal. Chem. 2010, 82, 4457–4463. [Google Scholar] [CrossRef]
- Dong, Y.H.; Zhang, X.F.; Soo, H.M.L.; Greenberg, E.P.; Zhang, L.H. The two-component response regulator PprB modulates quorum-sensing signal production and global gene expression in Pseudomonas aeruginosa. Mol. Microbiol. 2005, 56, 1287–1301. [Google Scholar] [CrossRef]
- Scales, B.S.; Dickson, R.P.; Lipuma, J.J.; Huffnagle, G.B. Microbiology, genomics, and clinical significance of the Pseudomonas fluorescens species complex, an unappreciated colonizer of humans. Clin. Microbiol. Rev. 2014, 27, 927–948. [Google Scholar] [CrossRef]
- Khan, S.R.; Mavrodi, D.V.; Jog, G.J.; Suga, H.; Farrand, S.K. Activation of the phz Operon of Pseudomonas uorescens 2-79 Requires the LuxR Homolog PhzR, N-(3-OH-Hexanoyl)-L-homoserine lactone produced by the LuxI homolog PhzI, and a cis-acting phz box. Society 2005, 187, 6517–6527. [Google Scholar]
- Riedel, K.; Hentzer, M.; Geisenberger, O.; Huber, B.; Steidle, A.; Wu, H.; Høiby, N.; Givskov, M.; Molin, S.; Eberl, L. N-acylhomoserine-lactone-mediated communication between Pseudomonas aeruginosa and Burkholderia cepacia in mixed biofilms. Microbiology 2001, 147, 3249–3262. [Google Scholar] [CrossRef] [PubMed]
- Gianino, E.; Miller, C.; Gilmore, J. Smart Wound Dressings for Diabetic Chronic Wounds. Bioengineering 2018, 5, 51. [Google Scholar] [CrossRef] [PubMed]
- Shiner, E.K.; Reddy, S.; Timmons, C.; Li, G.; Williams, S.C.; Rumbaugh, K.P. Construction of a bacterial autoinducer detection system in mammalian cells. Biol. Proced. Online 2004, 6, 268–276. [Google Scholar] [CrossRef] [PubMed]
- Tinafar, A.; Jaenes, K.; Pardee, K. Synthetic Biology Goes Cell-Free. BMC Biol. 2019, 17, 64. [Google Scholar] [CrossRef]
- Kawaguchi, T.; Yung, P.C.; Norman, R.S.; Decho, A.W. Rapid screening of quorum-sensing signal N-acyl homoserine lactones by an in vitro cell-free assay. Appl. Environ. Microbiol. 2008, 74, 3667–3671. [Google Scholar] [CrossRef]
- Chappell, J.; Jensen, K.; Freemont, P.S. Validation of an entirely in vitro approach for rapid prototyping of DNA regulatory elements for synthetic biology. Nucleic Acids Res. 2013, 41, 3471–3481. [Google Scholar] [CrossRef]
- Wen, K.Y.; Cameron, L.; Chappell, J.; Jensen, K.; Bell, D.J.; Kelwick, R.; Kopniczky, M.; Davies, J.C.; Filloux, A.; Freemont, P.S. A Cell-Free Biosensor for Detecting Quorum Sensing Molecules in P. aeruginosa-Infected Respiratory Samples. ACS Synth. Biol. 2017, 6, 2293–2301. [Google Scholar] [CrossRef]
- McClean, K.H.; Winson, M.K.; Fish, L.; Taylor, A.; Chhabra, S.R.; Camara, M.; Daykin, M.; Lamb, J.H.; Swift, S.; Bycroft, B.W.; et al. Quorum sensing and Chromobacterium violaceum: Exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology 1997, 143, 3703–3711. [Google Scholar] [CrossRef]
- Winzer, K.; Hardie, K.R.; Burgess, N.; Doherty, N.; Kirke, D.; Holden, M.T.G.; Linforth, R.; Cornell, K.A.; Taylor, A.J.; Hill, P.J.; et al. LuxS: Its role in central metabolism and the in vitro synthesis of 4-hydroxy-5-methyl-3(2H)-furanone. Microbiology 2002, 148, 909–922. [Google Scholar] [CrossRef]
- Zhao, Z.G.; Yu, Y.M.; Xu, B.Y.; Yan, S.S.; Xu, J.F.; Liu, F.; Li, G.M.; Ding, Y.L.; Wu, S.Q. Screening and anti-virulent study of N-acyl homoserine lactones DNA aptamers against Pseudomonas aeruginosa quorum sensing. Biotechnol. Bioprocess Eng. 2013, 18, 406–412. [Google Scholar] [CrossRef]
- Sismaet, H.J. Development and Optimization of Electrochemical Sensors to Detect Bacterial Pathogens for Point-Of-Care Applications. Ph.D. Thesis, Northeastern University, Boston, MA, USA, August 2017. [Google Scholar]
- Baldrich, E.; Muñoz, F.X.; García-Aljaro, C. Electrochemical detection of quorum sensing signaling molecules by dual signal confirmation at microelectrode arrays. Anal. Chem. 2011, 83, 2097–2103. [Google Scholar] [CrossRef] [PubMed]
- Stephenson, K.; Yamaguchi, Y.; Hoch, J.A. The mechanism of action of inhibitors of bacterial two-component signal transduction systems. J. Biol. Chem. 2000, 275, 38900–38904. [Google Scholar] [CrossRef] [PubMed]
- Teasdale, M.E.; Liu, J.; Wallace, J.; Akhlaghi, F.; Rowley, D.C. Secondary metabolites produced by the marine bacterium halobacillus salinus that Inhibit quorum sensing-controlled phenotypes in gram-negative bacteria. Appl. Environ. Microbiol. 2009, 75, 567–572. [Google Scholar] [CrossRef] [PubMed]
- Castillon, G.A.; Adames, N.R.; Rosello, C.H.; Seidel, H.S.; Longtine, M.S.; Cooper, J.A.; Heil-Chapdelaine, R.A. Septins Have a Dual Role in Controlling Mitotic Exit in Budding Yeast We assayed the spindle position checkpoint in these mutants with movies of living cells progressing through mitosis. The cells expressed GFP-Tub1p, allowing us to. Curr. Biol. 2003, 13, 654–658. [Google Scholar] [CrossRef]
- Geske, G.D.; Wezeman, R.J.; Siegel, A.P.; Blackwell, H.E. Small molecule inhibitors of bacterial quorum sensing and biofilm formation. J. Am. Chem. Soc. 2005, 127, 12762–12763. [Google Scholar] [CrossRef]
- Struss, A.K.; Nunes, A.; Waalen, J.; Lowery, C.A.; Pullanikat, P.; Denery, J.R.; Conrad, D.J.; Kaufmann, G.F.; Janda, K.D. Towards implementation of Quorum Sensing Autoinducers as Biomarkers for Infectious Disease States. Anal. Chem. 2013, 85, 3355–3362. [Google Scholar] [CrossRef]
- Barr, H.L.; Halliday, N.; Cámara, M.; Barrett, D.A.; Williams, P.; Forrester, D.L.; Simms, R.; Smyth, A.R.; Honeybourne, D.; Whitehouse, J.L.; et al. Pseudomonas aeruginosa quorum sensing molecules correlate with clinical status in cystic fibrosis. Eur. Respir. J. 2015, 46, 1046–1054. [Google Scholar] [CrossRef]
- Erickson, D.L.; Endersby, R.; Kirkham, A.; Stuber, K.; Vollman, D.D.; Rabin, H.R.; Mitchell, I.; Storey, D.G. Pseudomonas aeruginosa quorum-sensing systems may control virulence factor expression in the lungs of patients with cystic fibrosis. Infect. Immun. 2002, 70, 1783–1790. [Google Scholar] [CrossRef]
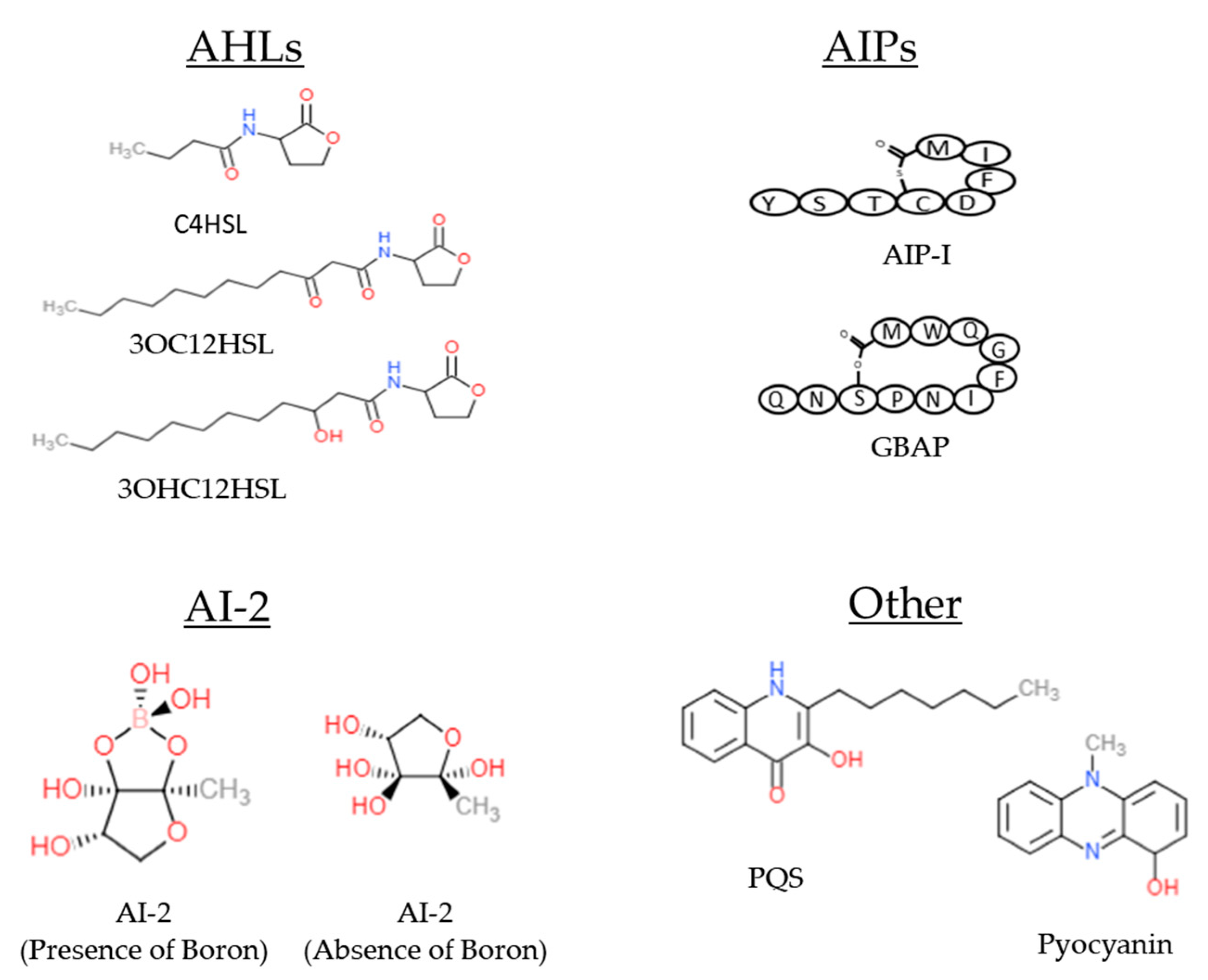


| Bacteria Strain | QS System | Main Signaling Molecules | Transcriptional Factor | QS Virulence Regulation | Ref. |
|---|---|---|---|---|---|
| S. aureus | Agr | AIP-I,II,III | AgrA | lipases, proteases, enterotoxins, superantigens, ureases | [7,20,21,48] |
| LuxS | AI-2 | LuxR-type | capsular polysaccharide synthesis | ||
| E. faecium/E. faecalis | Fsr | GBAP | FsrABCD | Cytolysin, gelatinase | [7] |
| S. pneumoniae | Com | CSP | ComE | polysaccharide capsule, pneumolysin | [7,29,30] |
| LuxS | AI-2 | LuxR-type | Biofilm formation | ||
| K. pneumoniae | LuxS | AI-2, 3OC10HSL, C8HSL | LuxR | Antibiotic resistance genes, biofilm formation | [7,39,40,41] |
| A. baumannii | Aba | 3OC12HSL, C12HSL, C10HSL, C14HSL, 3OC13HSL, C16HSL | AbaI/AbaR | Biofilms, siderophore, lipopolysaccharides, superoxide dismutase | [7,37,38] |
| P. aeruginosa | Las | 3OC12HSL | LasR/LasI | Elastase (lasB), staphylolysin (lasA), alkaline protease (aprA), exotoxin A (toxA), hydrogen cyanide synthase (hcnABC) | [7,35,36] |
| Rhl | C4HSL | RhlR/RhlI | Rhamnolipid synthase (rhlAB), type 1 lectin (lecA), type II lectin (lecB), hcnABC, pyocyanin | ||
| PQS | 2-heptyl-3hydroxy-4-quinolone (PQS) | PqsR | Pyocyanin, lecA, rhlAB, lasB | ||
| LuxS | AI-2 | LuxR-type | Biofilm formation | ||
| E. coli | LuxS | AI-2 | LsRB | Chemotaxis towards AI-2 | [47,49] |
| V. harveyi | LuxS | AI-2 | LuxP | bioluminescence | [7] |
| E. spp. | LuxR-type | C12HSL, short-chain (C6) HSL molecules | LuxR | Biofilm formation | [45,46] |
| Gram-Positive Detection Schemes | ||||
|---|---|---|---|---|
| Host Strain/Cell | Plasmid/Biorecognizing Element | Reporter System | Molecules | Detection Range/LOD |
| S. pneumoniae SMdC | pYH2-pOMZ47 | LacZ reporter/β-gal | CSP | Not reported [70,71] |
| E. faecalis JH2-2 | pSL101cylR2R1Pcyl | Bioluminescence | cytolysin | 640 CIU [72] |
| E. faecalis MMH594 | pREG696luxPfsrB45 and pREG696luxPgelE | Bioluminescence | GBAP | 320 GIU [72] |
| L. reuteri DSM20016 | pSIP409 | GusA | AIP-I | 10–1000 nM [77] |
| L. reuteri DSM20016 | pSIP409 (w/flipped slp-AgrCA) | GusA | AIP-I | 0.5–1000 nM [77] |
| S. aureus SH1000 | pAH1 (agr P3) | YFP, Cam | agr expression agr expression | Not reported [78,79] |
| pAH5 (SigB) | YFP, Cam | |||
| pAH6 (asp23) | mCherry, Cam | |||
| pAH7 (agr P3) | YFP, Erm | |||
| pAH8 (agr P3) | mCherry, Erm | |||
| pAH12 (sarAP1) | mCherry, Erm | |||
| pAH13 (tetracycline ind.) | GFP, Erm | |||
| pAH14 (sarAP1) | YFP, Erm | |||
| pAH15 (pAH14 w/SarA RBS) | YFP, Erm | |||
| pAH16 (pAH14 w/sod RBS) | YFP, Erm | |||
| pAH17 (pAH14 w/hld RBS) | YFP, Erm | |||
| pAH9 (sarA P1) | mCherry | |||
| Gram-Negative Detection Schemes | ||||
|---|---|---|---|---|
| Host Strain/Cell | Plasmid/Biorecognizing Element | Reporter System | Molecules | Detection Range |
| A. tumefaciens | pZLR4 | LacZ reporter/β-gal | C6HSL C8HSL C10HSL C12HSL C14HSL 3OC6HSL 3OC8HSL 3OC10HSL 3OC12HSL 3OHC8HSL 3OHC6HSL | Mostly quantitative [78,80,81,82,83] |
| A. tumefaciens A136 | pCF218-pMV26 | luxCDABE reporter (Luminescence-based assay) | C4HSL C6HSL C8HSL C10HSL C12HSL 3OC6HSL 3OC8HSL 3OC10HSL 3OC12HSL | C4HSL (25 nM) C6HSL (250 nM) C8HSL (0.25 nM) C10HSL (25 nM) C12HSL (250 nM) 3OC6HSL (20 pM) 3OC8HSL (0.2 pM) 3OC10HSL (0.02 pM) 3OC12HSL (0.02 pM) [79] |
| A. tumefaciens A136 | pCF218-pCF372 | LacZ reporter/β-gal | C6HSL 3OC6HSL 3OC8HSL | C6HSL (2.5 µM) 3OC6HSL (100 nM) 3OC8HSL (25 nM) [78] |
| A. tumefaciens KYC55 | pJZ384, pJZ410, and pJZ372 | LacZ reporter/β-gal | 3OC6HSL 3OC8HSL 3OC12HSL 3OHC6HSL 3OHC8HSL 3OHC10HSL C6HSL C8HSL C10HSL | 3OC6HSL (2.5 pM) 3OC8HSL (0.25 pM) 3OC12HSL (0.5 nM) 3hydroxylC6HSL (20 pM) 3hydroxylC8HSL (20 pM) 3hydroxylC10HSL (20 pM) C6HSL (100 pM) C8HSL (30 pM) C10HSL (40 pM) [85] |
| E. coli JM109 | psB1075 | luxCDABE/bioluminescent | C12HSL C14HSL C16HSL 3OC12HSL 3OC14HSL 3OC16HSL 3OHC12HSL | C12HSL (1 nM–50 µM) C14HSL (10 nM–50 µM) C16HSL (100 nM–5 µM) 3OC12HSL (1 nM–5 µM) 3OC14HSL (10 nM–50 µM) 3OC16HSL (10 nM–10 µM) 3OHC12HSL(10 µM–50 µM) [89] |
| E. coli K-12-Z1 | pSB1A2 | traI–luxCDABE | Synthetic HSL | 1µM [84] |
| E. coli | pUCGMA2T | mCherry | 3OC6HSL | 5 × 10−8–1 × 10−5 mol/L [90] |
| E. coli MT102 | pJBA132 | gfpmut3 | C4HSL C6HSL C8HSL 3OC6HSL 3OC10HSL | C4HSL (1 µM) C6HSL (10 nM) C8HSL (10 nM) 3OC6HSL (1 nM) 30C10HSL (10 nM) [91] |
| E. coli DH5α-T1 | pSDB1075 | LacZ reporter/β-gal w/X-gal immobilized on filter paper | C12HSL | 10 nM @ 90 min. 100 nM @ 60 min. [92] |
| P. aeruginosa PA14 | pUCP18 and pMS402 | luxiCDABE reporter | C4HSL 3OC12HSL | C4HSL (10 µM) 3OC12HSL (10 pM) [86] |
| P. aeruginosa M71LZ | pUCP19 | LacZ reporter/β-gal | C4HSL 3OC12HSL | C4HSL (1.0–100 µM) 3OC12HSL (0.01–100 µM) [94] |
| P. fluorescens 1855 | pSF105 and pSF107 | LacZ reporter/β-gal | 3OC6HSL | 10–10,000 nM [96] |
| P. aeruginosa PAO1-JP2 | pKRC12 | GFP | 3OC12HSL | 25 nM [97] |
| P. aeruginosa PAO1-JP2 | pASC8 | GFP | C8HSL | 25 nM [97] |
| monkey kidney COS-1 | LasBOX 1 sequence | Luciferase | C4HSL 3OC12HSL | Not reported [99] |
| A. tumefaciens NTL4 (cell-free) | pCF218 and pCF372 | β-gal | C6 HSL C7HSL C8HSL C10HSL C12HSL 3OC6HSL 3OC8HSL | C6 HSL (30 nM) C7HSL (25 nM) C8HSL (20 nM) C10HSL (100 nM) C12HSL (200 nM) 3OC6HSL (17 nM) 3OC8HSL (10 nM) [101] |
| E. coli extracts (cell-free) | pSB1A2 | GFP | 3OC12HSL | 5–100 nM [103] |
| C. violaceum CV026 | Sequenced on genome | violacein | C4HSL C6HSL C8HSL 3OC4HSL 3OC6HSL 3OC12HSL | C4HSL (1.8 nM) C6HSL (0.01 nM) C8HSL (0.44 nM) 3OC4HSL (73 nM) 3OC6HSL (0.14 nM) 3OC12HSL (0.83 nM) [105] |
| None | 3OC12HSL and C4HSL aptamers | electrochemically w/Methylene blue | C4HSL 3OC12HSL | 0.1–100 µM [107] |
| None | A. tumefaciens NTL4 reporter | β-gal and PAPG/PAP electrochemical detection | 3OC12HSL | 2.5 pM (2 h) 3.6 pM (5 h) [108] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miller, C.; Gilmore, J. Detection of Quorum-Sensing Molecules for Pathogenic Molecules Using Cell-Based and Cell-Free Biosensors. Antibiotics 2020, 9, 259. https://doi.org/10.3390/antibiotics9050259
Miller C, Gilmore J. Detection of Quorum-Sensing Molecules for Pathogenic Molecules Using Cell-Based and Cell-Free Biosensors. Antibiotics. 2020; 9(5):259. https://doi.org/10.3390/antibiotics9050259
Chicago/Turabian StyleMiller, Craig, and Jordon Gilmore. 2020. "Detection of Quorum-Sensing Molecules for Pathogenic Molecules Using Cell-Based and Cell-Free Biosensors" Antibiotics 9, no. 5: 259. https://doi.org/10.3390/antibiotics9050259
APA StyleMiller, C., & Gilmore, J. (2020). Detection of Quorum-Sensing Molecules for Pathogenic Molecules Using Cell-Based and Cell-Free Biosensors. Antibiotics, 9(5), 259. https://doi.org/10.3390/antibiotics9050259





