Abstract
Clostridium perfringens is a bacterial pathogen that causes necrotic enteritis in poultry and livestock, and is a source of food poisoning and gas gangrene in humans. As the agriculture industry eliminates the use of antibiotics in animal feed, alternatives to antibiotics will be needed. Bacteriophage endolysins are enzymes used by the virus to burst their bacterial host, releasing bacteriophage particles. This type of enzyme represents a potential replacement for antibiotics controlling C. perfringens. As animal feed is often heat-treated during production of feed pellets, thermostable enzymes would be preferred for use in feed. To create thermostable endolysins that target C. perfringens, thermophile endolysin catalytic domains were fused to cell wall binding domains from different C. perfringens prophage endolysins. Three thermostable catalytic domains were used, two from prophage endolysins from two Geobacillus strains, and a third endolysin from the deep-sea thermophilic bacteriophage Geobacillus virus E2 (GVE2). These domains harbor predicted L-alanine-amidase, glucosaminidase, and L-alanine-amidase activities, respectively and degrade the peptidoglycan of the bacterial cell wall. The cell wall binding domains were from C. perfringens prophage endolysins (Phage LYtic enzymes; Ply): PlyCP18, PlyCP10, PlyCP33, PlyCP41, and PlyCP26F. The resulting fifteen chimeric proteins were more thermostable than the native C. perfringens endolysins, and killed swine and poultry disease-associated strains of C. perfringens.
1. Introduction
Clostridium perfringens is a Gram-positive, spore forming, anaerobic bacterium, commonly present in the intestines of humans and animals. Spores of the pathogen can persist in soil [1], feces [2] or the environment [3]. The bacterium causes many severe infections of animals and humans, including food poisoning, gas gangrene, necrotic enteritis and non-foodborne gastrointestinal infections in humans [4]. C. perfringens has been estimated to be the third leading cause of foodborne illness in the U.S. [5]. C. perfringens strains have commonly been sorted into one of five types, A, B, C, D, and E, based on the types of toxins that the cells produce. Each type has been associated with gastro-intestinal disease in different animals [4].
In swine, C. perfringens most commonly causes disease in neonatal piglets [6]. C. perfringens type C can cause necrotic enteritis in piglets within the first five days of life, often resulting in high mortality rates in infected litters [7,8]. Additionally, piglets of a few days of age can suffer from diarrhea commonly caused by C. perfringens type A [9]. While this may not be fatal, the loss of weight gain can result in economic losses.
In chickens, C. perfringens type A is believed to be a causative agent of necrotic enteritis, the most common and financially devastating bacterial disease in modern broiler flocks [10]. Although the clinical illness is usually very short, mortality in an unprotected poultry flock can be devastating. Indeed, often the only sign of necrotic enteritis in a flock is a sudden increase in mortality. In addition to increased mortality, necrotic enteritis may present as birds with depression, ruffled feathers, and dark diarrhea. The disease can persist in a flock for several days, with mortality of 1% per day [11].
The use of antibiotics as animal growth promoters (AGP) has been banned in the European Union (EU), January 2006 (Regulation 1831/2003/EC), due to concerns about the practice leading to a rise in antibiotic resistance in bacteria. In 2017, the United States Food and Drug Administration (FDA) released guidelines limiting the use of antibiotics in animal feed for AGP purposes [12,13]. Without traditional antibiotics for the prevention of necrotic enteritis and other diseases caused by C. perfringens, such diseases could potentially become a far greater problem for the swine and poultry industry. Therefore, alternatives to traditional antibiotics are needed, which are effective in preventing and treating disease caused by C. perfringens, especially C. perfringens that affect swine and poultry. Additionally, should traditional antibiotics fail in a medical setting, these alternatives may be useful for treating infected humans.
Bacteriophage are viruses that infect bacteria, and produce lytic enzymes called endolysins (or lysins) during the infective process. The holin, another phage protein, facilitates endolysins crossing the inner membrane to reach the peptidoglycan of the cell wall. Endolysins are peptidoglycan hydrolases, and break open the host cells by degrading the peptidoglycan, resulting in osmolysis and the release of the replicated viral particles. Gram positive bacteria lack an outer membrane, and the peptidoglycan of the cell wall is exposed to the action of endolysins applied externally (reviewed in [14]). For these bacteria, endolysins can lyse them when exposed from the outside. Since the site of action for an endolysin is external to the pathogen, this avoids some of the intracellular drug resistance mechanisms, e.g., efflux pumps. Additionally, the phage and host have co-evolved, allowing the phage endolysin to recognize target sites in the cell wall that are difficult for the bacterium to mutate. Thus, it is believed that phage endolysins are highly refractory to resistance development [15]. This characteristic makes endolysins a good source of anti-bacterial agents against Gram-positive bacteria, like C. perfringens.
In the field of human therapeutics, two lysins, SAL200 and CF-301, are in clinical trials in the U.S. for the treatment of patients with Staphylococcus aureus bacteremia, or bloodstream infections. These trials are listed at ClinicalTrials.gov, with the identifier codes NCT03089697 for SAL200 and NCT03163446 for CF-301. These are drug add-on trials, where the patients receive standard of care treatment antibiotics with or without the lysin drug. Another lysin, named Staphefekt, has been tested in the Netherlands as a topical treatment for Staphylococcus aureus-associated atopic dermentitis [16]. No endolysin in animal feed studies were found from a search of PubMed at NCBI. However, another type of peptidoglycan hydrolase, hen egg white lysozyme, was tested in liquid feed (not feed pellets) for piglets, and shown to improve animal growth and to lower counts of pathogenic bacteria [17,18].
The production of animal feed pellets usually involves heat-treatment of the feed [19]. This treatment entails conditioning the mixture of feed ingredients with steam at temperatures up to 95 °C, with exposure times of 20 seconds to 4 min in a feed pellet mill [20]. Thus, for endolysins to be incorporated into animal feed pellet production, they should be thermostable or thermotolerant. It is known that Gram positive phage endolysins are modular proteins (usually), with N-terminal catalytic (CAT) and C-terminal cell wall binding (CWB) domains. CAT domains can often be redirected to target a different species by swapping CWB domains [21]. Previously, we have shown that the CAT domain from a thermostable endolysin of a different species could be re-targeted against C. perfringens [22]. Thermophilic bacteria produce thermostable enzymes whose CAT domains can potentially be re-targeted against C. perfringens. Thermophilic bacteria genomes were examined for prophages (bacteriophage genomes inserted into the bacterial genome). These prophage regions typically encode an endolysin gene. Two prophage endolysins from thermophilic Geobacillus bacteria, identified by a bioinformatics search, along with the previously identified phage lytic enzyme (Ply) PlyGVE2 endolysin, were used here. In this study, we report on a set of chimeric lysins created by fusing thermophile endolysin CAT domains with C. perfringens-targeting CWB domains.
2. Results
2.1. Peptidoglycan Structure and Endolysin Cleavage Sites
Bacterial peptidoglycan has a complex structure, with a sugar backbone of alternating units of N-acetyl glucosamine (GlcNac) and N-acetyl muramic acid (MurNac) residues, cross-linked by oligopeptide attachments (the peptide stem) at the MurNac, with peptide stems linked by a peptide cross-bridge. The C. perfringens peptidoglycan schematic is shown in Figure 1 [23]. Endolysins have evolved a modular design to deal with this complexity, with a C-terminal CWB domain and an N-terminal CAT domain. One protein can harbor one or two CAT domains, each with a different peptidoglycan digestion activity [14,24]. Endolysin CAT domains can be sorted into three broad categories, endopeptidase, glycosidase and amidase, that target different bonds in the peptidoglycan structure. Each CAT domain is small (70–200 amino acids). Any one of these domains is sufficient to lyse the bacterial target cell. Gram-positive cell walls have at least 10 to 20 layers of peptidoglycan, compared to one to three layers for Gram-negative cells [25].
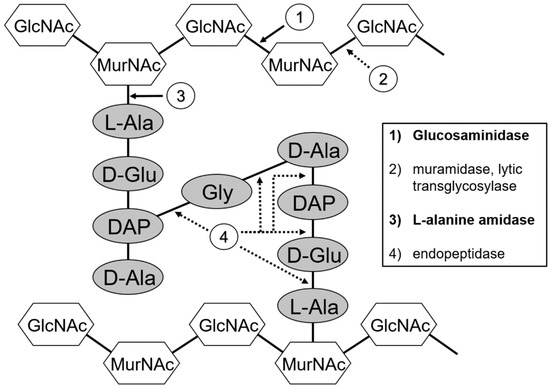
Figure 1.
Peptidoglycan structure and sites of hydrolase cleavage. (1) Glucosaminidases cleave between N-acetyl glucosamine (GlcNac) and N-acetyl muramic acid (MurNac). (2) Muramidases (lysozymes) and lytic transglycosylases cleave between MurNac and GlcNac. (3) L-alanine-amidases cleave between the MurNac and the first amino acid of the peptide stem, an alanine. (4) Endopeptidases cleave between the peptide bonds in the peptide stem, or in the cross-bridge that links the stems.
2.2. Discovery and Bioinformatic Analysis of Thermophile Endolysins
Gram-positive thermophilic bacteria genomes were examined at the Integrated Microbial Genomes (IMG) website (http://img.jgi.doe.gov/) for potential prophage endolysin sequences. Genes encoding potential endolysin proteins were identified adjacent to predicted bacteriophage genes. Two predicted endolysin genes were found in the genomes of Geobacillus strains Y412MC61 and Y4.1MC1. According to the IMG database, both strains were isolated from hot springs at the Yellowstone National Park, USA, and grown at 70 °C in the laboratory. As these thermophilic bacteria grow at elevated temperatures, it is likely that their proteins, including endolysins, are thermostable. Each endolysin was predicted by the NCBI Conserved Domain search engine [26] to have different enzymatic activities, and cleave a different bond in the peptidoglycan (Figure 1); one is an L-alanine amidase that targets the bond between the peptide stem and the glycan, and the other is a glucosaminidase that targets the glycan polymer. Both endolysins target peptidoglycan bonds that are highly conserved across bacteria species.
The first endolysin, named here as PlyGspY412 (IMG gene ID: 646372128; Genbank accession ACX77322), was found in the genome of Geobacillus sp. Y412MC61. It has an N-terminal L-alanine amidase domain, belonging to a subgroup or family defined by PFAM (http://pfam.xfam.org/) as amidase_3 (Figure 2), which cleaves the peptidoglycan bond between the MurNAc glycan residue and the alanine of the peptide stem (Figure 1). In the C-terminal half, there is a sporulation-related repeat (SPOR) domain, PFAM code PF05036, which is a peptidoglycan binding domain consisting of two 35-residue repeats, that is associated with proteins involved in sporulation, and is also found in some endolysin proteins. A BlastP search at NCBI website of the PlyGspY412 protein sequence against the Clostridium perfringens (taxid:1502) dataset yielded three hits at 34% identity (WP_115634784, WP_025647622, WP_070956814), all of which were L-alanine amidases with C-terminal SPOR domains.
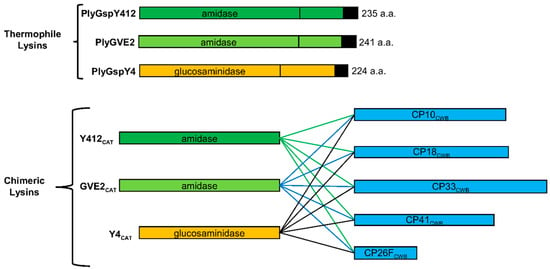
Figure 2.
Thermophile-derived chimeric recombinant lysins schematic. The thermophile endolysins, PlyGspY412, PlyGspY4, and PlyGVE2, supply the N-terminal catalytic (CAT) domains, GVE2CAT, Y412CAT and Y4CAT, respectively. The C-terminal CWB domain comes from one of several C. perfringens endolysins (PlyCP10, PlyCP18, PlyCP33, PlyCP41, PlyCP26F). The recombinant proteins have the amino acids LEHHHHHH added to the C-terminus to facilitate purification by nickel-affinity chromatography; those with CP18CWB have VEHHHHHH. GVE2CAT-CP26FCWB is the previously published lysin PlyGVE2CpCWB [22].
The other new endolysin, named here as PlyGspY4 (IMG gene ID: 649721886; Genbank accession ADP73474), was found in the genome of Geobacillus sp. Y4.1MC1. This bacteria was collected from a 88 °C water outflow from a hot spring [27]. PlyGspY4 has a predicted N-terminal glucosaminidase CAT domain (PFAM code PF01832) and a C-terminal LysM (PFAM code PF01476) peptidoglycan binding domain (Figure 2). BlastP analysis of PlyGspY4 protein sequence against the C. perfringens data set at NCBI yielded 88 hits, with the top three hits being glucosaminidase enzymes and having 39%–40% identity (WP_124044957, WP_110003240, WP_110021405).
A third endolysin comes from the Geobacillus virus E2, or GVE2, which was isolated from a deep-sea hydrothermal vent, and can infect Geobacillus sp. E263 [28]. The GVE2 phage endolysin, named here as PlyGVE2 (Genbank accession YP_001285830), was previously characterized and shown to be active at temperatures up to 80 °C [29]. PlyGVE2 provided the CAT domain for the fusion protein, PlyGVE2CpCWB, in a previous publication [22]. Similar to PlyGspY412, PlyGVE2 has an N-terminal amidase domain, and a C-terminal SPOR domain. Clustal Omega alignment of the protein sequences for PlyGVE2 and PlyGspY412 yielded a 60.7% identity between the two endolysins. BlastP analysis of PlyGVE2 against the C. perfringens data set at NCBI yielded 41 hits, with the top three hits having 37% identity, which were the same hits found for PlyGspY412.
Other predicted thermophile endolysins, initially included in our study, were removed due to absence of lytic activity against C. perfringens, or due to a failure to express soluble protein. The CAT domain from a predicted endopeptidase from Geobacillus vulcani PSS1 (Genbank accession WP_084177458) fused to CP18CWB was successfully expressed and purified, but did not have lytic activity against C. perfringens (data not shown). Additionally, constructs containing a CAT domain from a predicted L-alanine amidase endolysin from Clostridium thermocellum ATCC 27405 (Genbank accession YP_001038028) were insoluble when expressed in E. coli (data not shown).
2.3. Chimeric Lysins Description, Expression, and Purification
The thermophile endolysin gene constructs were synthesized (Genscript) with E. coli-codon bias. These synthesized genes served as templates for PCR, to amplify the sequences of the CAT domains and clone them into the pET21a plasmid for expression in BL21(DE3) E. coli. The E. coli-codon optimized sequences for the CWB domains from several C. perfringens prophage endolysins—PlyCP10, PlyCP18, PlyCP33, PlyCP41, and PlyCP26F—were then PCR amplified and cloned into the plasmids with the thermostable CAT domains (GVE2CAT, Y412CAT, and Y4CAT) sequences, to make the 15 lysin fusions (Figure 2) using standard molecular biology techniques. The Genbank accessions numbers for the C. perfringens lysins are as follows: PlyCP26F (AEA86246), PlyCP10 (AQS60701), PlyCP18 (AQS60703), PlyCP33 (AQS60704), and PlyCP41 (AQS60702). All five C. perfringens lysins target C. perfringens strain Cp39 (US patent application 20180195055; [30,31]). All recombinant plasmids were sequenced to confirm identity and cloning integrity. The peptide sequences for the chimeric lysins are available in Supplementary Figure S1.
Five different CWB domains were used to increase the likelihood of creating an active fusion protein. PlyCP26F has a SPOR domain (PFAM code PF05036) in its CWB; domain predicted by HMMScan [32]. The rest of these lysins have SH3_3-type binding domains (PFAM code PF08239) in their CWBs, with differences in their sequences and in the spacing of the domains within the CWB regions of the proteins. These CWBs potentially have a different binding affinity to C. perfringens, as the endolysins they came from have differences in activity against different strains of C. perfringens (US patent application 20180195055; [30,31]). Therefore, combining the three thermophile derived CAT domains with the five CWBs regions should increase the odds that one or more fusion proteins would be active, and potentially thermostable.
Proteins were expressed in BL21 (DE3) E. coli and purified by their His Tag, using nickel affinity chromatography. Purity of >95% is apparent from each of the recombinant proteins in the SDS-PAGE, (Figure 3, Figure 4 and Figure 5). PlyGspY412 and most of the chimeric proteins containing its CAT domain, Y412CAT-CP33CWB, migrated in the gel (Figure 3) one to three kilodaltons (kDa) faster than their predicted MW: Y412CAT-CP10CWB; 39.4 kDa, Y412CAT-CP18CWB; 40.5 kDa, Y412CAT-CP41CWB; 37 kDa, Y412CAT-CP26FCWB; 27.3 kDa, and PlyGspY412; 25.9 kDa. Y412CAT-CP33CWB migrated slightly slower, around 52 kDa, in the gel, versus the 49.2 kDa predicted from its sequence.
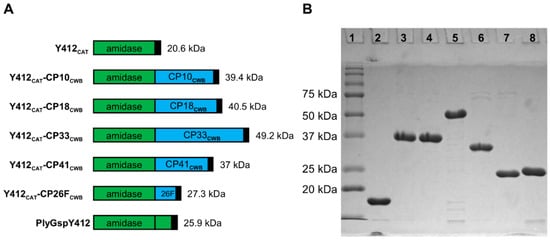
Figure 3.
PlyGspY412 derived recombinant lysins schematics and SDS-PAGE. (A) Schematics: PlyGspY412 supplied the N-terminal CAT domain (green), Y412CAT. C-terminal CWB domains (blue) were derived from one of five C. perfringens endolysins (PlyCP10, PlyCP18, PlyCP33, PlyCP41, PlyCP26F). Black box indicates the C-terminal 6 x His domain. Each recombinant protein has a C-terminal LEHHHHHH to facilitate purification by nickel-affinity chromatography; those with CP18CWB have VEHHHHHH (as described in Supplementary Data). (B) SDS-PAGE of Y412CAT fusion proteins purified by nickel chromatography. Lane 1 is Precision Plus Protein standard (Bio-Rad). The other lanes are as follows: lane 2, Y412CAT; lane 3, Y412CAT-CP10CWB; lane 4, Y412CAT-CP18CWB; lane5, Y412CAT-CP33CWB; lane 6, Y412CAT-CP41CWB; lane 7, Y412CAT-CP26FCWB; lane 8, PlyGspY412.
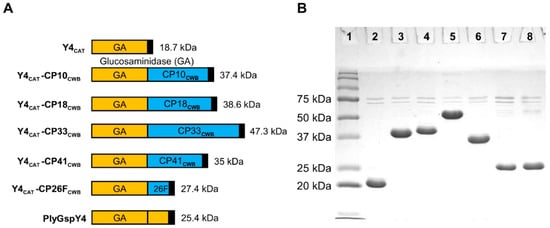
Figure 4.
PlyGspY4-derived recombinant lysins purity via SDS-PAGE. (A) Schematics: PlyGspY4 supplied the N-terminal CAT domain (orange), Y4CAT. The C-terminal CWB domain (blue) is derived from one of five C. perfringens endolysins (PlyCP10, PlyCP18, PlyCP33, PlyCP41, PlyCP26F). Black box indicates the C-terminal 6 x His domain. (B) SDS-PAGE of Y4CAT fusion proteins purified by nickel chromatography. Lane 1 is Precision Plus Protein standard (Bio-Rad). The other lanes are as follows: lane 2, Y4CAT; lane 3, Y4CAT-CP10CWB; lane 4, Y4CAT-CP18CWB; lane5, Y4CAT-CP33CWB; lane 6, Y4CAT-CP41CWB; lane 7, Y4CAT-CP26FCWB; lane 8, PlyGspY4.
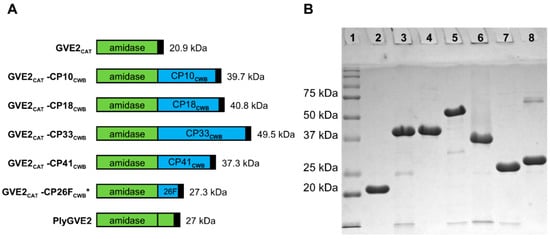
Figure 5.
PlyGVE2-derived recombinant lysins schematics and SDS-PAGE. (A) Schematics, PlyGVE2 supplied the N-terminal CAT domain (green), GVE2CAT, and the C-terminal CWB domain (blue) comes from one of several C. perfringens endolysins (PlyCP10, PlyCP18, PlyCP33, PlyCP41, PlyCP26F). Black box indicates the C-terminal 6 x His domain. GVE2CAT-CP26FCWB * is the previously published lysin PlyGVE2CpCWB. (B) SDS-PAGE of GVE2CAT fusion proteins purified by nickel chromatography. Lane 1 is Precision Plus Protein standard (Bio-Rad). The other lanes are as follows: lane 2, GVE2CAT; lane 3, GVE2CAT-CP10CWB; lane 4, GVE2CAT-CP18CWB; lane5, GVE2CAT-CP33CWB; lane 6, GVE2CAT-CP41CWB; lane 7, GVE2CAT-CP26FCWB; lane 8, PlyGVE2.
As seen in Figure 4, all Y4CAT-containing proteins, except Y4CAT-CP33CWB, migrated in the gel within one to three kDa of their predicted MW: Y4CAT; 18.7 kDa, Y4CAT-CP10CWB; 37.4 kDa, Y4CAT-CP18CWB; 38.6 kDa, Y4CAT-CP41CWB; 35 kDa, Y4CAT-CP26FCWB; 27.4 kDa, and PlyGspY4; 25.4 kDa. Y4CAT-CP33CWB ran closer to 55 kDa on the gel, larger than the sequence prediction of 47.3 kDa.
In Figure 5, all purified GVE2CAT-containing proteins, except GVE2CAT-CP33CWB, migrated in the gel within one to three kDa of their predicted MW: GVE2CAT; 20.9 kDa, GVE2CAT-CP10CWB; 39.7 kDa, GVE2CAT-CP18CWB; 40.8 kDa, GVE2CAT-CP41CWB; 37.3 kDa, GVE2CAT-CP26FCWB; 27.3 kDa, and PlyGVE2; 27 kDa. GVE2CAT-CP33CWB has a predicted size of 49.5 kDa, but ran closer to 55 kDa on the gel.
2.4. Lysis of C. perfringens by the Chimeric Lysins
Testing for lytic activity of the recombinant lysins was done by turbidity reduction assay. Lysins were mixed with mid-log phase Clostridium perfringens cells (strain Cp39), and the optical density, the turbidity, was reduced by the lysis of the cells by the recombinant proteins in phosphate buffer. The cells in buffer alone (buffer control), have decreasing turbidity overtime, likely because of lysis from the handling to put cells into buffer, exposure to buffer, and/or exposure to air, as these bacteria are anaerobic. The greater loss of turbidity that seen for the buffer control is indicative of lytic activity. Figure 6A shows the degree of lytic activity of the recombinant lysins derived from PlyGspY412, harboring the Y412CAT (the amidase CAT domain). Y412CAT-CP33CWB had the steepest curve, indicating the highest rate of turbidity reduction. Lytic activity was as follows (highest to lowest): Y412CAT-CP33CWB > Y412CAT-CP18CWB > Y412CAT-CP10CWB = Y412CAT-CP41CWB > Y412CAT-CP26FCWB Figure 6A. PlyGspY412 and Y412CAT were inactive against Cp39 cells (see Supplementary Figure S2, Table S1).
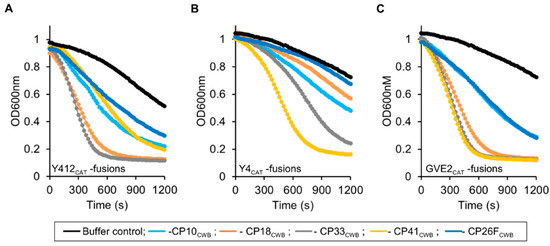
Figure 6.
Turbidity reduction assay activity of Y412CAT, Y4CAT, GVE2CAT, and their fusions. (A) Y412CAT and fusions. (B) Y4CAT and fusions. (C) GVE2CAT and fusions. Lines are as follows: buffer, black; fusion to CP10cwb, light blue; fusion to CP18cwb, orange; fusion to CP33cwb, gray; fusion to CP41cwb, yellow; fusion to CP26Fcwb dark blue. Enzymes were diluted into 50 mM NaH2PO4 pH 7.0 and mixed 1:1 with Clostridium perfringens cells in water. Turbidity was read as optical density (OD) at 600 nm shown over 20 min. The decrease in optical density (OD) represents lysis of cells in suspension. Final enzyme concentration was 0.005 mg/mL. Representative (single well) data shown.
Testing of the PlyGspY4 derived CAT domain, Y4CAT, and its fusion proteins was done by standard turbidity reduction assay. As seen in Figure 6B, lytic activity was as follows (highest to lowest): Y4CAT-CP41CWB > Y4CAT-CP33CWB > Y4CAT-CP10CWB > Y4CAT-CP18CWB. Y4CAT-CP26FCWB was inactive against Cp39 cells, as seen by its line adjacent to the buffer control line at the top of the plot in Figure 6B. Interestingly, Y4CAT-CP41CWB appears to be more active compared to the other Y4CAT-fusions, whereas the Y412CAT-CP41CWB fusion falls into the middle of the Y412CAT-fusions in terms of activity in this assay. PlyGspY4 and Y4CAT were inactive against Cp39 cells (see Supplementary Figure S2, Table S1).
The degree of lytic activity of the recombinant lysins derived from PlyGVE2, harboring the GVE2CAT (the amidase CAT domain), are shown in Figure 6C. GVE2CAT-CP41CWB (Gve2-41) and GVE2CAT-CP33CWB (Gve2-33) had the steepest curves, indicating the highest rate of turbidity reduction. Lytic activity was as follows (highest to lowest): GVE2CAT-CP41CWB = GVE2CAT-CP33CWB > GVE2CAT-CP18CWB > GVE2CAT-CP10CWB = GVE2CAT-CP26FCWB. GVE2CAT and PlyGVE2 had trace activity against Cp39 cells (see Supplementary Figure S2, Table S1).
Due to the fact that all enzymes do not yield quantitatively equivalent activity in all peptidoglycan hydrolase assays [33], it is useful to show similar relative activity between enzymes in more than one assay. Another test of lytic activity is the plate lysis assay. All chimeric lysins containing a C. perfringens-targeting CWB domain had activity and created a clear zone on the plate (Figure 7, rows A–C for columns 1–5). Fusions to the binding domains from PlyCP18, PlyCP33 and PlyCP41 appeared most active, (Figure 7, columns 2, 3, and 4, respectively), whereas fusions to the binding domain from PlyCP26F had low to miniscule activity in this assay, and made faint to very faint clearings (Figure 7, column 5). However, the CAT domains without a binding domain were not active and did not create clear zones (Figure 7, column 6). The GVE2CAT-CP10CWB fusion (Figure 7, row A column 1) displayed a white discoloration on the plate, which may represent protein precipitation. It is apparent from these experiments, plate lysis and turbidity reduction assays (TRA), that not all CWB domains are equivalent in their ability to redirect the thermophile lysin CAT domain. The chimeric lysins, with CWB domains from PlyCP41, PlyCP33, and PlyCP18, were more active against C. perfringens than those using the binding domains from PlyCP10 or PlyCP26F. Interestingly, Y412CAT-CP41CWB appears more active in this assay, relative to the other Y412CAT-containing lysins (Figure 7, row C), than it did in the TRA assay (Figure 6A).
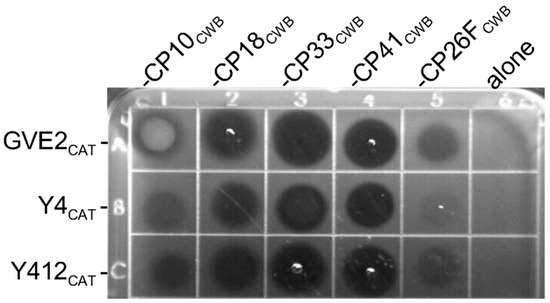
Figure 7.
Plate lysis assay of the recombinant chimeric lysins. C. perfringens Cp39 cells were embedded in semisolid BYC agar, and then 10 micrograms of each lysin was spotted on top of the solidified agar and allowed to dry. The plate was incubated at 37 °C for 2 h. Clear zones indicate lytic activity of the lysins. The CAT domains alone, not fused to a binding domain, did not display activity (final column). Row A represents the chimeric lysins based on the PlyGVE2 (GVE2CAT) CAT domain fused to the CWB from PlyCP10, PlyCP18, PlyCP33, PlyCP41, and PlyCP26F. Row B represents the chimeric lysins based on the CAT domain from PlyGspY4 (Y4CAT) fused to the same binding domains. Row C represents the chimeric lysins based on the CAT domain from PlyGspY412 (Y412CAT) fused to the same binding domains.
2.5. Thermostability of the Chimeric Lysins
Most of the thermophile fusion proteins are more tolerant of heating than PlyCP18 (Figure 8). PlyCP18 lost activity after incubation at 60 °C. All the Y412CAT fusions retained substantial activity after the 60 °C heat challenge (Figure 8A), and Y412CAT-CP26FCWB retained activity after heat treatment at 70 °C. Of the Y4CAT derived fusion proteins, only Y4CAT-CP41CWB retain substantial activity after incubation at 60 °C (Figure 8B). The GVE2CAT derived fusion proteins showed substantial residual activity after being exposed to elevated temperatures (Figure 8C). All the GVE2CAT fusions showed activity after incubation at 60 °C while GVE2CAT-CP10CWB, GVE2CAT-CP18CWB, GVE2CAT-CP41CWB, and the published fusion PlyGVE2CpCWB (GVE2CAT-CP26FCWB) showed activity after 70 °C heat treatment. The improved thermostability of the fusion proteins might make them more tolerant of heat treatments used in production of animal feed pellets. After being kept at 4 °C, all the new fusion proteins had greater activity than the published fusion protein, PlyGVE2CpCWB, which under current nomenclature is GVE2CAT-CP26FCWB. The thermophile derived lysins and their corresponding CAT domains had little to no activity against Cp39 cells (Supplementary Figure S2).
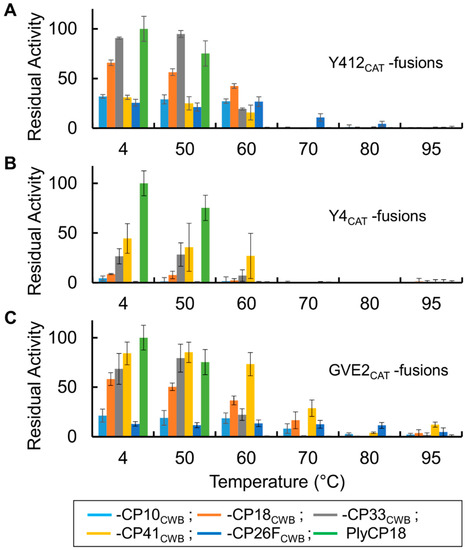
Figure 8.
Thermostability of lysin fusion proteins. (A) PlyGspY412 (Y412CAT) derived fusion proteins compared to PlyCP18. (B) PlyGspY4 (Y4CAT) derived fusion proteins. (C) PlyGVE2 (GVE2CAT) derived fusion proteins. Bars are as follows: fusion to CP10CWB, light blue; fusion to CP18CWB, orange; fusion to CP33CWB, gray; fusion to CP41CWB, yellow; fusion to CP26FCWB (PlyGVE2CpCWB), dark blue; and PlyCP18, green. Proteins were incubated 15 min at the indicated temperature (X-axis), cooled on ice, and then assayed at 40 °C for residual activity. On the Y-axis is the residual activity relative to the activity of PlyCP18 (4 °C data point).
2.6. Activity of the Chimeric Lysins against Pathogenic C. perfringens
Examination of the activity of these lysins against both chicken (Cp509 and Cp734) and swine (JGS1504 and JGS1659) pathogenic C. perfringens can be seen in Table 1. The strains Cp509 and Cp734 were isolated from chickens with necrotic enteritis, while the JGS1504 and JGS1659 strains were isolated from pigs with enteritis. The PlyGVE2 (parental thermophile lysin) and GVE2CAT (CAT domain alone) proteins had little to no activity against the five tested C. perfringens strains. The addition of the CWB domains to this CAT domain in fusion proteins drastically improved the killing of C. perfringens. Interestingly, while PlyGVE2 and GVE2CAT had some activity against non-C. perfringens bacteria, the fusion proteins with the C. perfringens endolysin CWB domains did not kill any of the other species tested, suggesting that addition of these CWB domains increased the activity against C. perfringens, and reduced non-specific activity against other bacterial species. Three of the four new fusions, all had greater killing activity against the five C. perfringens strains than the published fusion PlyGVE2CpCWB (GVE2CAT-CP26FCWB). The sole exception to this was GVE2CAT-CP10CWB, which had activity similar to PlyGVE2CpCWB, which could weakly kill this strain under these conditions (lysin at 0.005 mg/mL).

Table 1.
Activity of lysins on C. perfringens strains and other bacteria.
The CAT domain of PlyGspY4 is a glucosaminidase domain (by homology screening), and is predicted to cleave a different peptidoglycan bond than the CAT domains of PlyGVE2 and PlyGspY412, which have L-alanine amidase domain homology. The Y4CAT fusions displayed overall reduced activity, compared to the other fusion proteins. The fusions Y4CAT-CP10CWB, Y4CAT-CP18CWB, Y4CAT-CP33CWB, and Y4CAT-CP41CWB all had activity against the five C. perfringens strains tested. Y4CAT-CP26FCWB had poor lytic activity against Cp509, and was not active against the other four C. perfringens strains (lysin at 0.005 mg/mL).
A minimal inhibitory concentration (MIC) test of the more active chimeric lysins was performed by microbroth serial dilution. Interestingly, Y4CAT-CP41CWB, which was not the most active chimeric lysin by turbidity reduction assay, was the most active by the MIC assay against Cp39 bacteria (Table 2), with MIC values ranging from 1.6 to 6.3 µg/mL. Additionally, the other Y4CAT-containing lysin tested, Y4CAT-CP33CWB had MIC values ranging from 25 to 100 µg/mL, whereas the Y412CAT-containing lysins tested, Y412CAT-CP33CWB and Y412CAT-CP41CWB had 100 to >100 µg/mL MIC values in independent assays.

Table 2.
Summary of Minimal Inhibitory Concentration (MIC) of chimeric lysins against Cp39 bacteria.
3. Discussion
We were able to identify several endolysins from the genomes of thermophilic bacteria in the IMG database. Two of these, PlyGspY412 and PlyGspY4, along with the previously identified PlyGVE2 [29,34], provided the CAT domains that we fused to C. perfringens CWB domains from previously reported enzymes PlyCP10, PlyCP41, PlyCP33, PlyCP18, and PlyCP26F ([30,31] and US patent application 20180195055). All fusions did not show equal activity or thermostability. Those thermophile CAT domains fused to CP41CWB, CP33CWB, and CP18CWB were more active than the other chimeras in lysing C. perfringens (Figure 6 and Figure 7). This activity is similar to what is seen for the native C. perfringens lysins, with PlyCP10 having less activity than PlyCP41 [31], or than PlyCP33 and PlyCP18 (data not shown, U.S. patent application 20180195055).
Thermostability was assessed by incubation at elevated temperatures for 15 min, then cooling, followed by assaying residual activity at 40 °C, a temperature close to the body temperature of chickens (40–43 °C), pigs (39–40 °C), and cows (38–39 °C) [35]. Steam conditioning of animal feed ingredients at 95 °C is less than 15 min, and is reported to be between 20 s and 4 min by one vendor of feed pellet milling machinery [20]. The more stringent 15 min exposure used in this study reflects the steam conditioning plus the cooling down period afterwards. After incubation at 60 °C, PlyCP18 lost activity. In previous studies of the other native C. perfringens lysins, PlyCP10, PlyCP41, and PlyCP26F were inactivated after exposure to temperatures between 50 °C and 60 °C [22,31]. PlyCP33 was inactivated after a 60 °C incubation (data not shown). Another native CP lysin, PlyCM was reported to have retained activity after a 30 min incubation at 50 °C, but to have lost activity after incubation at 55 °C [36]. The GVE2CAT- and Y412CAT-containing lysin chimeras showed the greatest increases in thermostability over the native endolysins. Of the Y4CAT-containing chimeras, only Y4CAT-CP33CWB and Y4CAT-CP41CWB were more stable than PlyCP18. Only the GVE2CAT-fusions were able to retain some low-level activity after a 15 min incubation at the more elevated temperatures of 80 °C and 95 °C, common in feed pellet processing. In fact, GVE2CAT-CP26FCWB had fairly good stability across the board (Figure 8C), but it suffered from low activity at physiological temperatures (Figure 6 and Figure 7), whereas, GVE2CAT-CP41CWB had good activity at physiological temperatures (Figure 6 and Figure 7), but activity decreased as treatment temperatures increased (Figure 8C), though it still retained >14% of its own activity after incubation at 95 °C (Supplementary Table S1). There are other thermostable endolysins besides those presented here, and some are very thermostable, like the Ts2631 endolysin, from Thermus scotoductus phage vB_Tsc2631, which retains > 80% activity after 30 min at 95 °C, and has a thermal melt-point (Tm), representing 50% unfolded, of 99.8 °C [37]. However, this endolysin was ineffective against Clostridium sporogenes, and had relatively low activity against other mesophilic Gram-positive bacteria when compared to its activity against Thermus scotoductus. Additionally, it is less active at physiological temperatures. The non-thermophile bacteriophage T5 endolysin is also very thermostable, retaining ~65% of its activity after a 30 min incubation at 90 °C [38]. Recently, a report of a new C. perfringens lysin, LysCPS2, indicates that LysCPS2 had 30% residual activity after incubation at 95 °C for 10 min [39]. While this incubation time was 33% shorter than our work, its stability is still impressive for a native C. perfringens lysin.
It is known by the work of Kusuma et al., 2005 [33], that peptidoglycan hydrolase assays do not always agree quantitatively. We too find that relative activity of our fusions is not always maintained between turbidity reduction vs. plate lysis vs. MIC assays. Although this can be confounding when analyzing the data, it has the advantage of identifying candidate enzymes for further study that might otherwise have been overlooked. Both plate lysis and TRA did not suggest that Y4CAT-CP41CWB was going to stand out, with almost 10-fold increased relative activity in the MIC assay. In fact, both Y4CAT-CP41CWB and Y4CAT-CP33CWB chimeras showed substantially more activity than the other lysins. Because of this MIC data, the Y4CAT-CP41CWB lysin and, to a lesser extent, the Y4CAT-CP33CWB lysin could still have potential as a therapeutic drug for the treatment of animals or humans. The absence of activity in the MIC assay for the GVE2CAT-containing lysins, GVE2CAT-CP33CWB and GVE2CAT-CP41CWB, was very interesting when compared to how active they are in the TRA and the plate lysis assay. It could be that they are inhibited under the conditions of the MIC assay. Unlike PlyGVE2 and PlyGspY412, which have an L-alanine amidase CAT domain, the CAT domain of PlyGspY4 is predicted to be a glucosaminidase. Possibly, this targeting of a different bond in the peptidoglycan is why chimeras containing this domain were more effective in the MIC assay. No glucosaminidase-containing endolysins were found in a 2011 study which analyzed nine public C. perfringens genomes, and found 45 endolysin-like enzymes [36]. This suggests the Y4CAT fusions may represent a rare lytic activity against C. perfringens strains.
The goal of this study was to identify optimal fusions for further testing in animal/human model systems. Thus, relative activity levels are sufficient for the goals of this work. Our rationale for performing the peptidoglycan hydrolase assays with µg/mL enzyme concentrations, rather than molar concentrations, was two-fold. First, so we would generate results that are more readily comparable between assays and, second, to follow convention (the MIC assay results are always expressed in µg/mL). This raises the caveat that, if the size of the enzymes are highly variable, this could yield results that might be misleading (e.g., if comparing enzymes with 10× different MW: at equal µg/mL concentrations, there are 10× more enzyme molecules of the smaller construct than the 10× larger MW constructs). The validity of our approach to identify our top candidates is supported by the fact that our constructs are all similarly sized [none of the constructs have more than 2–3-fold difference in MW (see MWs in Figure 3, Figure 4 and Figure 5)], and the fact that the most active constructs in each series were the larger MW constructs (which represent the smaller number of enzyme molecules).
Unexpectedly, the native thermophile lysins and their CAT domains without CWBs were active against Bacillus cereus (Table 1). The fusion of C. perfringens CWBs to the CAT domains eliminated this activity. This phenomenon suggests that these native thermophile endolysins might be targeted against B. cereus or related species without a CWB domain swap. Random or directed point mutagenesis may prove sufficient to increase the effectiveness of these lysins against B. cereus.
The ability of these thermophile-derived CAT domains to lyse C. perfringens strains when fused to a variety CWB domains from C. perfringens endolysins suggests that they would also be active when fused to other binding domains that target different species of Gram-positive bacteria, though several binding domains may need to be screened to find effective CAT–CWB fusions.
4. Materials and Methods
4.1. Endolysin Synthetic Genes, Chimeric Lysins, and Cloning Vector
The genes for the Geobacillus thermophile endolysins, PlyGsPY412 (Genbank ACX77322; predicted N-acetylmuramoyl-L-alanine amidase enzyme activity, EC 3.5.1.28), PlyGspY4 (ADP73474; predicted endo-beta-N-acetylglucosamidase enzyme activity, EC 3.2.1.96), and PlyGVE2 (YP_001285830; predicted N-acetylmuramoyl-L-alanine amidase enzyme activity, EC 3.5.1.28), were synthesized as E. coli-codon optimized DNA sequences and cloned into NdeI and XhoI sites of the E. coli expression vector pET-21a(+) by GenScript (Piscataway, NJ, USA). This resulted in the addition of a C-terminal His Tag (LEHHHHHH) to the recombinant protein sequence. The DNA sequences for the CAT domains of these enzymes and the DNA sequences for the CWB domains from the Clostridium perfringens endolysins PlyCP10, PlyCP18, PlyCP33, PlyCP41, and PlyCP26F were PCR amplified and ligated into the pET-21a (+) vector. The Genbank accessions numbers for the C. perfringens lysins are as follows: PlyCP26F (AEA86246), PlyCP10 (AQS60701), PlyCP18 (AQS60703), PlyCP33 (AQS60704), and PlyCP41 (AQS60702). All plasmids were sequenced to confirm identity and cloning integrity. Due to the presence of an XhoI site internal to the PlyCP18 sequence, its CWB was cloned, using a SalI site, to the plasmids XhoI site, which resulted in its C-terminal His Tag peptide being VEHHHHHH. The peptide sequences for the chimeric lysins are available in Supplementary Figure S1 (and in a patent application, pending at the United States Patent and Trademark Office).
4.2. Bacteria Strains, Growth, and Protein Expression
C. perfringens strains Cp39, Cp509, Cp734, and Bacillus cereus strain Bc17 were from Bruce Seal, Poultry Microbiology Safety Research Unit, Agricultural Research Service, U.S. Department of Agriculture (USDA), Athens, GA. C. perfringens strains JGS1504 and JGS1659 were from Nancy Cornick, Veterinary Microbiology and Preventive Medicine, Iowa State University, Ames, IA. Enterococcus faecalis strain EF-17 was provided by Paul Hyman (Biology & Toxicology Department, Ashland University, Ashland, OH, USA). Clostridium difficile strain ATCC 700057 was from the American Type Culture Collection (Manassas, VA, USA). The Streptococcus agalactiae and Staphylococcus aureus strains were from Max J. Paape, USDA, Beltsville, MD.
NEB-5α E. coli were transformed for the purposes of cloning and plasmid DNA production. The pET-21a (+) variants were transformed into BL21(DE3) E. coli (Invitrogen) for the purpose of recombinant protein expression. BL21(DE3) transformants were grown in 1 liter modified Luria-Bertani (mLB) medium (15 g/L tryptone, 8 g/L yeast extract, 5 g/L NaCl) [40] with 150 µg/mL ampicillin, to exponential phase, A600 = 0.6, and expression was induced with 1 mM isopropyl-β-D-1-thiogalactopyranoside (IPTG) at 10 °C with shaking overnight. Cells were pelleted by centrifugation and lysed in Lysis Buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, 30% glycerol, pH 8.0) by sonication. The lysate was then centrifuged 9000× g for 30 min and the supernatant was then applied to 1 mL of nickel-NTA Superflow resin (Qiagen, Valencia, CA, USA) in a column. The column was washed with 20 mL of Lysis buffer and with 40 mL Wash Buffer (50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole, 30% glycerol, pH 8.0). Recombinant protein was eluted with Elution buffer (50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole, 30% glycerol, pH 8.0). The purified proteins were stored at 4 °C (one week) and at −80 °C (long term storage). Protein purity was examined by SDS-PAGE, using 15% gels and Coomassie Blue staining.
4.3. Determination of Lytic Activity of the Recombinant Proteins
The turbidity reduction assay (TRA) measures the rate of reduction in the optical density (OD) or absorbance (A) at 600 nm of a suspension of C. perfringens bacteria, due to the lytic activity of the lysin proteins. The recombinant lysins were diluted into 50 mM NaH2PO4 pH 7.0 and mixed 1:1 with mid-log phase C. perfringens cells (strain Cp39) in water. The assay was conducted in a 96-well flat-bottom polystyrene plate, at 40 °C, using a Molecular Devices (Molecular Devices, LLC, San Jose, CA, USA) plate reader. Turbidity was read every 20 seconds as optical density (OD) at 600 nm over 20 min. The decrease in OD represents lysis of cells in suspension. TRA data presented is the reduction in OD after subtracting the drop in OD by cells alone, in just buffer. Each species has a small, but measurable, reduction in OD in the TRA buffer alone, presumably due to species-specific physiology, related to osmotolerance of the TRA buffer. Final enzyme concentration in the assay was 0.005 mg/mL. Activity is defined as the maximal rate of reduction, V, in the linear portion of the graph trace. Relative activity is set to the maximal rate determined for each experiment. Data represents duplicate samples in each of the three independent experiments.
The thermostability assay measures residual activity of lysins incubated at target temperatures, prior to cooling and testing by TRA. Enzymes were incubated at 4 °C, 50 °C, 60 °C, 70 °C, 80 °C, or 95 °C for 15 min, then placed on ice for 10 min, followed by mixing the enzymes 1:1 with Cp39 cells. Residual enzyme activity was assayed by turbidity reduction in the plate reader at 40 °C, as described above.
The plate lysis assay, also known as the spot lysis assay, was performed essentially as described by Becker et al. [41]. C. perfringens cultures were grown to mid-log phase (OD600 = 0.4–0.6) in 100 mL BYC broth (37 g/L brain heart infusion, 5 g/L yeast extract, 0.5 g/L L-cysteine). Cells were pelleted by centrifugation, concentrated 50-fold in PBS 25% glycerol, and stored at −80 °C until needed. The frozen cells were thawed on ice and washed with 10 mL sterile H2O, and then twice more with lysin buffer A (50 mM NH4OAc, 10 mM CaCl2, 1 mM DTT, pH 6.2). The cells were then pelleted and suspended in 1 mL lysin buffer A. Twelve milliliters of melted 50 °C semisolid BYC agar (BYC with 7 g agar per liter, autoclaved) were added to the cells and then the mixture was poured into a sterile square petri dish. After solidifying, 10 µg of purified chimeric recombinant lysin, in a 5 µL volume, was spotted onto the plate. The plate was incubated in an anaerobic chamber for 2 h at 37 °C before scoring for clear zones. Active lysins create a clear zone in the turbidity of the embedded cells.
The Minimal Inhibitory Concentration (MIC) assays was done to test the effectiveness of the lysins to inhibit growth of the bacteria in rich medium. Clostridium perfringens strain Cp39 was grown to mid-log phase in BYCT broth (37 g/L brain heart infusion, 5 g/L yeast extract, 0.5 g/L L-cysteine, 0.5 g/L sodium thioglycolate). Thioglycolate was added to the broth to stabilize C. perfringens for the course of the assay. Chimeric lysins in PBS containing 25% glycerol (filter sterile) were two-fold serial diluted with BYCT broth (0.1 mL + 0.1 mL) across the row of a 96-well plate. Cp39 cells were diluted to ~8 × 105 cells/mL in BYCT broth and 0.1 mL cells were added to lysins. The highest lysin concentration tested was 100 micrograms per milliliter, with 0.4 mg/mL lysin diluted four-fold after the addition of cells in the first well of each row. Serial diluted PBS containing 25% glycerol served as the buffer control. The 96-well plate was incubated in an anaerobic chamber at 37 °C for 20–24 h, before being read for absorbance at 600 nm (A600) in a plate reader. The MIC value is the lowest concentration of lysin that creates a visually clear well in the dilution series. Three independent assays were used to determine the MIC values for the lysins.
5. Conclusions
Until recent bans, the practice of feeding antibiotic growth promotants to swine and poultry was the industry norm, and replacements for those antibiotics are sorely needed. These lytic enzyme fusions represent an alternative to the use of antibiotics in the treatment of C. perfringens in swine, poultry, or other animals, either as a growth promotant or as a therapeutic. Fusing the thermophile endolysin CAT domains with the C. perfringens CWB domains produced chimeric lysins that were both active against C. perfringens and more thermostable than the native C. perfringens lysins that provided the CWB domains. While all the tested chimeric lysins had activity against C. perfringens, some were more active than others (see summary in Table S1). The GVE2CAT- and Y412CAT-fusions showed improved thermostability compared to PlyCP18, and some of these, like GVE2CAT-CP41CWB, could be candidates for testing in animal feed pelleting conditions. Surprisingly, Y4CAT-CP41CWB was the best enzyme by the MIC assay, almost 10× more active than any of the other fusions tested and is thus the top candidate for use as a therapeutic agent in animal and human studies, where thermostability is less of an issue. In this study, we report the successful development of thermophile chimeric lysins, with potential uses as growth promotants in animal feed or as novel therapeutics against C. perfringens.
6. Patents
This work includes material present in a pending U.S. patent application.
Supplementary Materials
The following are available online at https://www.mdpi.com/2079-6382/8/4/214/s1, Figure S1: Protein sequences of native lysins and recombinant fusion lysins. Figure S2: Turbidity reduction assay activity of PlyGspY412, PlyGspY4, PlyGVE2, and their catalytic domains. Table S1. Summary of Data.
Author Contributions
Conceptualization, S.M.S. and D.M.D.; formal analysis, S.M.S. and K.P.R.; investigation, S.M.S. and K.P.R.; writing—original draft preparation, S.M.S.; writing—review and editing, S.M.S., D.M.D. and T.G.R.; supervision, project administration, funding acquisition, D.M.D. and T.G.R.
Funding
Support for the research was provided by the U.S. Department of Agriculture, Agricultural Research Service (ARS CRIS project #1245-31630-001-00D and ARS CRIS project # 8042-31000-103).
Acknowledgments
Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the Agriculture Research Service, United States Department of Agriculture.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Smith, L.D.; Gardner, M.V. The occurrence of vegetative cells of Clostridium perfringens in soil. J. Bacteriol. 1949, 58, 407. [Google Scholar] [PubMed]
- Matches, J.R.; Liston, J.; Curran, D. Clostridium perfringens in the environment. Appl. Microbiol. 1974, 28, 655–660. [Google Scholar] [PubMed]
- Tschirdewahn, B.; Notermans, S.; Wernars, K.; Untermann, F. The presence of enterotoxigenic Clostridium perfringens strains in faeces of various animals. Int. J. Food Microbiol. 1991, 14, 175–178. [Google Scholar] [CrossRef]
- Songer, J.G. Clostridial enteric diseases of domestic animals. Clin. Microbiol. Rev. 1996, 9, 216–234. [Google Scholar] [CrossRef] [PubMed]
- Scallan, E.; Griffin, P.M.; Angulo, F.J.; Tauxe, R.V.; Hoekstra, R.M. Foodborne illness acquired in the United States-unspecified agents. Emerg. Infect. Dis. 2011, 17, 16–22. [Google Scholar] [CrossRef]
- Songer, J.G.; Uzal, F.A. Clostridial enteric infections in pigs. J. Vet. Diagn. Investig. 2005, 17, 528–536. [Google Scholar] [CrossRef]
- Jaggi, M.; Wollschlager, N.; Abril, C.; Albini, S.; Brachelente, C.; Wyder, M.; Posthaus, H. Retrospective study on necrotizing enteritis in piglets in Switzerland. Schweiz. Arch. Tierheilkd. 2009, 151, 369–375. [Google Scholar] [CrossRef]
- Moon, H.W.; Bergeland, M.E. Clostridium perfringens type C enterotoxemia of the newborn pig. Can. Vet. J. 1965, 6, 159–161. [Google Scholar]
- Collins, J.E.; Bergeland, M.E.; Bouley, D.; Ducommun, A.L.; Francis, D.H.; Yeske, P. Diarrhea associated with Clostridium perfringens type A enterotoxin in neonatal pigs. J. Vet. Diagn. Investig. 1989, 1, 351–353. [Google Scholar] [CrossRef]
- Cooper, K.K.; Songer, J.G.; Uzal, F.A. Diagnosing clostridial enteric disease in poultry. J. Vet. Diagn. Investig. 2013, 25, 314–327. [Google Scholar] [CrossRef]
- Kaldhusdal, M.; Løvland, A. The economical impact of Clostridium perfringens is greater than anticipated. World Poult. 2000, 16, 50–51. [Google Scholar]
- FDA. Guidance for Industry #213, New Animal Drugs and New Animal Drug Combination Products Administered in or on Medicated Feed or Drinking Water of Food-Producing Animals: Recommendations for Drug Sponsors for Voluntarily Aligning Product Use Conditions with GFI #209; U.S. Food & Drug Administration: Rockville, MD, USA, 2013.
- FDA. FDA Announces Implementation of GFI #213, Outlines Continuing Efforts to Address Antimicrobial Resistance. Available online: https://www.fda.gov/AnimalVeterinary/NewsEvents/CVMUpdates/ucm535154.htm (accessed on 29 October 2018).
- Schmelcher, M.; Donovan, D.M.; Loessner, M.J. Bacteriophage endolysins as novel antimicrobials. Future Microbiol. 2012, 7, 1147–1171. [Google Scholar] [CrossRef] [PubMed]
- Fischetti, V.A. Bacteriophage endolysins: A novel anti-infective to control gram-positive pathogens. Int. J. Med. Microbiol. 2010, 300, 357–362. [Google Scholar] [CrossRef] [PubMed]
- Totte, J.; de Wit, J.; Pardo, L.; Schuren, F.; van Doorn, M.; Pasmans, S. Targeted anti-staphylococcal therapy with endolysins in atopic dermatitis and the effect on steroid use, disease severity and the microbiome: study protocol for a randomized controlled trial (MAAS trial). Trials 2017, 18, 404. [Google Scholar] [CrossRef] [PubMed]
- May, K.D.; Wells, J.E.; Maxwell, C.V.; Oliver, W.T. Granulated lysozyme as an alternative to antibiotics improves growth performance and small intestinal morphology of 10-day-old pigs. J. Anim. Sci. 2012, 90, 1118–1125. [Google Scholar] [CrossRef] [PubMed]
- Oliver, W.T.; Wells, J.E. Lysozyme as an alternative to growth promoting antibiotics in swine production. J. Anim. Sci. Biotechnol. 2015, 6, 35. [Google Scholar] [CrossRef]
- Cutlip, S.E.; Hott, J.M.; Buchanan, N.P.; Rack, A.L.; Latshaw, J.D.; Moritz, J.S. The Effect of steam-conditioning practices on pellet quality and growing broiler nutritional value. J. Appl. Poult. Res. 2008, 17, 249–261. [Google Scholar] [CrossRef]
- Buhler-AG. Pelleting. Available online: https://www.buhlergroup.com/content/buhlergroup/global/en/products/pellet_mill.html (accessed on 7 November 2019).
- Mao, J.; Schmelcher, M.; Harty, W.J.; Foster-Frey, J.; Donovan, D.M. Chimeric Ply187 endolysin kills Staphylococcus aureus more effectively than the parental enzyme. FEMS Microbiol. Lett. 2013, 342, 30–36. [Google Scholar] [CrossRef]
- Swift, S.M.; Seal, B.S.; Garrish, J.K.; Oakley, B.B.; Hiett, K.; Yeh, H.Y.; Woolsey, R.; Schegg, K.M.; Line, J.E.; Donovan, D.M. A thermophilic phage endolysin fusion to a Clostridium perfringens-specific cell wall binding domain creates an anti-clostridium antimicrobial with improved thermostability. Viruses 2015, 7, 3019–3034. [Google Scholar] [CrossRef]
- Leyh-Bouille, M.; Bonaly, R.; Ghuysen, J.M.; Tinelli, R.; Tipper, D. LL-diaminopimelic acid containing peptidoglycans in walls of Streptomyces sp. and of Clostridium perfringens (type A). Biochemistry 1970, 9, 2944–2952. [Google Scholar] [CrossRef]
- Oliveira, H.; Melo, L.D.; Santos, S.B.; Nobrega, F.L.; Ferreira, E.C.; Cerca, N.; Azeredo, J.; Kluskens, L.D. Molecular aspects and comparative genomics of bacteriophage endolysins. J. Virol. 2013, 87, 4558–4570. [Google Scholar] [CrossRef] [PubMed]
- Scheffers, D.J.; Pinho, M.G. Bacterial cell wall synthesis: new insights from localization studies. Microbiol. Mol. Biol. Rev. 2005, 69, 585–607. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Bryant, S.H. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 2004, 32, 327–331. [Google Scholar] [CrossRef] [PubMed]
- Brumm, P.; Land, M.L.; Hauser, L.J.; Jeffries, C.D.; Chang, Y.-J.; Mead, D.A. Complete genome sequences of Geobacillus sp. Y412MC52, a xylan-degrading strain isolated from obsidian hot spring in Yellowstone National Park. Stand Genom. Sci. 2015, 10, 81. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Wu, S.; Song, Q.; Zhang, X.; Xie, L. Two novel bacteriophages of thermophilic bacteria isolated from deep-sea hydrothermal fields. Curr. Microbiol. 2006, 53, 163–166. [Google Scholar] [CrossRef] [PubMed]
- Ye, T.; Zhang, X. Characterization of a lysin from deep-sea thermophilic bacteriophage GVE2. Appl. Microbiol. Biotechnol. 2008, 78, 635–641. [Google Scholar] [CrossRef]
- Simmons, M.; Donovan, D.M.; Siragusa, G.R.; Seal, B.S. Recombinant expression of two bacteriophage proteins that lyse Clostridium perfringens and share identical sequences in the C-terminal cell wall binding domain of the molecules but are dissimilar in their N-terminal active domains. J. Agric. Food Chem. 2010, 58, 10330–10337. [Google Scholar] [CrossRef]
- Swift, S.M.; Waters, J.J.; Rowley, D.T.; Oakley, B.B.; Donovan, D.M. Characterization of two glycosyl hydrolases, putative prophage endolysins, that target Clostridium perfringens. FEMS Microbiol. Lett. 2018, 365. [Google Scholar] [CrossRef]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; Lopez, R.; Finn, R.D. HMMER web server: 2018 update. Nucleic Acids Res. 2018, 46, 200–204. [Google Scholar] [CrossRef]
- Kusuma, C.; Kokai-Kun, J. Comparison of four methods for determining lysostaphin susceptibility of various strains of Staphylococcus aureus. Antimicrob. Agents Chemother. 2005, 49, 3256–3263. [Google Scholar] [CrossRef]
- Jin, M.; Ye, T.; Zhang, X. Roles of bacteriophage GVE2 endolysin in host lysis at high temperatures. Microbiology 2013, 159, 1597–1605. [Google Scholar] [CrossRef] [PubMed]
- Robertshaw, D. Temperature regulation and thermal environment. In Dukes’ Physiology of Domestic Animals; Reece, W.O., Ed.; Cornell University Press: Ithaca, NY, USA, 2004. [Google Scholar]
- Schmitz, J.E.; Ossiprandi, M.C.; Rumah, K.R.; Fischetti, V.A. Lytic enzyme discovery through multigenomic sequence analysis in Clostridium perfringens. Appl. Microbiol. Biotechnol. 2011, 89, 1783–1795. [Google Scholar] [CrossRef] [PubMed]
- Plotka, M.; Kaczorowska, A.K.; Morzywolek, A.; Makowska, J.; Kozlowski, L.P.; Thorisdottir, A.; Skirnisdottir, S.; Hjorleifsdottir, S.; Fridjonsson, O.H.; Hreggvidsson, G.O.; et al. Biochemical characterization and validation of a catalytic site of a highly thermostable Ts2631 endolysin from the thermus scotoductus phage vB_Tsc2631. PLoS ONE 2015, 10, e0137374. [Google Scholar] [CrossRef] [PubMed]
- Shavrina, M.S.; Zimin, A.A.; Molochkov, N.V.; Chernyshov, S.V.; Machulin, A.V.; Mikoulinskaia, G.V. In vitro study of the antibacterial effect of the bacteriophage T5 thermostable endolysin on Escherichia coli cells. J. Appl. Microbiol. 2016, 121, 1282–1290. [Google Scholar] [CrossRef]
- Ha, E.; Son, B.; Ryu, S. Clostridium perfringens virulent bacteriophage CPS2 and its thermostable endolysin LysCPS2. Viruses 2018, 10, 251. [Google Scholar] [CrossRef]
- Schmelcher, M.; Powell, A.M.; Becker, S.C.; Camp, M.J.; Donovan, D.M. Chimeric phage lysins act synergistically with lysostaphin to kill mastitis-causing Staphylococcus aureus in murine mammary glands. Appl. Environ. Microbiol. 2012, 78, 2297–2305. [Google Scholar] [CrossRef]
- Becker, S.C.; Dong, S.; Baker, J.R.; Foster-Frey, J.; Pritchard, D.G.; Donovan, D.M. LysK CHAP endopeptidase domain is required for lysis of live staphylococcal cells. FEMS Microbiol. Lett. 2009, 294, 52–60. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).