Application of Response Surface Methodology to Evaluate Photodynamic Inactivation Mediated by Eosin Y and 530 nm LED against Staphylococcus aureus
Abstract
1. Introduction
2. Results and Discussion
2.1. Light Doses
2.2. Photodynamic Inactivation of Staphylococcus aureus
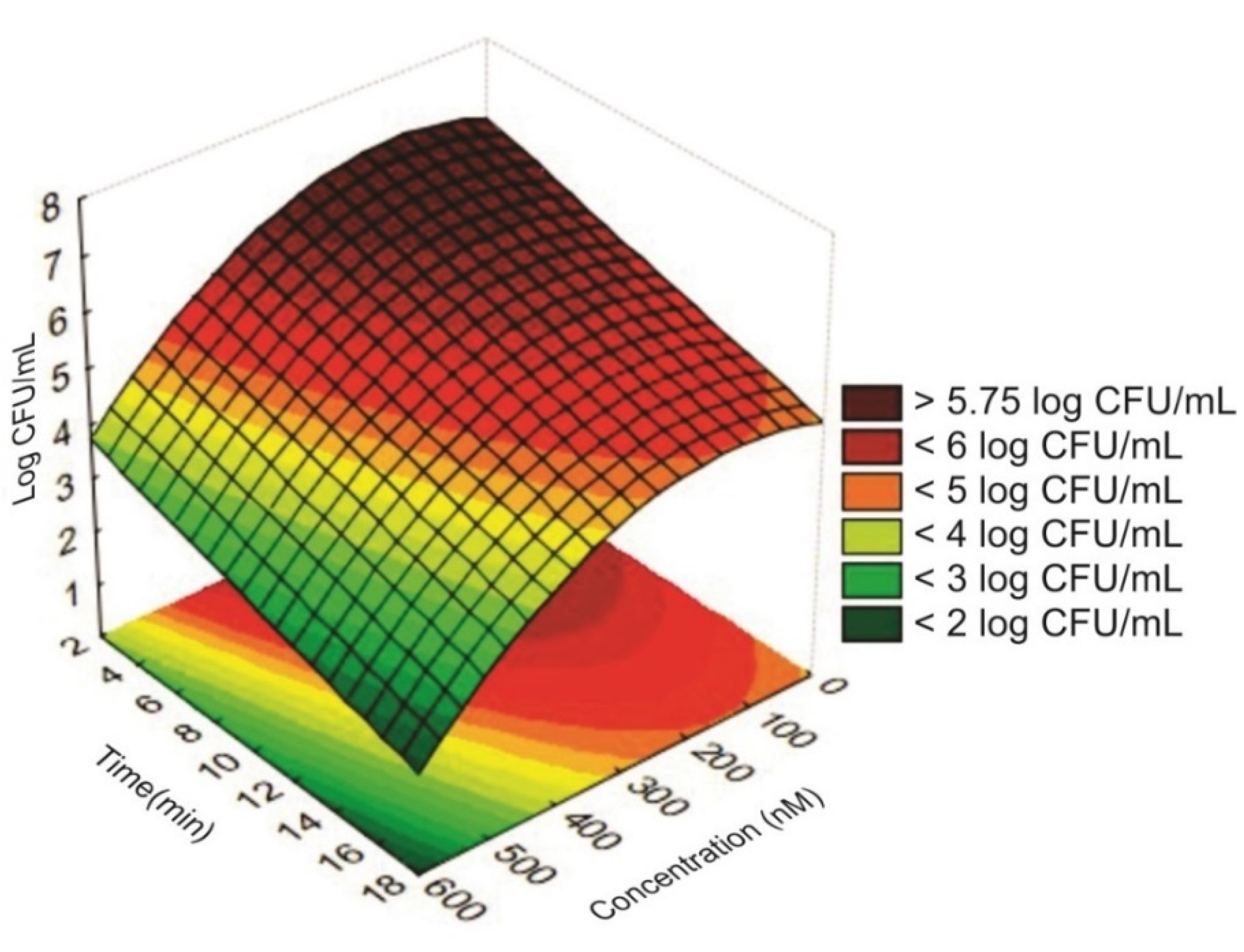
2.3. Determination of Photoinhibitory Activity of Eosin Y and Green LED Light Using the Statistical Experimental Design
2.4. Model Validation
3. Materials and Methods
3.1. Bacterial Strains and Culture Conditions
3.2. Photosensitizers and LED Light Source
3.3. Photodynamic Inactivation of Staphylococcus aureus
3.4. Determination of Photoinhibitory Activity of Eosin Y Using the Statistical Experimental Design
3.5. Statistical Analysis
3.6. Model Validation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Food Safety. Available online: http://www.who.int/en/news-room/fact-sheets/detail/food-sagety (accessed on 29 September 2019).
- Centers for Disease Control and Prevention. National Outbreak Reporting System (NORS). Available online: https://wwwn.cdc.gov/norsdashboard/ (accessed on 29 September 2019).
- Bonin, E.; Dos Santos, A.R.; Fiori da Silva, A.; Ribeiro, L.H.; Favero, M.E.; Campanerut-Sa, P.A.Z.; de Freitas, C.F.; Caetano, W.; Hioka, N.; Mikcha, J.M.G. Photodynamic inactivation of foodborne bacteria by eosin Y. J. Appl. Microbiol. 2018, 124, 1617–1628. [Google Scholar] [CrossRef]
- Alves, E.; Faustino, M.A.F.; Neves, M.G.P.M.S.; Cunha, Â.; Nadais, H.; Almeida, A. Potential applications of porphyrins in photodynamic inactivation beyond the medical scope. J. Photoch. Photobio. C 2015, 22, 34–57. [Google Scholar] [CrossRef]
- Penha, C.B.; Bonin, E.; da Silva, A.F.; Hioka, N.; Zanqueta, É.B.; Nakamura, T.U.; de Abreu Filho, B.A.; Campanerut-Sá, P.A.Z.; Mikcha, J.M.G. Photodynamic inactivation of foodborne and food spoilage bacteria by curcumin. LWT-Food Sci. Technol. 2017, 76, 198–202. [Google Scholar] [CrossRef]
- Silva, A.F.; Dos Santos, A.R.; Trevisan, D.A.C.; Bonin, E.; Freitas, C.F.; Batista, A.F.P.; Hioka, N.; Simoes, M.; Graton Mikcha, J.M. Xanthene Dyes and Green LED for the Inactivation of Foodborne Pathogens in Planktonic and Biofilm States. Photoch. Photobio. 2019. [Google Scholar] [CrossRef] [PubMed]
- Yassunaka, N.; Fabiano de Freitas, C.; Ribeiro Rabello, B.; Regina Santos, P.; Caetano, W.; Hioka, N.; Ueda Nakamura, T.; Alves de Abreu Filho, B.; Martha Graton Mikcha, J. Photodynamic Inactivation Mediated by Erythrosine and its Derivatives on Foodborne Pathogens and Spoilage Bacteria. Curr. Microbiol. 2015, 71. [Google Scholar] [CrossRef] [PubMed]
- Tao, R.; Zhang, F.; Tang, Q.-J.; Xu, C.-S.; Ni, Z.-J.; Meng, X.-H. Effects of curcumin-based photodynamic treatment on the storage quality of fresh-cut apples. Food Chem. 2019, 274, 415–421. [Google Scholar] [CrossRef]
- Bartolomeu, M.; Rocha, S.; Cunha, A.; Neves, M.G.; Faustino, M.A.; Almeida, A. Effect of Photodynamic Therapy on the Virulence Factors of Staphylococcus aureus. Front. Microbial. 2016, 7, 267. [Google Scholar] [CrossRef]
- Marciel, L.; Mesquita, M.Q.; Ferreira, R.; Moreira, B.; Neves, M.G.P.; Faustino, M.A.F.; Almeida, A. An efficient formulation based on cationic porphyrins to photoinactivate Staphylococcus aureus and Escherichia coli. Future Med. Chem. 2018, 10, 1821–1833. [Google Scholar] [CrossRef]
- Martins, D.; Mesquita, M.Q.; Neves, M.; Faustino, M.A.F.; Reis, L.; Figueira, E.; Almeida, A. Photoinactivation of Pseudomonas syringae pv. actinidiae in kiwifruit plants by cationic porphyrins. Planta 2018, 248, 409–421. [Google Scholar] [CrossRef]
- Vieira, C.; Santos, A.; Mesquita, M.Q.; Gomes, A.T.P.C.; Neves, M.G.P.M.S.; Faustino, M.A.F.; Almeida, A. Advances in aPDT based on the combination of a porphyrinic formulation with potassium iodide: Effectiveness on bacteria and fungi planktonic/biofilm forms and viruses. J. Porphyr. Phthalocya. 2019, 23, 534–545. [Google Scholar] [CrossRef]
- Hu, J.; Lin, S.; Tan, B.K.; Hamzah, S.S.; Lin, Y.; Kong, Z.; Zhang, Y.; Zheng, B.; Zeng, S. Photodynamic inactivation of Burkholderia cepacia by curcumin in combination with EDTA. Food Res. Int. 2018, 111, 265–271. [Google Scholar] [CrossRef] [PubMed]
- Estevão, B.M.; Pellosi, D.S.; de Freitas, C.F.; Vanzin, D.; Franciscato, D.S.; Caetano, W.; Hioka, N. Interaction of eosin and its ester derivatives with aqueous biomimetic micelles: Evaluation of photodynamic potentialities. J. Photochem. Photobiol. A 2014, 287, 30–39. [Google Scholar] [CrossRef]
- Silva, A.F.; Borges, A.; Giaouris, E.; Graton Mikcha, J.M.; Simoes, M. Photodynamic inactivation as an emergent strategy against foodborne pathogenic bacteria in planktonic and sessile states. Crit. Rev. Microbiol. 2018, 44, 667–684. [Google Scholar] [CrossRef] [PubMed]
- Almeida, A.; Faustino, M.A.F.; Tomé, J.P.C. Photodynamic inactivation of bacteria: Finding the effective targets. Future Med. Chem. 2015, 7, 1221–1224. [Google Scholar] [CrossRef] [PubMed]
- de Freitas, C.F.; Pellosi, D.S.; Estevao, B.M.; Calori, I.R.; Tsubone, T.M.; Politi, M.J.; Caetano, W.; Hioka, N. Nanostructured Polymeric Micelles Carrying Xanthene Dyes for Photodynamic Evaluation. Photochem. Photobiol. 2016, 92, 790–799. [Google Scholar] [CrossRef]
- Luksiene, Z.; Brovko, L. Antibacterial photosensitizationbased treatment for food safety. Food Eng. Rev. 2013, 5, 185–199. [Google Scholar] [CrossRef]
- Kato, H.; Komagoe, K.; Nakanishi, Y.; Inoue, T.; Katsu, T. Xanthene dyes induce membrane permeabilization of bacteria and erythrocytes by photoinactivation. Photochem. Photobiol. 2012, 88, 423–431. [Google Scholar] [CrossRef]
- Fracalossi, C.; Nagata, J.Y.; Pellosi, D.S.; Terada, R.S.S.; Hioka, N.; Baesso, M.L.; Sato, F.; Rosalen, P.L.; Caetano, W.; Fujimaki, M. Singlet oxygen production by combining erythrosine and halogen light for photodynamic inactivation of Streptococcus mutans. Photodiagn. Photodyn. 2016, 15, 127–132. [Google Scholar] [CrossRef]
- dos Santos, A.R.; da Silva, A.F.; de Freitas, C.F.; da Silva, M.V.; Bona, E.; Nakamura, C.V.; Hioka, N.; Mikcha, J.M.G. Response surface methodology can be used to predict photoinactivation of foodborne pathogens using Rose Bengal excited by 530 nm LED. J. Food Safety 2019, e12736. [Google Scholar] [CrossRef]
- Brancaleon, L.; Moseley, H. Laser and non-laser light sources for photodynamic therapy. Lasers Med. Sci. 2002, 17, 173–186. [Google Scholar] [CrossRef]
- Amaral, L.S.; Azevedo, E.B.; Perussi, J.R. The response surface methodology speeds up the search for optimal parameters in the photoinactivation of E. coli by photodynamic therapy. Photodiagn. Photodyn. 2018, 22, 26–33. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Chen, T.; Yang, S.; Wang, X.; Guo, H.J.A.M. Response surface methodology to design a selective enrichment broth for rapid detection of Salmonella spp. by SYBR Green Ι real-time PCR. Appl. Microbiol. Biot. 2013, 97, 4149–4158. [Google Scholar] [CrossRef] [PubMed]
- Iliadis, I.; Daskalopoulou, A.; Simoes, M.; Giaouris, E. Integrated combined effects of temperature, pH and sodium chloride concentration on biofilm formation by Salmonella enterica ser. Enteritidis and Typhimurium under low nutrient food-related conditions. Food Res. Int. 2018, 107, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Gerola, A.P.; Semensato, J.; Pellosi, D.S.; Batistela, V.R.; Rabello, B.R.; Hioka, N.; Caetano, W. Chemical determination of singlet oxygen from photosensitizers illuminated with LED: New calculation methodology considering the influence of photobleaching. J. Photochem. Photobiol. A 2012, 232, 14–21. [Google Scholar] [CrossRef]
- Brovko, L.; Anany, H.; Bayoumi, M.; Giang, K.; Kunkel, E.; Lim, E.; Naboka, O.; Rahman, S.; Li, J.; Filipe, C.D.M.; et al. Antimicrobial light-activated materials: Towards application for food and environmental safety. J Appl. Microbiol. 2014, 117, 1260–1266. [Google Scholar] [CrossRef]
- Johnson, G.A.; Muthukrishnan, N.; Pellois, J.P. Photoinactivation of Gram positive and Gram negative bacteria with the antimicrobial peptide (KLAKLAK)(2) conjugated to the hydrophilic photosensitizer eosin Y. Bioconjugate Chem. 2013, 24, 114–123. [Google Scholar] [CrossRef]
- Doria Filho, U. Introdução à Bioestatística para Simples Mortais, 3rd ed.; Nagócio: Rio de Janeiro, Spain, 2001. [Google Scholar]
- Oscar, T.P. Response Surface Models for Effects of Temperature, pH, and Previous Growth pH on Growth Kinetics of Salmonella Typhimurium in Brain Heart Infusion Broth. J. Food Protect. 1999, 62, 106–111. [Google Scholar] [CrossRef]
 498 nM;
498 nM;  440 nM;
440 nM;  300 nM;
300 nM;  160 nM; and
160 nM; and  102 nM).
102 nM).
 498 nM;
498 nM;  440 nM;
440 nM;  300 nM;
300 nM;  160 nM; and
160 nM; and  102 nM).
102 nM).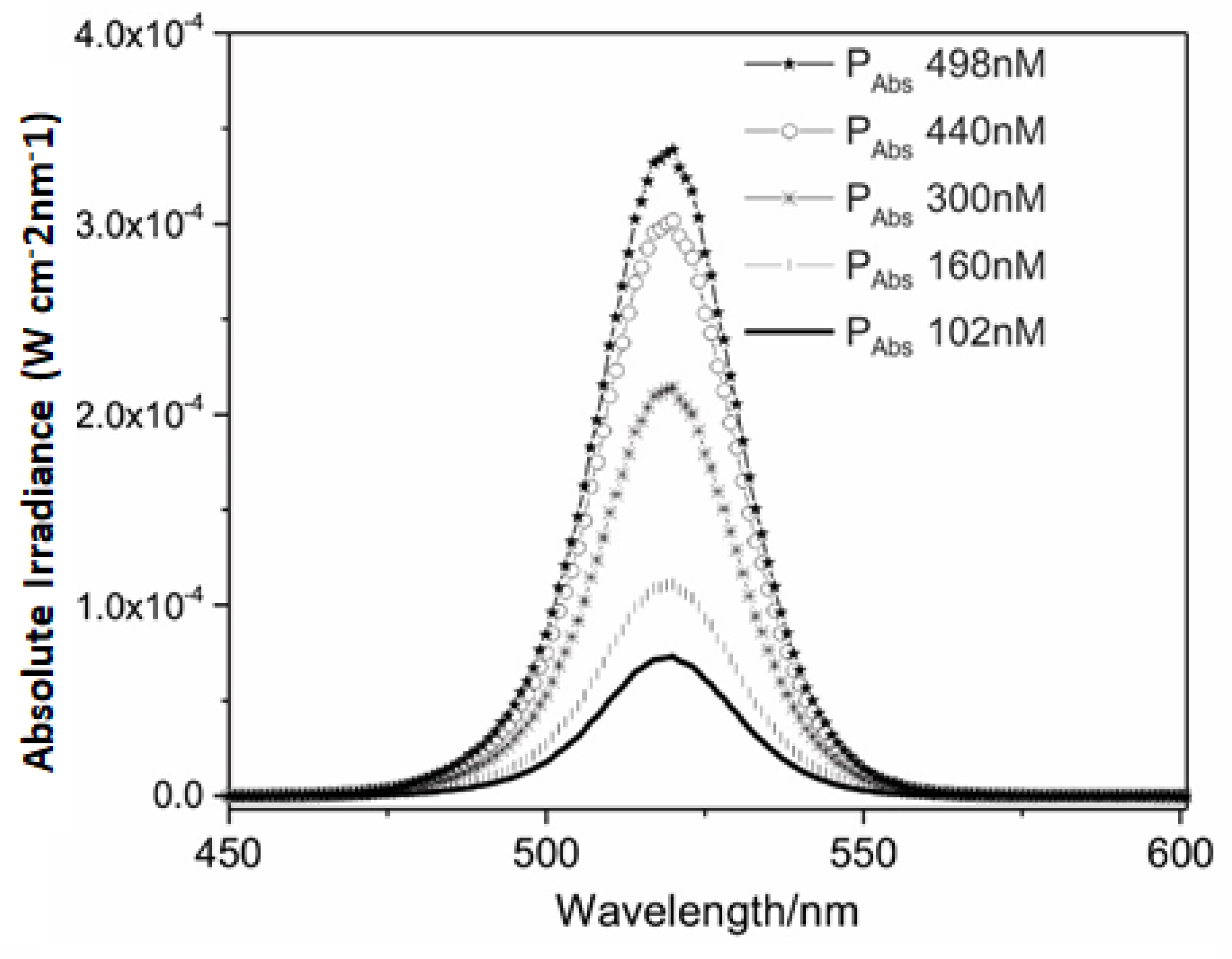
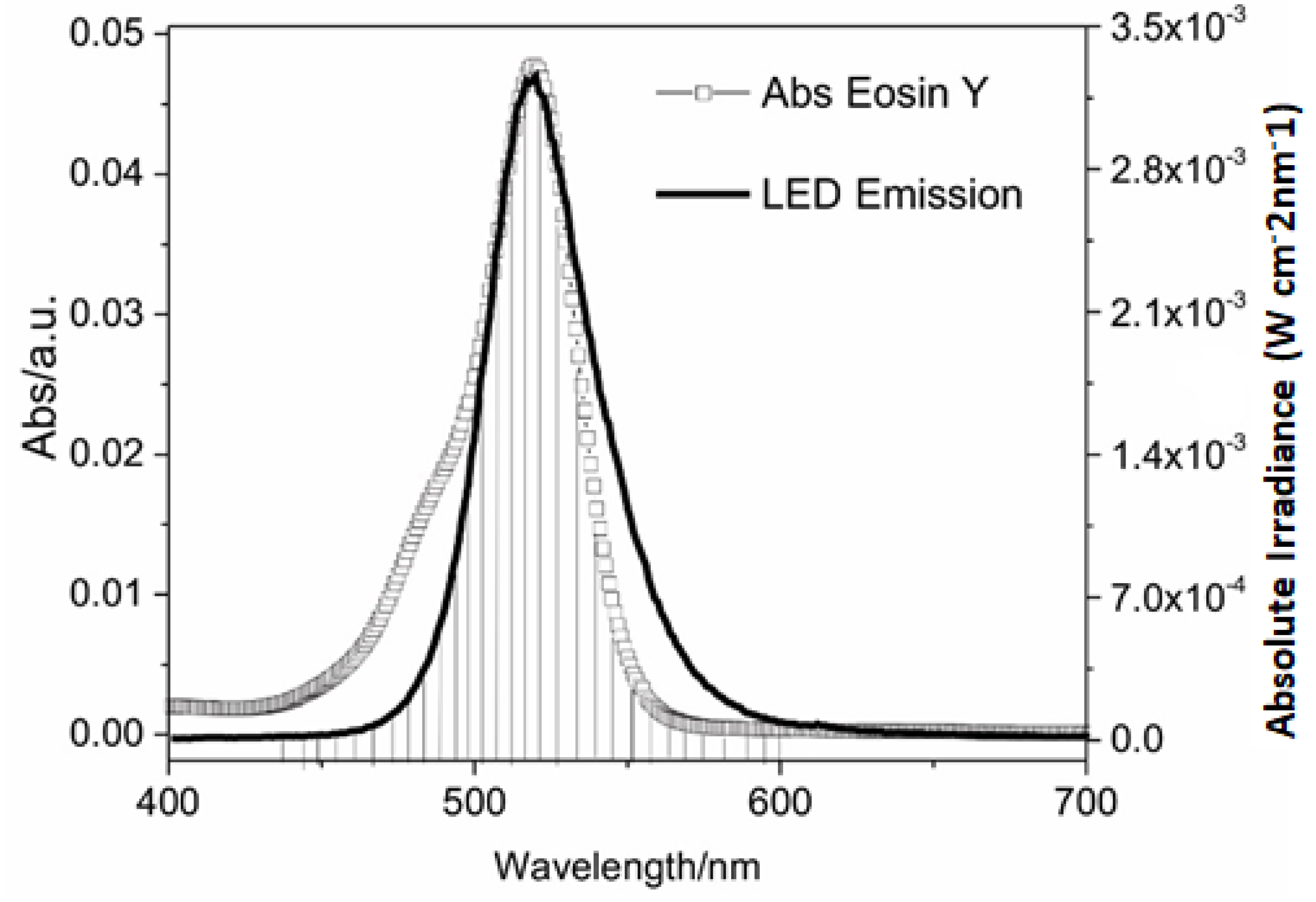
| Experiments | Coded Values | Real Values | ||||
|---|---|---|---|---|---|---|
| X1 | X2 | Concentration (nM) | Time (min) | Light Doses (J/cm2 [15]) | Cell Viability (Log CFU/mL) ** | |
| Control (PS−L−) * | -- | -- | 0 | 0 | 0 | 6.23 ± 0.06 |
| 1 | −1.00000 | −1.00000 | 160 | 6.00 | 1.96 | 6.24 ± 0.07 |
| 2 | −1.00000 | 1.00000 | 160 | 14.00 | 4.56 | 5.17 ± 0.02 |
| 3 | 1.00000 | −1.00000 | 440 | 6.00 | 5.32 | 5.17 ± 0.06 |
| 4 | 1.00000 | 1.00000 | 440 | 14.00 | 12.43 | 4.14 ± 0.03 |
| 5 | −1.41421 | 0.00000 | 102 | 10.00 | 2.13 | 6.20 ± 0.02 |
| 6 | 1.41421 | 0.00000 | 498 | 10.00 | 9.98 | 3.91 ± 0.02 |
| 7 | 0.00000 | −1.41421 | 300 | 4.34 | 2.72 | 6.30 ± 0.07 |
| 8 | 0.00000 | 1.41421 | 300 | 15.65 | 9.84 | 5.11 ± 0.05 |
| 9 | 0.00000 | 0.00000 | 300 | 10.00 | 6.30 | 5.49 ± 0.01 |
| 10 | 0.00000 | 0.00000 | 300 | 10.00 | 6.30 | 5.31 ± 0.02 |
| 11 | 0.00000 | 0.00000 | 300 | 10.00 | 6.30 | 5.53 ± 0.06 |
| 12 | 0.00000 | 0.00000 | 300 | 10.00 | 6.30 | 5.78 ± 0.04 |
| Responses | Cell Viability (Log CFU/mL) | ||
|---|---|---|---|
| Experiment 1 | Experiment 2 | Experiment 3 | |
| Predicted | 4.70 | 5.25 | 3.36 |
| PCILL-95% a | 4.23 | 4.78 | 2.89 |
| PCIUL-95% b | 5.16 | 5.71 | 3.82 |
| Observed | 5.13 ± 0.11 | 5.09 ± 0.24 | 4.08 ± 0.28 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santos, A.R.; da Silva, A.F.; Batista, A.F.P.; Freitas, C.F.; Bona, E.; Sereia, M.J.; Caetano, W.; Hioka, N.; Mikcha, J.M.G. Application of Response Surface Methodology to Evaluate Photodynamic Inactivation Mediated by Eosin Y and 530 nm LED against Staphylococcus aureus. Antibiotics 2020, 9, 125. https://doi.org/10.3390/antibiotics9030125
Santos AR, da Silva AF, Batista AFP, Freitas CF, Bona E, Sereia MJ, Caetano W, Hioka N, Mikcha JMG. Application of Response Surface Methodology to Evaluate Photodynamic Inactivation Mediated by Eosin Y and 530 nm LED against Staphylococcus aureus. Antibiotics. 2020; 9(3):125. https://doi.org/10.3390/antibiotics9030125
Chicago/Turabian StyleSantos, Adriele R., Alex F. da Silva, Andréia F. P. Batista, Camila F. Freitas, Evandro Bona, Maria J. Sereia, Wilker Caetano, Noburu Hioka, and Jane M. G. Mikcha. 2020. "Application of Response Surface Methodology to Evaluate Photodynamic Inactivation Mediated by Eosin Y and 530 nm LED against Staphylococcus aureus" Antibiotics 9, no. 3: 125. https://doi.org/10.3390/antibiotics9030125
APA StyleSantos, A. R., da Silva, A. F., Batista, A. F. P., Freitas, C. F., Bona, E., Sereia, M. J., Caetano, W., Hioka, N., & Mikcha, J. M. G. (2020). Application of Response Surface Methodology to Evaluate Photodynamic Inactivation Mediated by Eosin Y and 530 nm LED against Staphylococcus aureus. Antibiotics, 9(3), 125. https://doi.org/10.3390/antibiotics9030125





