Molecular Epidemiology of Antibiotic-Resistant Escherichia coli from Farm-to-Fork in Intensive Poultry Production in KwaZulu-Natal, South Africa
Abstract
1. Introduction
2. Results
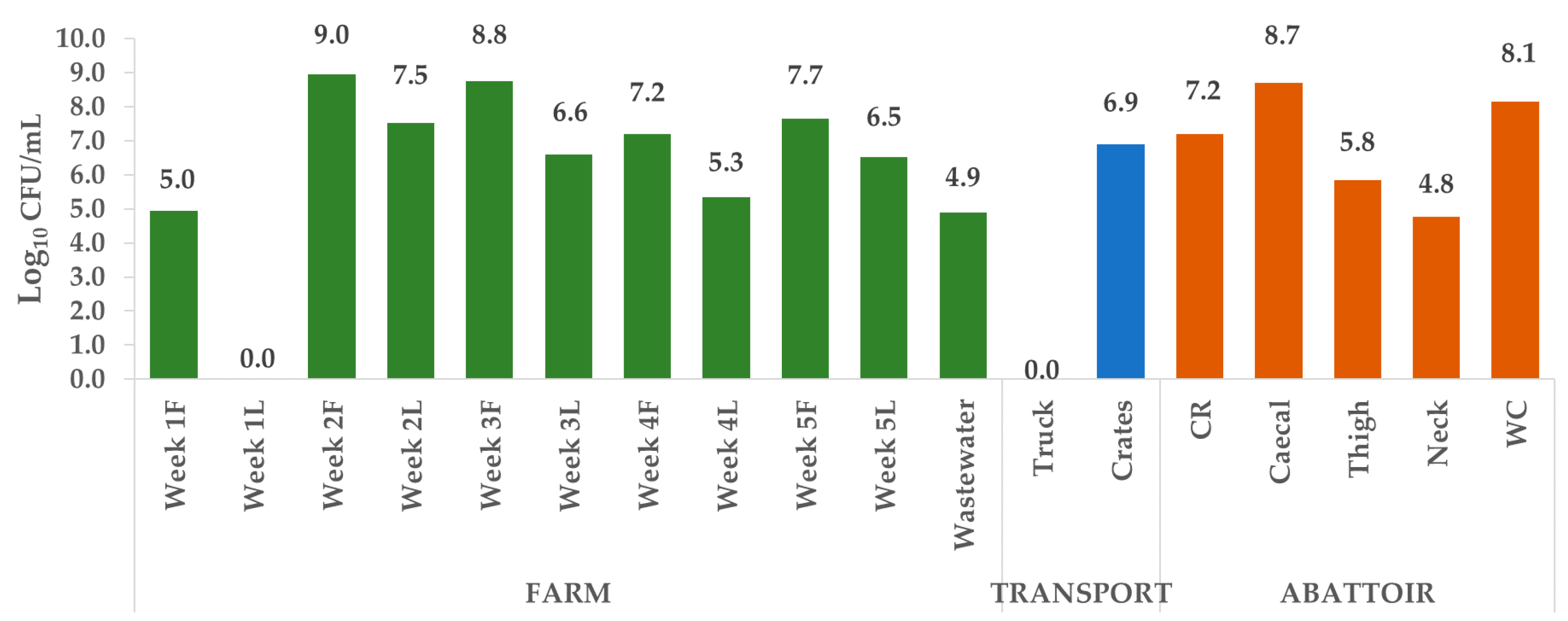
2.1. Enumeration of Escherichia coli
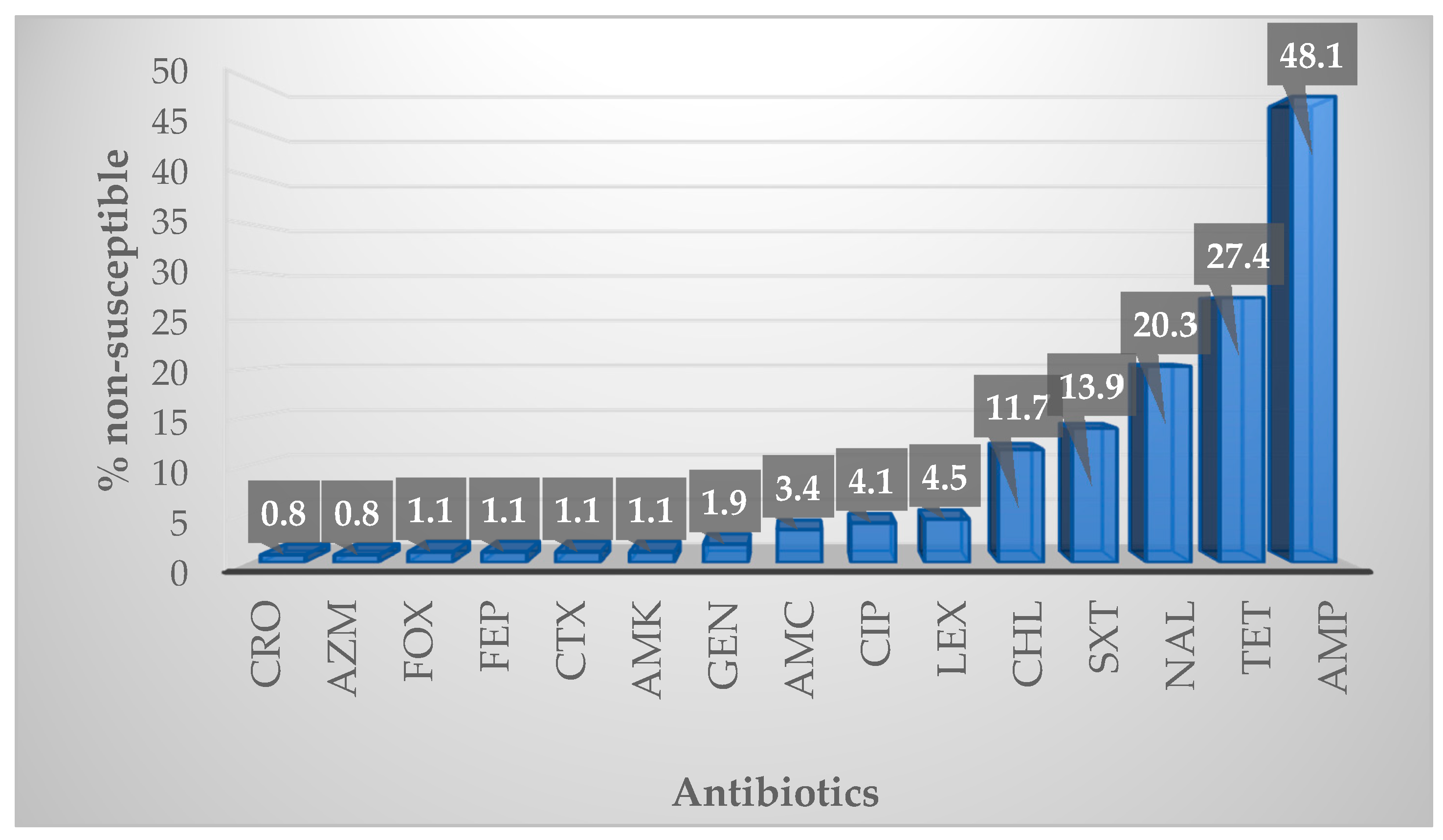
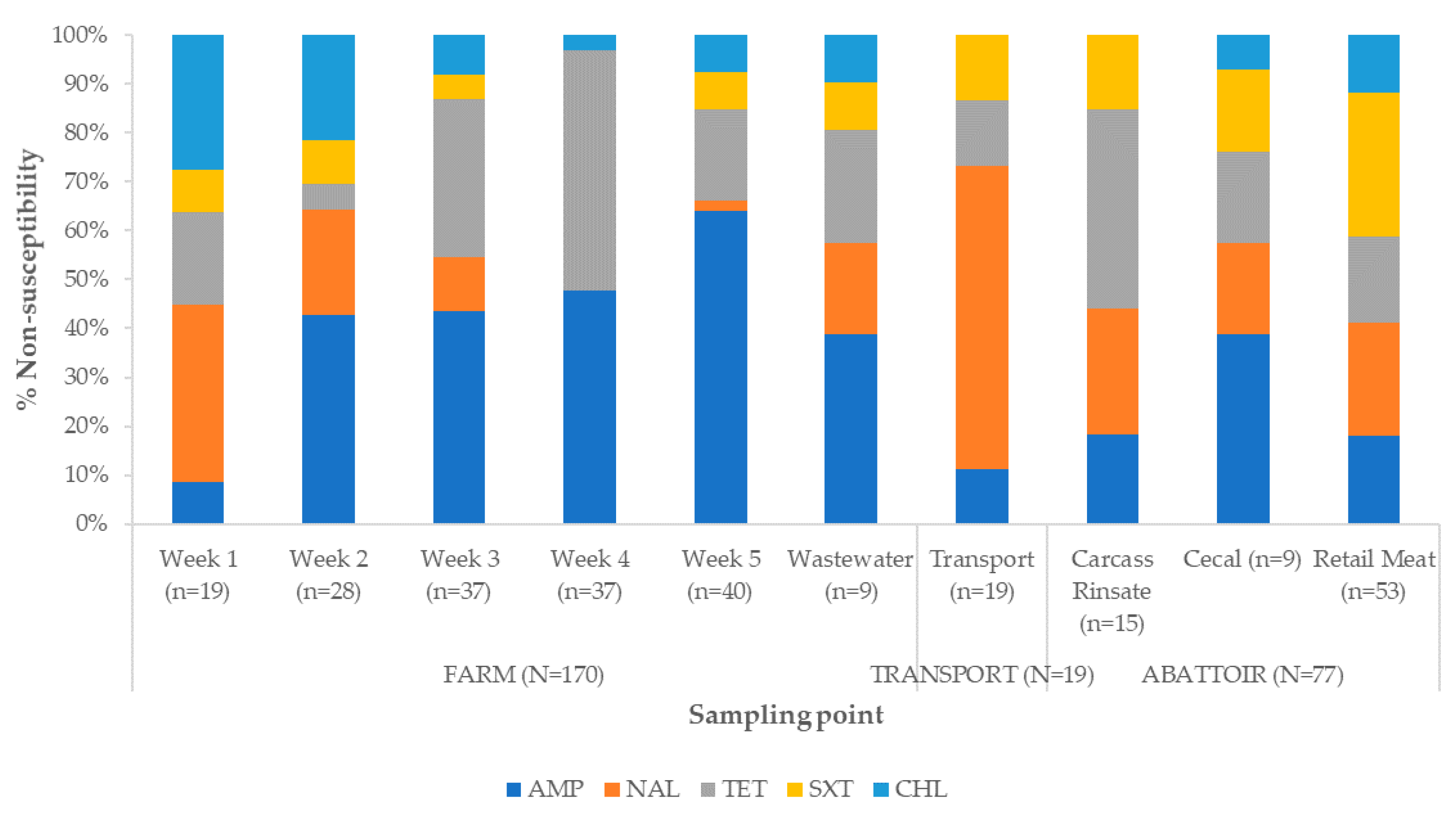
2.2. Antibiotic Susceptibility
2.2.1. Overall Susceptibility Profile
2.2.2. Antibiotic Susceptibility Profiles and Multidrug Resistance Patterns
2.2.3. Detection of Antibiotic Resistance Genes
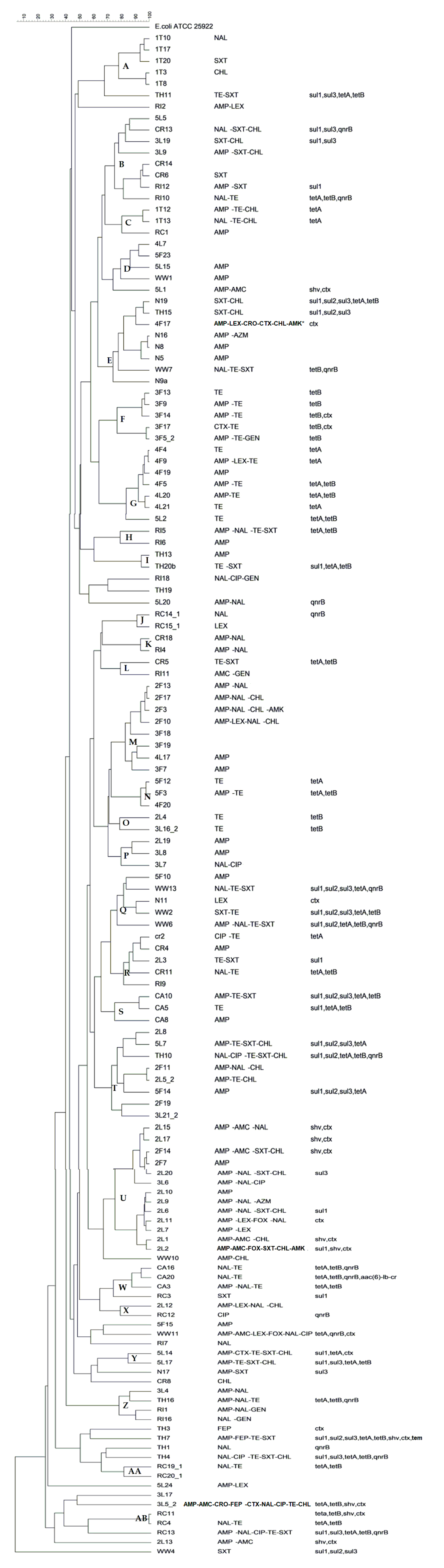
2.3. Determination of Clonality
3. Discussion
3.1. Enumeration of Escherichia coli
3.2. Antibiotic Susceptibility
3.3. Determination of Clonality
4. Materials and Methods
4.1. Ethical Considerations
4.2. Study Population, Sampling, and Sample Processing
4.3. Bacterial Isolation, Purification, and Identification
4.4. DNA Extraction and Molecular Confirmation of Isolates
4.5. Antibiotics Susceptibility Testing (AST)
4.6. Antibiotic Resistance Genes Detection
4.7. Determination of Clonality
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Appendix A
| Sample | Putative E. coli | Confirmed E. coli |
|---|---|---|
| Week 1 | 41 | 19 |
| Week 2 | 40 | 28 |
| Week 3 | 40 | 37 |
| Week 4 | 39 | 37 |
| Week 5 | 50 | 40 |
| Truck | 0 | 0 |
| Crate | 20 | 19 |
| Carcass rinsate | 20 | 15 |
| Cecal | 20 | 9 |
| Retail meat | 62 | 53 |
| Wastewater | 13 | 9 |
| Total | 345 | 266 |
| Target | Gene | Primers Sequence 5′–3′ | Control Strain 1 | Reference |
|---|---|---|---|---|
| E. coli | uidA-F uidA-R | AAAACGGCAAGAAAAAGCAG ACGCGTGGTTAACAGTCTTGCG | E. coli ATCC 25922 | [52] |
| Resistance genes | tetA-F tetA-R | GTAATTCTGAGCACTGTCGC CTGCCTGGACAACATTGCTT | Klebsiella pneumonia strain GCKP12 | [53] |
| tetB-F tetB-R | CTCAGTATTCCAAGCCTTTG ACTCCCCTGAGCTTGAGGGG | E. coli strain PN091E1Il | [53] | |
| qnrB-F qnrB-R | GGAATCGAAATTGGCCACTG TTTGCCGTTCGCCAGTCGAA | K. pneumonia strain KP224 | [31] | |
| qnrS-F qnrS-R | CACTTTGATGTCGCAGAT CAACATACCCAGTGCTT | K. pneumonia strain KP230 | [31] | |
| aac(6)-lb-cr-F aac(6)-lb-cr-R | GATGCTCTATGGGTGGCTAA GGTCCGTTTGGATCTTGGTGA | K. pneumonia strain GCKP12 | [31] | |
| sul1-F sul1-R | CTTCGATGAGAGCCGGCGGC GCAAGGCGGAAACCGCGCC | C. freundii | [54] | |
| sul2-F sul2-R | TCGTCAACATAACCTCGGACAC GTTGCGTTTGATACCGGCAC | C. freundii | [54] | |
| sul3-F sul3-R | GAGCAAGATTTTTGGAATCG CATCTGCAGCTAACCTAGGGCTTTGGA | C. freundii | [54] | |
| SHV-F SHV-R | TTAACTCCCTGTTAGCCA GATTTGCTGATTTCGCCC | K. pneumoniae (950117510) | [55] | |
| CTXM-F CTXM-R | GGTTAAAAAATCACTGCGTC TTGGTGACGATTTTAGCCGC | K. pneumoniae (945169659) | [55] | |
| TEM-F TEM-R | AAAATTCTTGAAGACG TTACCAATGCTTAATCA | K. pneumoniae (945169659) | [55] | |
| Clonality | ERIC 1 ERIC 2 | ATGTAAGCTCCTGGGGATTCAC AAGTAAGTGACTGGGGTGAGCG | E. coli ATCC 25922 | [56] |
References
- Graham, J.P.; Boland, J.J.; Silbergeld, E. Growth Promoting Antibiotics in Food Animal Production: An Economic Analysis. Public Health Rep. 2007, 122, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Mehdi, Y.; Létourneau-Montminy, M.P.; Gaucher, M.L.; Chorfi, Y.; Gayatri, S.; Rouissi, T.; Brar, S.K.; Côté, C.; Ramirez, A.A.; Godbout, S. Use of antibiotics in broiler production: Global impacts and alternatives. Anim. Nutr. 2018, 4, 170–178. [Google Scholar] [CrossRef] [PubMed]
- Filho, H.C.K.; Brito, K.C.T.; Cavalli, L.S.; Brito, B.G. Avian Pathogenic Escherichia coli (APEC) - an update on the control. In The Battle Against Microbial Pathogens: Basic Science, Technological Advances and Educational Programs, 5th ed.; Méndez-Vilas, A., Ed.; Formatex Research Center: Badajoz, Spain, 2015; pp. 598–618. [Google Scholar]
- Singer, R. Therapeutics and antibiotic resistance. In 19th World Veterinary Poultry Association Congress in South Africa; World Veterinary Association: Bruxelles, Belgium, 2015; pp. 1–7. [Google Scholar]
- Blaak, H.; Van Hoek, A.H.A.M.; Hamidjaja, R.A.; Van Der Plaats, R.Q.J.; Kerkhof-De Heer, L.; De Roda Husman, A.M.; Schets, F.M. Distribution, numbers, and diversity of ESBL-producing E. coli in the poultry farm environment. PLoS ONE 2015, 10, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Von Eugen, K.; Nordquist, R.E.; Zeinstra, E.; van der Staay, F.J. Stocking density affects stress and anxious behavior in the laying hen chick during rearing. Animals 2019, 9, 53. [Google Scholar] [CrossRef] [PubMed]
- Gomes, A.V.S.; Quinteiro-Filho, W.M.; Ribeiro, A.; Ferraz-de-Paula, V.; Pinheiro, M.L.; Baskeville, E.; Akamine, A.T.; Astolfi-Ferreira, C.S.; Ferreira, A.J.P.; Palermo-Neto, J. Overcrowding stress decreases macrophage activity and increases Salmonella Enteritidis invasion in broiler chickens. Avian Pathol. 2014, 43, 82–90. [Google Scholar] [CrossRef]
- Souillard, R.; Répérant, J.M.; Experton, C.; Huneau-Salaun, A.; Coton, J.; Balaine, L.; Le Bouquin, S. Husbandry practices, health, and welfare status of organic broilers in France. Animals 2019, 9, 97. [Google Scholar] [CrossRef] [PubMed]
- Founou, L.L.; Founou, R.C.; Essack, S.Y. Antibiotic resistance in the food chain: A developing country-perspective. Front. Microbiol. 2016, 7, 1–19. [Google Scholar] [CrossRef]
- Chang, Q.; Wang, W.; Regev-Yochay, G.; Lipsitch, M.; Hanage, W.P. Antibiotics in agriculture and the risk to human health: How worried should we be? Evol. Appl. 2015, 8, 240–247. [Google Scholar] [CrossRef] [PubMed]
- Antimicrobial Resistance: Tackling a Crisis for the Health and Wealth of Nations. Available online: https://amr-review.org/sites/default/files/AMR%20Review%20Paper%20-%20Tackling%20a%20crisis%20for%20the%20health%20and%20wealth%20of%20nations_1.pdf (accessed on 6 November 2020).
- Olonitola, O.S.; Fahrenfeld, N.; Pruden, A. Antibiotic resistance profiles among mesophilic aerobic bacteria in Nigerian chicken litter and associated antibiotic resistance genes. Poult. Sci. 2015, 94, 867–874. [Google Scholar] [CrossRef]
- Han, X.; Guan, X.; Zeng, H.; Li, J.; Huang, X.; Wen, Y.; Zhao, Q.; Huang, X.; Yan, Q.; Huang, Y.; et al. Prevalence, antimicrobial resistance profiles and virulence-associated genes of thermophilic Campylobacter spp. isolated from ducks in a Chinese slaughterhouse. Food Control. 2019, 104, 157–166. [Google Scholar] [CrossRef]
- Cyoia, P.S.; Koga, V.L.; Nishio, E.K.; Houle, S.; Dozois, C.M.; De Brito, K.C.T.; De Brito, B.G.; Nakazato, G.; Kobayashi, R.K.T. Distribution of ExPEC virulence factors, blaCTX-M, fosA3, and mcr-1 in escherichia coli isolated from commercialized chicken carcasses. Front. Microbiol. 2019, 10, 1–9. [Google Scholar] [CrossRef]
- Whole-Genome Sequencing for Surveillance of Antimicrobial Resistance. Available online: https://www.who.int/publications/i/item/9789240011007 (accessed on 6 November 2020).
- Mellata, M. Human and Avian Extraintestinal Pathogenic Escherichia coli: Infections, Zoonotic Risks, and Antibiotic Resistance Trends. Foodborne Pathog. Dis. 2013, 10, 916–932. [Google Scholar] [CrossRef]
- Aarestrup, F.M. Monitoring of antimicrobial resistance in animals: Principles and Limitations. J. Vet. Med. 2004, 51, 380–388. [Google Scholar] [CrossRef]
- Yatsuyanagi, J.; Saito, S.; Sato, H.; Miyajima, Y.; Amano, K.I.; Enomoto, K. Characterization of enteropathogenic and enteroaggregative Escherichia coli isolated from diarrheal outbreaks. J. Clin. Microbiol. 2002, 40, 294–297. [Google Scholar] [CrossRef]
- Theobald, S.; Etter, E.M.C.; Gerber, D.; Abonlik, C. Antibiotic Resistance Trends in Escherichia coli in South African Poultry:2009–2015. Foodborne Pathog. Dis. 2019, 16, 652–660. [Google Scholar] [CrossRef] [PubMed]
- Ewers, C.; Antão, E.M.; Diehl, I.; Philipp, H.C.; Wieler, L.H. Intestine and environment of the chicken as reservoirs for extraintestinal pathogenic Escherichia coli strains with zoonotic potential. Appl. Environ. Microbiol. 2009, 75, 184–192. [Google Scholar] [CrossRef]
- Nauta, M.; Van Der Fels-Klerx, I.; Havelaar, A. A poultry-processing model for quantitative microbiological risk assessment. Risk Anal. 2005, 25, 85–98. [Google Scholar] [CrossRef] [PubMed]
- Kelly, L.A.; Hartnett, E.; Gettinby, G.; Fazil, A.; Snary, E.; Wooldridge, M. Microbiological safety of poultry meat: Risk assessment as a way forward. Worlds. Poult. Sci. J. 2003, 59, 495–508. [Google Scholar] [CrossRef]
- Oguttu, J.W.; Veary, C.M.; Picard, J.A. Antimicrobial drug resistance of Escherichia coli isolated from poultry abattoir workers at risk and broilers on antimicrobials. J. S. Afr. Vet. Assoc. 2008, 79, 161–166. [Google Scholar] [CrossRef][Green Version]
- Johnson, T.J.; Logue, C.M.; Johnson, J.R.; Kuskowski, M.A.; Sherwood, J.S.; Barnes, H.J.; DebRoy, C.; Wannemuehler, Y.M.; Obata-Yasuoka, M.; Spanjaard, L.; et al. Associations Between Multidrug Resistance, Plasmid Content, and Virulence Potential Among Extraintestinal Pathogenic and Commensal Escherichia coli from Humans and Poultry. Foodborne Pathog. Dis. 2012, 9, 37–46. [Google Scholar] [CrossRef]
- Thibodeau, A.; Quessy, S.; Guévremont, E.; Houde, A.; Topp, E.; Diarra, M.S.; Letellier, A. Antibiotic resistance in Escherichia coli and Enterococcus spp. isolates from commercial broiler chickens receiving growth-promoting doses of bacitracin or virginiamycin. Can. J. Vet. Res. 2008, 72, 129–136. [Google Scholar]
- Winkler, M.L.; Papp-Wallace, K.M.; Hujer, A.M.; Domitrovic, T.N.; Hujer, K.M.; Hurless, K.N.; Tuohy, M.; Hall, G.; Bonomo, R.A. Unexpected challenges in treating multidrug-resistant gram-negative bacteria: Resistance to ceftazidime-avibactam in archived isolates of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2015, 59, 1020–1029. [Google Scholar] [CrossRef]
- Rychen, G.; Aquilina, G.; Azimonti, G.; Bampidis, V.; de Lourdes Bastos, M.; Bories, G.; Chesson, A.; Cocconcelli, P.S.; Flachowsky, G.; Kolar, B.; et al. Safety and efficacy of Sacox® microGranulate (salinomycin sodium) for chickens for fattening and chickens reared for laying. EFSA J. 2017, 15, 1–39. [Google Scholar]
- Diarra, M.S.; Silversides, F.G.; Diarrassouba, F.; Pritchard, J.; Masson, L.; Brousseau, R.; Bonnet, C.; Delaquis, P.; Bach, S.; Skura, B.J.; et al. Impact of Feed Supplementation with Antimicrobial Agents on Growth Performance of Broiler Chickens, Clostridium perfringens and Enterococcus Counts, and Antibiotic Resistance Phenotypes and Distribution of Antimicrobial Resistance Determinants in Escherichia coli Isolates. Appl. Environ. Microbiol. 2007, 73, 6566–6576. [Google Scholar]
- Adelowo, O.O.; Fagade, O.E.; Agersø, Y. Antibiotic resistance and resistance genes in Escherichia coli from poultry farms, southwest Nigeria. J. Infect. Dev. Ctries. 2014, 8, 1103–1112. [Google Scholar] [CrossRef] [PubMed]
- Awad, A.; Arafat, N.; Elhadidy, M. Genetic elements associated with antimicrobial resistance among avian pathogenic Escherichia coli. Ann. Clin. Microbiol. Antimicrob. 2016, 15, 59. [Google Scholar] [CrossRef]
- Li, L.; Wang, B.; Feng, S.; Li, J.; Wu, C.; Wang, Y.; Ruan, X.; Zeng, M. Prevalence and characteristics of extended-spectrum β-lactamase and plasmid-mediated fluoroquinolone resistance genes in Escherichia coli isolated from chickens in Anhui Province, China. PLoS ONE 2014, 9, 1–6. [Google Scholar] [CrossRef]
- Grossman, T.H. Tetracycline antibiotics and resistance. Cold Spring Harb. Perspect. Med. 2016, 6, a025387. [Google Scholar] [CrossRef]
- Dessie, H.K.; Bae, D.H.; Lee, Y.J. Characterization of integrons and their cassettes in Escherichia coli and Salmonella isolates from poultry in Korea. Poult. Sci. 2013, 92, 3036–3043. [Google Scholar] [CrossRef]
- Redgrave, L.S.; Sutton, S.B.; Webber, M.A.; Piddock, L.J.V. Fluoroquinolone resistance: Mechanisms, impact on bacteria, and role in evolutionary success. Trends Microbiol. 2014, 22, 438–445. [Google Scholar] [CrossRef]
- Hooper, D.C.; Jacoby, G.A. Mechanisms of drug resistance: Quinolone resistance. Ann. N. Y. Acad. Sci. 2015, 1354, 12–31. [Google Scholar] [CrossRef]
- Eibach, D.; Dekker, D.; Boahen, K.G.; Akenten, C.W.; Sarpong, N.; Campos, C.B.; Berneking, L.; Aepfelbacher, M.; Krumkamp, R.; Owusu-Dabo, E.; et al. Extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae in local and imported poultry meat in Ghana. Vet. Microbiol. 2018, 217, 7–12. [Google Scholar] [CrossRef]
- Niero, G.; Bortolaia, V.; Vanni, M.; Intorre, L.; Guardabassi, L.; Piccirillo, A. High diversity of genes and plasmids encoding resistance to third-generation cephalosporins and quinolones in clinical Escherichia coli from commercial poultry flocks in Italy. Vet. Microbiol. 2018, 216, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Geornaras, I.; Hastings, J.W.; Von Holy, A. Genotypic Analysis of Escherichia coli Strains from Poultry Carcasses and Their Susceptibilities to Antimicrobial Agents. Appl. Environ. Microbiol. 2001, 67, 1940–1944. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fair, R.J.; Tor, Y. Perspectives in Medicinal Chemistry Antibiotics and Bacterial Resistance in the 21st Century. Perspect. Medicin. Chem. 2014, 6, 25–64. [Google Scholar] [CrossRef]
- Stokes, H.W.; Gillings, M.R. Gene flow, mobile genetic elements and the recruitment of antibiotic resistance genes into Gram-negative pathogens. FEMS Microbiol. Rev. 2011, 35, 790–819. [Google Scholar] [CrossRef]
- Saleha, A.; Myaing, T.T.; Ganapathy, K.; Zulkifli, I.; Raha, R.; Arifah, K. Possible Effect of Antibiotic-Supplemented Feed and Environment on the Occurrence of Multiple Antibiotic Resistant Escherichia coli in Chickens. Int. J. Poult. Sci. 2009, 8, 28–31. [Google Scholar] [CrossRef]
- Baron, S.; Jouy, E.; Larvor, E.; Eono, F.; Bougeard, S.; Kempf, I. Impact of third-generation-cephalosporin administration in hatcheries on fecal Escherichia coli antimicrobial resistance in broilers and layers. Antimicrob. Agents Chemother. 2014, 58, 5428–5434. [Google Scholar] [CrossRef]
- Molechan, C.; Amoako, D.G.; Abia, A.L.K.; Somboro, A.M.; Bester, L.A.; Essack, S.Y. Molecular epidemiology of antibiotic-resistant Enterococcus spp. from the farm-to-fork continuum in intensive poultry production in KwaZulu-Natal, South Africa. Sci. Total Environ. 2019, 692, 868–878. [Google Scholar] [CrossRef]
- Pillay, S.; Amoako, D.G.; Abia, A.L.K.; Somboro, A.M.; Shobo, C.O.; Perrett, K.; Bester, L.A.; Essack, S.Y. Characterisation of Campylobacter spp. isolated from poultry in Kwazulu-Natal, South Africa. Antibiotics 2020, 9, 42. [Google Scholar] [CrossRef]
- Langerhuus, S.N.; Ingvartsen, K.L.; Bennedsgaard, T.W.; Røntved, C.M. Gram-typing of mastitis bacteria in milk samples using flow cytometry. J. Dairy Sci. 2013, 96, 267–277. [Google Scholar] [CrossRef]
- Moffat, J.; Chalmers, G.; Reid-Smith, R.; Mulvey, M.R.; Agunos, A.; Calvert, J.; Cormier, A.; Ricker, N.; Weese, J.S.; Boerlin, P. Resistance to extended-spectrum cephalosporins in Escherichia coli and other Enterobacterales from Canadian turkeys. PLoS ONE 2020, 15, e0236442. [Google Scholar] [CrossRef]
- Soepranianondo, K.; Wardhana, D.K.; Budiarto; Diyantoro. Analysis of bacterial contamination and antibiotic residue of beef meat from city slaughterhouses in East Java Province, Indonesia. Vet. World 2019, 12, 243–248. [Google Scholar] [CrossRef]
- Ebomah, K.E.; Adefisoye, M.A.; Okoh, A.I. Pathogenic Escherichia coli Strains Recovered from Selected Aquatic Resources in the Eastern Cape, South Africa, and Its Significance to Public Health. Int. J. Environ. Res. Public Health 2018, 15, 1506. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 27th ed.; Approved Standard M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2017. [Google Scholar]
- EUCAST. Breakpoint Tables for Interpretation of MICs and Zone Diameters. Available online: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_7.1_Breakpoint_Tables.pdf (accessed on 6 November 2020).
- WHO. Integrated Surveillance of Antimicrobial Resistance in Foodborne Bacteria. Available online: https://apps.who.int/iris/bitstream/handle/10665/255747/9789241512411-eng.pdf;jsessionid=9047EE3FB9B6F665B7107EB74BD3E917?sequence=1 (accessed on 6 November 2020).
- Dungeni, M.; van der Merwe, R.; Momba, M.N.B. Abundance of pathogenic bacteria and viral indicators in chlorinated effluents produced by four wastewater treatment plants in the Gauteng Province, South Africa. Water SA 2010, 36, 607–614. [Google Scholar] [CrossRef]
- Sengeløv, G.; Halling-Sørensen, B.; Aarestrup, F.M. Susceptibility of Escherichia coli and Enterococcus faecium isolated from pigs and broiler chickens to tetracycline degradation products and distribution of tetracycline resistance determinants in E. coli from food animals. Vet. Microbiol. 2003, 95, 91–101. [Google Scholar] [CrossRef]
- Byrne-Bailey, K.G.; Gaze, W.H.; Kay, P.; Boxall, A.B.A.; Hawkey, P.M.; Wellington, E.M.H. Prevalence of sulfonamide resistance genes in bacterial isolates from manured agricultural soils and pig slurry in the United Kingdom. Antimicrob. Agents Chemother. 2009, 53, 696–702. [Google Scholar] [CrossRef]
- Chirindze, L.M.; Zimba, T.F.; Sekyere, J.O.; Govinden, U.; Chenia, H.Y.; Sundsfjord, A.; Essack, S.Y.; Simonsen, G.S. Faecal colonization of E. coli and Klebsiella spp. producing extended-spectrum beta-lactamases and plasmid-mediated AmpC in Mozambican university students. BMC Infect. Dis. 2018, 18, 1–8. [Google Scholar] [CrossRef]
- Versalovic, J.; Koeuth, T.; Lupski, J.R. Distribution of repetitive DNA sequences in eubacteria and application to finerpriting of bacterial genomes. Nucleic Acids Res. 1991, 19, 6823–6831. [Google Scholar] [CrossRef]
| Antibiotic Susceptibility Profiles | Farm (n = 170) | Transport (n = 19) | Abattoir (n = 77) | Total (n = 266) |
|---|---|---|---|---|
| Susceptible | 50 | 9 | 28 | 87 |
| AMP | 47 | 1 | 8 | 56 |
| AZM | 1 | 0 | 1 | 2 |
| CHL | 1 | 1 | 0 | 2 |
| CIP | 0 | 0 | 1 | 1 |
| GEN | 0 | 0 | 2 | 2 |
| LEX | 2 | 0 | 2 | 4 |
| NAL | 2 | 0 | 7 | 9 |
| SXT | 2 | 1 | 2 | 5 |
| TET | 33 | 4 | 1 | 38 |
| AMP-AMK | 1 | 0 | 0 | 1 |
| AMP-CHL | 3 | 0 | 0 | 3 |
| AMP-LEX | 2 | 0 | 1 | 3 |
| AMP-NAL | 2 | 1 | 0 | 3 |
| AMP-SXT | 0 | 0 | 1 | 1 |
| AMP-TET | 1 | 0 | 0 | 1 |
| LEX-TET | 1 | 0 | 0 | 1 |
| NAL-TET | 0 | 1 | 9 | 10 |
| SXT-CHL | 6 | 1 | 2 | 9 |
| TET-CHL | 1 | 0 | 0 | 1 |
| TET-GEN | 1 | 0 | 0 | 1 |
| TET-SXT | 1 | 0 | 4 | 5 |
| AMP-LEX-CHL | 1 | 0 | 0 | 1 |
| AMP-NAL-GEN | 0 | 0 | 1 | 1 |
| AMP-NAL-TET | 0 | 0 | 2 | 2 |
| AMP-TET-SXT | 1 | 0 | 2 | 3 |
| NAL-TET-SXT | 3 | 0 | 2 | 5 |
| AMP-TET-SXT-CHL | 2 | 0 | 0 | 2 |
| NAL-CIP-TET-SXT | 0 | 0 | 1 | 1 |
| AMP-CTX-TET-SXT-CHL | 1 | 0 | 0 | 1 |
| AMP-LEX-CRO-CTX-CHL | 1 | 0 | 0 | 1 |
| AMP-AMC-FOX-SXT-CHL-AMK | 1 | 0 | 0 | 1 |
| AMP-AMC-LEX-FOX-NAL-CIP | 1 | 0 | 0 | 1 |
| AMP-LEX-CRO-CTX-NAL-TET-CHL | 1 | 0 | 0 | 1 |
| ARG | Farm | Wastewater | Transport | Carcass Rinsate | Cecal | Retail Meat | Total |
|---|---|---|---|---|---|---|---|
| tetA | 14 (58%) | 4 (80%) | 3 (100%) | 2 (67%) | 5 (100%) | 11 (100%) | 39 (77%) |
| tetB | 13 (56%) | 3 (60%) | 2 (67%) | 2 (67%) | 5 (100%) | 11 (100%) | 36 (71%) |
| qnrB | 1 (5.6%) | 4 (100%) | 1 (33%) | 1 (17%) | 2 (67%) | 7 (78%) | 16 (37%) |
| qnrS | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) |
| aac(6)-lb-cr | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 1 (33%) | 0 (0%) | 1 (2%) |
| sul1 | 7 (64%) | 4 (80%) | 1 (50%) | 1 (50%) | 2 (100%) | 9 (100%) | 24 (80%) |
| sul2 | 2 (20%) | 4 (80%) | 0 (0%) | 0 (0%) | 1 (50%) | 4 (44%) | 11 (37%) |
| sul3 | 5 (50%) | 3 (60%) | 1 (50%) | 0 (0%) | 1 (50%) | 6 (67%) | 16 (53%) |
| SHV | 8 (62%) | 0 (0%) | NT | 1 (100%) | NT | 2 (50%) | 11 (58%) |
| CTX-M | 13 (100%) | 1 (100%) | NT | 1 (100%) | NT | 4 (100%) | 19 (100%) |
| TEM | 0 (0%) | 0 (0%) | NT | 0 (0%) | NT | 1 (25%) | 1 (5%) |
| Per Sample | 61 (36%) | 23 (58%) | 8 (38%) | 8 (25%) | 17 (68%) | 56 (64%) | 174 (46%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
McIver, K.S.; Amoako, D.G.; Abia, A.L.K.; Bester, L.A.; Chenia, H.Y.; Essack, S.Y. Molecular Epidemiology of Antibiotic-Resistant Escherichia coli from Farm-to-Fork in Intensive Poultry Production in KwaZulu-Natal, South Africa. Antibiotics 2020, 9, 850. https://doi.org/10.3390/antibiotics9120850
McIver KS, Amoako DG, Abia ALK, Bester LA, Chenia HY, Essack SY. Molecular Epidemiology of Antibiotic-Resistant Escherichia coli from Farm-to-Fork in Intensive Poultry Production in KwaZulu-Natal, South Africa. Antibiotics. 2020; 9(12):850. https://doi.org/10.3390/antibiotics9120850
Chicago/Turabian StyleMcIver, Katherine S., Daniel Gyamfi Amoako, Akebe Luther King Abia, Linda A. Bester, Hafizah Y. Chenia, and Sabiha Y. Essack. 2020. "Molecular Epidemiology of Antibiotic-Resistant Escherichia coli from Farm-to-Fork in Intensive Poultry Production in KwaZulu-Natal, South Africa" Antibiotics 9, no. 12: 850. https://doi.org/10.3390/antibiotics9120850
APA StyleMcIver, K. S., Amoako, D. G., Abia, A. L. K., Bester, L. A., Chenia, H. Y., & Essack, S. Y. (2020). Molecular Epidemiology of Antibiotic-Resistant Escherichia coli from Farm-to-Fork in Intensive Poultry Production in KwaZulu-Natal, South Africa. Antibiotics, 9(12), 850. https://doi.org/10.3390/antibiotics9120850








