Comparison of Gene Expression Profiles of Uropathogenic Escherichia Coli CFT073 after Prolonged Exposure to Subinhibitory Concentrations of Different Biocides
Abstract
1. Introduction
2. Results
2.1. Subinhibitory Concentrations of Biocides
2.2. Global Gene Expression after Exposure to Biocides
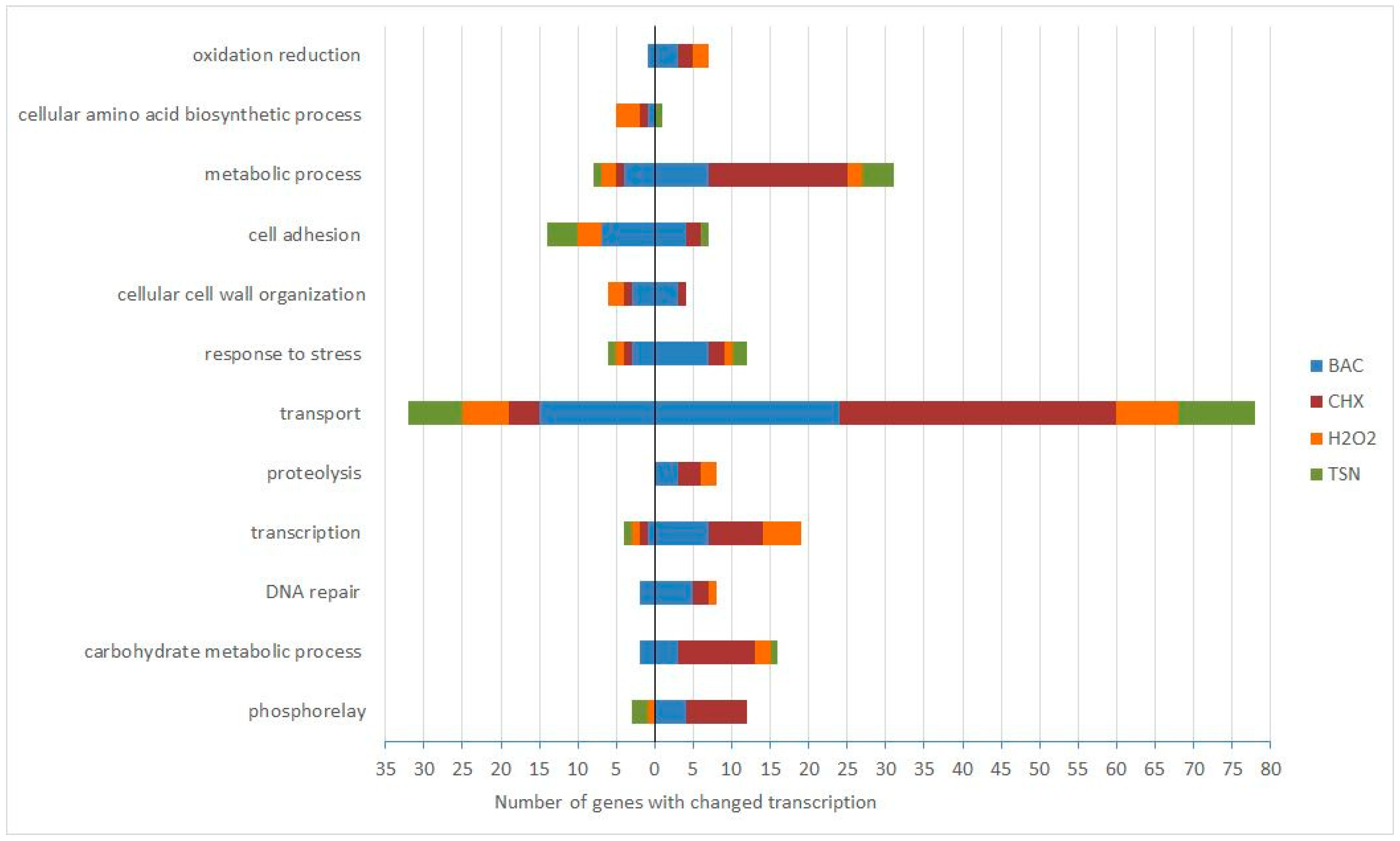
2.3. Functional Analysis of Genes Affected after Biocide Exposure
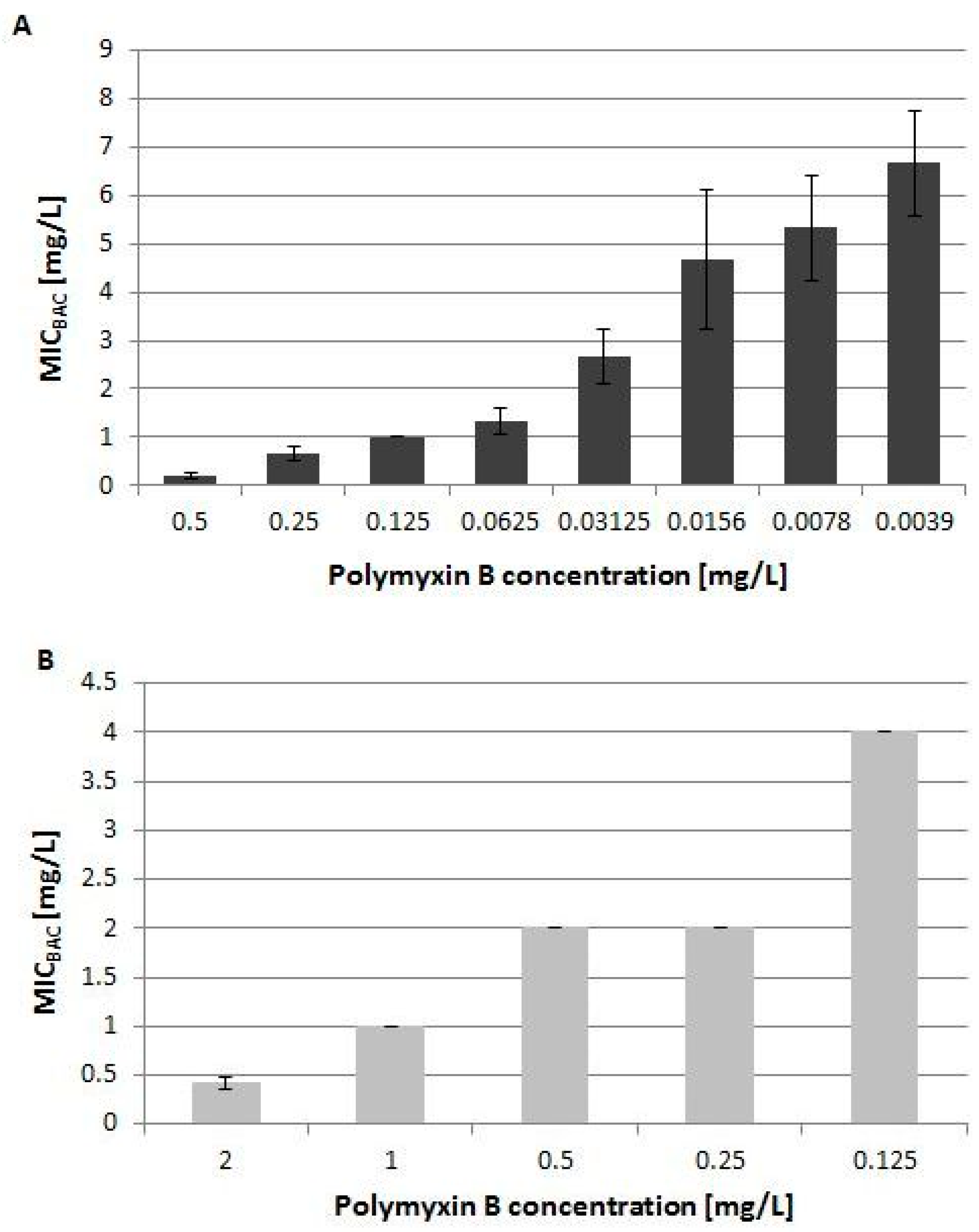
2.3.1. Polymyxin Resistance Induced by Benzalkonium Chloride on the Transcriptional Level
2.3.2. Transport Genes are the Most Affected in Response to Prolonged Biocide Treatment
2.3.3. Variable Expression of Genes Encoding Fimbriae
2.3.4. Transcription of Cryptic Phage Genes Increased in Response to Hydrogen Peroxide
2.4. Biocide Specific Response
3. Discussion
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. Bacterial Strains and Growth Conditions
4.3. Determination of MIC Values of Biocides Using the Broth Microdilution Method
4.4. Collecting RNA Samples for Microarray Analysis
4.5. Samples Preparation for Microarray Analysis
4.6. Microarray Data Analysis
4.7. Quantitative Real Time PCR
4.8. Polymyxin B and Biocide Cross-Resistance
4.9. Microarray Data Accession Number
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Maillard, J.-Y. Antimicrobial biocides in the healthcare environment: Efficacy, usage, policies, and perceived problems. Ther. Clin. Risk Manag. 2005, 1, 307–320. [Google Scholar] [PubMed]
- SCENIHR SCENIHR (Scientific Committee on Emerging and Newly Identified Health Risks). Assessment of the Antibiotic Resistance Effects of Biocides, 19 January 2009; 1–87.
- McDonnell, G.; Russell, D.A. Antiseptics and disinfectants: Activity, action, and resistance. Clin. Microbiol. Rev. 1999, 12, 147–179. [Google Scholar] [CrossRef] [PubMed]
- Maillard, J.-Y. Bacterial target sites for biocide action. J. Appl. Microbiol. Symp. Suppl. 2002, 92, 16S–27S. [Google Scholar] [CrossRef]
- McMurry, L.M.; Oethinger, M.; Levy, S.B. Triclosan targets lipid synthesis. Nature 1998, 394, 531–532. [Google Scholar] [CrossRef] [PubMed]
- Heath, R.J.; Rubin, J.R.; Holland, D.R.; Zhang, E.; Snow, M.E.; Rock, C.O. Mechanism of Triclosan Inhibition of Bacterial Fatty Acid Synthesis. J. Biol. Chem. 1999, 274, 11110–11114. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, P.; McBain, A.J. Potential Impact of Increased Use of Biocides in Consumer Products on Prevalence of Antibiotic Resistance. Clin. Microbiol. Rev. 2003, 16, 189–208. [Google Scholar] [CrossRef] [PubMed]
- Denyer, S.P. Mechanisms of Action of Biocides. Int. Biodeterior. 1990, 26, 89–100. [Google Scholar] [CrossRef]
- Russell, A.D. Mechanisms of antimicrobial action of antiseptics and disinfectants: An increasingly important area of investigation. J. Antimicrob. Chemother. 2002, 49, 597–599. [Google Scholar] [CrossRef] [PubMed]
- Russell, A.D. Biocide use and antibiotic resistance: The relevance of laboratory findings to clinical and environmental situations. Lancet Infect. Dis. 2003, 3, 794–803. [Google Scholar] [CrossRef]
- Condell, O.; Iversen, C.; Cooney, S.; Power, K.A.; Walsh, C.; Burgess, C.; Fanning, S. Efficacy of Biocides Used in the Modern Food Industry To Control Salmonella enterica and Links between Biocide Tolerance and Resistance to Clinically Relevant Antimicrobial Compounds. Appl. Environ. Microbiol. 2012, 78, 3087–3097. [Google Scholar] [CrossRef] [PubMed]
- Moen, B.; Rudi, K.; Bore, E.; Langsrud, S. Subminimal Inhibitory Concentrations of the Disinfectant Benzalkonium Chloride Select for a Tolerant Subpopulation of Escherichia coli with Inheritable Characteristics. Int. J. Mol. Sci. 2012, 13, 4101–4123. [Google Scholar] [CrossRef] [PubMed]
- Thomas, L.; Maillard, J.-Y.; Lambert, R.J.W.; Russell, A.D. Development of resistance to chlorhexidine diacetate in Pseudomonas aeruginosa and the effect of a “residual” concentration. J. Hosp. Infect. 2000, 46, 297–303. [Google Scholar] [CrossRef] [PubMed]
- Braoudaki, M.; Hilton, A.C. Adaptive Resistance to Biocides in Salmonella enterica and Escherichia coli O157 and Cross-Resistance to Antimicrobial Agents. J. Clin. Microbiol. 2004, 42, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Kampf, G. Biocidal agents used for disinfection can enhance antibiotic resistance in gram-negative species. Antibiotics 2018, 7, 110. [Google Scholar] [CrossRef] [PubMed]
- Kastbjerg, V.G.; Larsen, M.H.; Gram, L.; Ingmer, H. Influence of Sublethal Concentrations of Common Disinfectants on Expression of Virulence Genes in Listeria monocytogenes. Appl. Environ. Microbiol. 2010, 76, 303–309. [Google Scholar] [CrossRef] [PubMed]
- de las Heras, A.; Cain, R.J.; Bielecka, M.K.; Vázquez-Boland, J.A. Regulation of Listeria virulence: PrfA master and commander. Curr. Opin. Microbiol. 2011, 14, 118–127. [Google Scholar] [CrossRef] [PubMed]
- Pizarro-Cerdá, J.; Kühbacher, A.; Cossart, P. Entry of Listeria monocytogenes in Mammalian Epithelial Cells: An Updated View. Cold Spring Harb. Perspect. Med. 2012, 2, a010009. [Google Scholar] [CrossRef] [PubMed]
- Morita, Y.; Murata, T.; Mima, T.; Shiota, S.; Kuroda, T.; Mizushima, T.; Gotoh, N.; Nishino, T.; Tsuchiya, T. Induction of mexCD-oprJ operon for a multidrug efflux pump by disinfectants in wild-type Pseudomonas aeruginosa PAO1. J. Antimicrob. Chemother. 2003, 51, 991–994. [Google Scholar] [CrossRef] [PubMed]
- Huet, A.A.; Raygada, J.L.; Mendiratta, K.; Seo, S.M.; Kaatz, G.W. Multidrug efflux pump overexpression in Staphylococcus aureus after single and multiple in vitro exposures to biocides and dyes. Microbiology 2008, 154, 3144–3153. [Google Scholar] [CrossRef]
- Sanchez, P.; Moreno, E.; Martinez, J.L. The Biocide Triclosan Selects Stenotrophomonas maltophilia Mutants That Overproduce the SmeDEF Multidrug Efflux Pump. Antimicrob. Agents Chemother. 2005, 49, 781–782. [Google Scholar] [CrossRef]
- Henly, E.L.; Dowling, J.A.R.; Malngay, J.B.; Lacey, M.M.; Smith, T.J.; Forbes, S. Biocide exposure induces changes in susceptibility, pathogenicity, and biofilm formation in uropathogenic Escherichia coli. Antimicrob. Agents Chemother. 2019, 63. [Google Scholar] [CrossRef] [PubMed]
- Bailey, A.M.; Constantinidou, C.; Ivens, A.; Garvey, M.I.; Webber, M.A.; Coldham, N.; Hobman, J.L.; Wain, J.; Woodward, M.J.; Piddock, L.J. V Exposure of Escherichia coli and Salmonella enterica serovar Typhimurium to triclosan induces a species-specific response, including drug detoxification. J. Antimicrob. Chemother. 2009, 64, 973–985. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Deng, K.; Zaremba, S.; Deng, X.; Lin, C.; Wang, Q.; Tortorello, M.L.; Zhang, W. Transcriptomic Response of Escherichia coli O157:H7 to Oxidative Stress. Appl. Environ. Microbiol. 2009, 75, 6110–6123. [Google Scholar] [CrossRef] [PubMed]
- Small, D.A.; Chang, W.; Toghrol, F.; Bentley, W.E. Comparative global transcription analysis of sodium hypochlorite, peracetic acid, and hydrogen peroxide on Pseudomonas aeruginosa. Appl. Microbiol. Biotechnol. 2007, 76, 1093–1105. [Google Scholar] [CrossRef] [PubMed]
- Riordan, J.T.; Dupre, J.M.; Cantore-Matyi, S.A.; Kumar-Singh, A.; Song, Y.; Zaman, S.; Horan, S.; Helal, N.S.; Nagarajan, V.; Elasri, M.O.; et al. Alterations in the transcriptome and antibiotic susceptibility of Staphylococcus aureus grown in the presence of diclofenac. Ann. Clin. Microbiol. Antimicrob. 2011, 10, 30. [Google Scholar] [CrossRef] [PubMed]
- Berry, D.; Holder, D.; Xi, C.; Raskin, L. Comparative transcriptomics of the response of Escherichia coli to the disinfectant monochloramine and to growth conditions inducing monochloramine resistance. Water Res. 2010, 44, 4924–4931. [Google Scholar] [CrossRef] [PubMed]
- Moen, B.; Janbu, A.O.; Langsrud, S.; Langsrud, Ø.; Hobman, J.L.; Constantinidou, C.; Kohler, A.; Rudi, K. Global responses of Escherichia coli to adverse conditions determined by microarrays and FT-IR spectroscopy. Can. J. Microbiol. 2009, 55, 714–728. [Google Scholar] [CrossRef] [PubMed]
- Harding, G.K.M.; Ronald, A.R. The management of urinary infections: What have we learned in the past decade? Int. J. Antimicrob. Agents 1994, 4, 83–88. [Google Scholar] [CrossRef]
- Foxman, B. Epidemiology of Urinary Tract Infections: Incidence, Morbidity, and Economic Costs. Disease-A-Month 2003, 49, 53–70. [Google Scholar] [CrossRef] [PubMed]
- Foxman, B. The epidemiology of urinary tract infection. Nat. Rev. Urol. 2010, 7, 653–660. [Google Scholar] [CrossRef]
- Wassenaar, T.; Gaastra, W. Bacterial virulence: Can we draw the line? FEMS Microbiol. Lett. 2001, 201, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Neidhardt, F.C.; Bloch, P.L.; Smith, D.F. Culture Medium for Enterobacteria. J. Bacteriol. 1974, 119, 736–747. [Google Scholar] [PubMed]
- Vaara, M.; Vaara, T.; Jensen, M.; Helander, I.; Nurminen, M.; Rietschel, E.T.; Makela, P.H. Characterization of the lipopolysaccharide from the polymyxin-resistant pmrA mutants of Salmonella typhimurium. FEBS Lett. 1981, 129, 145–149. [Google Scholar] [CrossRef]
- Groisman, E.A.; Kayser, J.; Soncini, F.C. Regulation of Polymyxin Resistance and Adaptation to Low-Mg 2+ Environments. J. Bacteriol. 1997, 179, 7040–7045. [Google Scholar] [CrossRef]
- Trent, M.S.; Ribeiro, A.A.; Doerrler, W.T.; Lin, S.; Cotter, R.J.; Raetz, C.R.H. Accumulation of a Polyisoprene-linked Amino Sugar in Polymyxin-resistant Salmonella typhimurium and Escherichia coli: Structural characterization and transfer to lipid A in the periplasm. J. Biol. Chem. 2001, 276, 43132–43144. [Google Scholar] [CrossRef]
- Berger, E.A.; Heppel, L.A. A Binding Protein Involved in the Transport of Cystine and Diaminopimelic Acid in Escherichia coli. J. Biol. Chem. 1972, 247, 7684–7694. [Google Scholar] [PubMed]
- Deutch, C.E.; Spahija, I.; Wagner, C.E. Susceptibility of Escherichia coli to the toxic L-proline analogue L-selenaproline is dependent on two L-cystine transport systems. J. Appl. Microbiol. 2014, 117, 1487–1499. [Google Scholar] [CrossRef] [PubMed]
- Sugiyama, Y.; Nakamura, A.; Matsumoto, M.; Kanbe, A.; Sakanaka, M.; Higashi, K.; Igarashi, K.; Katayama, T.; Suzuki, H.; Kurihara, S. A novel putrescine exporter SapBCDF of Escherichia coli. J. Biol. Chem. 2016, 291, 26343–26351. [Google Scholar] [CrossRef]
- Clegg, S.J.; Jia, W.; Cole, J.A. Role of the Escherichia coli nitrate transport protein, NarU, in survival during severe nutrient starvation and slow growth. Microbiology 2006, 152, 2091–2100. [Google Scholar] [CrossRef]
- Wu, L.F.; Mandrand-Berthelot, M.A. A family of homologous substrate-binding proteins with a broad range of substrate specificity and dissimilar biological functions. Biochimie 1995, 77, 744–750. [Google Scholar] [CrossRef]
- Mu, X.-Q.; Bullitt, E. Structure and assembly of P-pili: A protruding hinge region used for assembly of a bacterial adhesion filament. Proc. Natl. Acad. Sci. USA 2006, 103, 9861–9866. [Google Scholar] [CrossRef]
- Welch, R.A.; Burland, V.; Plunkett III, G.; Redford, P.; Roesch, P.; Rasko, D.; Buckles, E.L.; Liou, S.-R.; Boutin, A.; Hackett, J.; et al. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 2002, 99, 17020–17024. [Google Scholar] [CrossRef]
- Brzuszkiewicz, E.; Brüggemann, H.; Liesegang, H.; Emmerth, M.; Olschläger, T.; Nagy, G.; Albermann, K.; Wagner, C.; Buchrieser, C.; Emody, L.; et al. How to become a uropathogen: Comparative genomic analysis of extraintestinal pathogenic Escherichia coli strains. Proc. Natl. Acad. Sci. USA 2006, 103, 12879–12884. [Google Scholar] [CrossRef]
- Ishikawa, S.; Matsumura, Y.; Katoh-Kubo, K.; Tsuchido, T. Antibacterial activity of surfactants against Escherichia coli cells is influenced by carbon source and anaerobiosis. J. Appl. Microbiol. 2002, 93, 302–309. [Google Scholar] [CrossRef]
- Boman, H.G.; Monner, D.A. Characterization of Lipopolysaccharides from Escherichia coli K-12 mutants. J. Bacteriol. 1975, 121, 455–464. [Google Scholar]
- Raetz, C.E.H.; Whitfield, C. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 2002, 71, 635–700. [Google Scholar] [CrossRef]
- Newton, B.A. The Properties and Mode of Action of the Polymyxins. Bacteriol. Rev. 1956, 20, 14–27. [Google Scholar]
- Falagas, M.E.; Rafailidis, P.I.; Matthaiou, D.K. Resistance to polymyxins: Mechanisms, frequency and treatment options. Drug Resist. Updat. 2010, 13, 132–138. [Google Scholar] [CrossRef] [PubMed]
- Yan, A.; Guan, Z.; Raetz, C.R.H. An Undecaprenyl Phosphate-Aminoarabinose Flippase Required for Polymyxin Resistance in Escherichia coli. J. Biol. Chem. 2007, 282, 36077–36089. [Google Scholar] [CrossRef]
- Kline, T.; Trent, M.S.; Stead, C.M.; Lee, M.S.; Sousa, M.C.; Felise, H.B.; Nguyen, H.V.; Miller, S.I. Synthesis of and Evaluation of Lipid A Modification by 4-Substituted 4-Deoxy Arabinose Analogs as Potential Inhibitors of Bacterial Polymyxin Resistance. Bioorg. Med. Chem. Lett. 2008, 18, 1507–1510. [Google Scholar] [CrossRef] [PubMed]
- Breazeale, S.D.; Ribeiro, A.A.; McClerren, A.L.; Raetz, C.R.H. A Formyltransferase Required for Polymyxin Resistance in Escherichia coli and the Modification of Lipid A with 4-amino-4-deoxy-l-arabinose: Identification and function of UDP-4-deoxy-4-formamido-l-arabinose. J. Biol. Chem. 2005, 280, 14154–14167. [Google Scholar] [CrossRef]
- Gunn, J.S.; Lim, K.B.; Krueger, J.; Kim, K.; Guo, L.; Hackett, M.; Miller, S.I. PmrA-PmrB-regulated genes necessary for 4-aminoarabinose lipid A modification and polymyxin resistance. Mol. Microbiol. 1998, 27, 1171–1182. [Google Scholar] [CrossRef]
- Gunn, J.S.; Ryan, S.S.; Van Velkinburgh, J.C.; Ernst, R.K.; Miller, S.I. Genetic and Functional Analysis of a PmrA-PmrB-Regulated Locus Necessary for Lipopolysaccharide Modification, Antimicrobial Peptide Resistance, and Oral Virulence of Salmonella enterica Serovar Typhimurium. Infect. Immun. 2000, 68, 6139–6146. [Google Scholar] [CrossRef]
- Tamayo, R.; Ryan, S.S.; Mccoy, A.J.; Gunn, J.S. Identification and Genetic Characterization of PmrA-Regulated Genes and Genes Involved in Polymyxin B Resistance in Salmonella enterica Serovar Typhimurium. Infect. Immun. 2002, 70, 6770–6778. [Google Scholar] [CrossRef]
- Kim, W.; Killam, T.; Sood, V.; Surette, M.G. Swarm-Cell Differentiation in Salmonella enterica Serovar Typhimurium Results in Elevated Resistance to Multiple Antibiotics. J. Bacteriol. 2003, 185, 3111–3117. [Google Scholar] [CrossRef]
- Baev, M.V.; Baev, D.; Jansco Radek, A.; Campbell, J.W. Growth of Escherichia coli MG1655 on LB medium: Monitoring utilization of amino acids, peptides, and nucleotides with transcriptional microarrays. Appl. Microbiol. Biotechnol. 2006, 71, 317–322. [Google Scholar] [CrossRef]
- Zimmer, D.P.; Soupene, E.; Lee, H.L.; Wendisch, V.F.; Khodursky, A.B.; Peter, B.J.; Bender, R.A.; Kustu, S. Nitrogen regulatory protein C-controlled genes of Escherichia coli: Scavenging as a defense against nitrogen limitation. Proc. Natl. Acad. Sci. USA 2000, 97, 14674–14679. [Google Scholar] [CrossRef]
- Wurpel, D.J.; Totsika, M.; Allsopp, L.P.; Webb, R.I.; Moriel, D.G.; Schembri, M.A. Comparative proteomics of uropathogenic Escherichia coli during growth in human urine identify UCA-like (UCL) fimbriae as an adherence factor involved in biofilm formation and binding to uroepithelial cells. J. Proteom. 2016, 131, 177–189. [Google Scholar] [CrossRef]
- Miller-Fleming, L.; Olin-Sandoval, V.; Campbell, K.; Ralser, M. Remaining Mysteries of Molecular Biology: The Role of Polyamines in the Cell. J. Mol. Biol. 2015, 427, 3389–3406. [Google Scholar] [CrossRef]
- Schneider, B.L.; Hernandez, V.J.; Reitzer, L. Putrescine catabolism is a metabolic response to several stresses in Escherichia coli. Mol. Microbiol. 2013, 88, 537–550. [Google Scholar] [CrossRef]
- Groisman, E.A.; Parra-Lopez, C.; Salcedo, M.; Lipps, C.J.; Heffron, F. Resistance to host antimicrobial peptides is necessary for Salmonella virulence. Proc. Natl. Acad. Sci. USA 1992, 89, 11939–11943. [Google Scholar] [CrossRef]
- Parra-Lopez, C.; Baer, M.T.; Groisman, E.A. Molecular genetic analysis of a locus required for resistance to antimicrobial peptides in Salmonella typhimurium. EMBO J. 1993, 12, 4053–4062. [Google Scholar] [CrossRef]
- Shelton, C.L.; Raffel, F.K.; Beatty, W.L.; Johnson, S.M.; Mason, K.M. Sap transporter mediated import and subsequent degradation of antimicrobial peptides in Haemophilus. PLoS Pathog. 2011, 7. [Google Scholar] [CrossRef]
- Subashchandrabose, S.; Smith, S.N.; Spurbeck, R.R.; Kole, M.M.; Mobley, H.L.T. Genome-wide detection of fitness genes in uropathogenic Escherichia coli during systemic infection. PLoS Pathog. 2013, 9, e1003788. [Google Scholar] [CrossRef]
- Scott, C.; Hilton, M.E.; Coppin, C.W.; Russell, R.J.; Oakeshott, J.G.; Sutherland, T.D. A global response to sulfur starvation in Pseudomonas putida and its relationship to the expression of low-sulfur-content proteins. FEMS Microbiol. Lett. 2007, 267, 184–193. [Google Scholar] [CrossRef][Green Version]
- Ritz, D.; Beckwith, J. Roles of Thiol-Redox Pathways in Bacteria. Annu. Rev. Microbiol. 2001, 55, 21–48. [Google Scholar] [CrossRef]
- Green, J.; Paget, M.S. Bacterial redox sensors. Nat. Rev. Microbiol. 2004, 2, 954–966. [Google Scholar] [CrossRef]
- Sekowska, A.; Kung, H.-F.; Danchin, A. Sulfur metabolism in Escherichia coli and related bacteria: Facts and fiction. J. Mol. Microbiol. Biotechnol. 2000, 2, 145–177. [Google Scholar]
- Miller, E.N.; Jarboe, L.R.; Turner, P.C.; Pharkya, P.; Yomano, L.P.; York, S.W.; Nunn, D.; Shanmugam, K.T.; Ingram, L.O. Furfural Inhibits Growth by Limiting Sulfur Assimilation in Ethanologenic Escherichia coli Strain LY180. Appl. Environ. Microbiol. 2009, 75, 6132–6141. [Google Scholar] [CrossRef]
- Spurbeck, R.R.; Mobley, H.L.T. Uropathogenic Escherichia coli. In Escherichia coli; Donnenberg, M.S., Ed.; Elsevier: Amsterdam, The Netherlands, 2013; pp. 275–304. ISBN 9780123970480. [Google Scholar]
- Johnson, J.R. Virulence Factors in Escherichia coli Urinary Tract Infection. Clin. Microbiol. Rev. 1991, 4, 80–128. [Google Scholar] [CrossRef]
- Kaper, J.B.; Nataro, J.P.; Mobley, H.L. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2004, 2, 123–140. [Google Scholar] [CrossRef]
- Snyder, J.A.; Haugen, B.J.; Lockatell, C.V.; Maroncle, N.; Hagan, E.C.; Johnson, D.E.; Welch, R.A.; Mobley, H.L.T. Coordinate Expression of Fimbriae in Uropathogenic Escherichia coli. Infect. Immun. 2005, 73, 7588–7596. [Google Scholar] [CrossRef]
- Gally, D.L.; Bogan, J.A.; Eisenstein, B.I.; Blomfield, I.C. Environmental Regulation of the fim Switch Controlling Type 1 Fimbrial Phase Variation in Escherichia coli K-12: Effects of Temperature and Media. J. Bacteriol. 1993, 175, 6186–6193. [Google Scholar] [CrossRef]
- Schwan, W.R.; Lee, J.L.; Lenard, F.A.; Brian, T.; Beck, M.T.; Matthews, B.T. Osmolarity and pH Growth Conditions Regulate fim Gene Transcription and Type 1 Pilus Expression in Uropathogenic Escherichia coli. Infect. Immun. 2002, 70, 1391–1402. [Google Scholar] [CrossRef]
- Chew, B.H.; Cadieux, P.A.; Reid, G.; Denstedt, J.D. In-Vitro Activity of Triclosan-Eluting Ureteral Stents against Common Bacterial Uropathogens. J. Endourol. 2006, 20, 949–958. [Google Scholar] [CrossRef]
- Korea, C.G.; Badouraly, R.; Prevost, M.C.; Ghigo, J.M.; Beloin, C. Escherichia coli K-12 possesses multiple cryptic but functional chaperone-usher fimbriae with distinct surface specificities. Environ. Microbiol. 2010, 12, 1957–1977. [Google Scholar] [CrossRef]
- Łoś, J.M.; Łoś, M.; Wegrzyn, A.; Wegrzyn, G. Hydrogen peroxide-mediated induction of the Shiga toxin-converting lambdoid prophage ST2-8624 in Escherichia coli O157:H7. FEMS Immunol. Med. Microbiol. 2010, 58, 322–329. [Google Scholar] [CrossRef]
- Banks, D.J.; Lei, B.; Musser, J.M. Prophage Induction and Expression of Prophage-Encoded Virulence Factors in Group A Streptococcus Serotype M3 Strain MGAS315. Infect. Immun. 2003, 71, 7079–7086. [Google Scholar] [CrossRef]
- Frye, J.G.; Porwollik, S.; Blackmer, F.; Cheng, P.; McClelland, M. Host Gene Expression Changes and DNA Amplification during Temperate Phage Induction. J. Bacteriol. 2005, 187, 1485–1492. [Google Scholar] [CrossRef]
- Wang, X.; Kim, Y.; Ma, Q.; Hong, S.H.; Pokusaeva, K.; Sturino, J.M.; Wood, T.K. Cryptic prophages help bacteria cope with adverse environments. Nat. Commun. 2010, 1, 147. [Google Scholar] [CrossRef]
- Russell, A.D. Similarities and differences in the responses of microorganisms to biocides. J. Antimicrob. Chemother. 2003, 52, 750–763. [Google Scholar] [CrossRef]
- Mobley, H.L.T.; Green, D.M.; Trifillis, A.L.; Johnson, D.E.; Chippendale, G.R.; Lockatell, C.V.; Jones, B.D.; Warren, J.W. Pyelonephritogenic Escherichia coli and Killing of Cultured Human Renal Proximal Tubular Epithelial Cells: Role of Hemolysin in Some Strains. Infect. Immun. 1990, 58, 1281–1289. [Google Scholar]
- Clinical and Laboratory Standards Institute. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically; Approved Standard—Ninth Edition M07-A9; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2012; Volume 32, ISBN 1562387839. [Google Scholar]
- Li, C.; Wong, W.H. Model-based analysis of oligonucleotide arrays: Expression index computation and outlier detection. Proc. Natl. Acad. Sci. USA 2001, 98, 31–36. [Google Scholar] [CrossRef]
- Li, C.; Wong, W.H. Model-based analysis of oligonucleotide arrays: Model validation, design issues and standard error application. Genome Biol. 2001, 2. [Google Scholar] [CrossRef]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—New capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Bio-Rad. Real-Time PCR Applications Guide; Bio-Rad: Hercules, CA, USA, 2006; Volume Bulletin 5. [Google Scholar]

| Group | Biocide | MIC | Sub-MIC |
|---|---|---|---|
| QAC | Benzalkonium chloride (BAC) | 8 mg/L | 2 mg/L |
| Biguanide | Chlorhexidine (CHX) | 0.25 mg/L | 0.0625 mg/L |
| Peroxide | Hydrogen peroxide (H2O2) | 0.004% | 0.001% |
| Phenol | Triclosan (TSN) | 2.5 mg/L | 0.3 mg/L |
| Biocide | Number of Total Genes | Up | Down | ||
|---|---|---|---|---|---|
| Number | % | Number | % | ||
| BAC | 407 | 238 | 58.5 | 169 | 41.5 |
| CHX | 389 | 339 | 87.1 | 50 | 12.9 |
| H2O2 | 233 | 171 | 73.4 | 62 | 26.6 |
| TSN | 117 | 63 | 53.8 | 54 | 46.2 |
| BAC | TSN | |||||
|---|---|---|---|---|---|---|
| Gene | Microarray | qPCR | SD | Microarray | qPCR | SD |
| arnT | 2.38 | −0.82 | 2.14 | − | - | − |
| kgtP | − | − | − | −1.96 | −2.14 | 7.24 |
| papA | −4.25 | −4.57 | 2.96 | −5.98 | −17.31 | 18.97 |
| papH | −2.35 | −3.75 | 1.54 | −2.41 | −6.93 | 5.55 |
| Gene | BAC | TSN | Gene Product |
|---|---|---|---|
| arnA | 1.74 | − | fused UDP-l-Ara4N formyltransferase/UDP-GlcA C-4′-decarboxylase |
| arnD | 2.30 | 2.52 | Undecaprenyl phosphate-alpha-l-ara4FN deformylase |
| arnT | 2.38 | − | 4-amino-4-deoxy-l-arabinose transferase |
| Gene | Fold Change | Pathway(s) or Processes |
|---|---|---|
| cysH | H2O2 (2.43), TSN (6.79) | Superpathway of sulfate assimilation and cysteine biosynthesis; Sulfate reduction I (assimilatory) |
| cysI | TSN (11.39) | |
| cysJ cysN | BAC (1.72) TSN (3.44) | |
| cysD | CHX (4.32), H2O2 (9.28), TSN (18.58) | Sulfate activation for sulfonation |
| sbp(c4868) | H2O2 (1.67) | Sulfate/thiosulfate/selenite transport |
| cysA | TSN (3.25) | |
| cysP | H2O2 (4.72), TSN (3.12) |
| Biocide | Intracellular | Membrane | Unclassified | Total Number of Biocide Specific Genes | ||||
|---|---|---|---|---|---|---|---|---|
| Up | Down | Up | Down | Up | Down | Up | Down | |
| BAC | 55% | 29% | 35% | 38% | 10% | 33% | 86 | 70 |
| CHX | 29% | 38% | 24% | 31% | 47% | 31% | 181 | 16 |
| H2O2 | 53% | 20% | 7% | 35% | 40% | 45% | 83 | 20 |
| TSN | 67% | 16% | 33% | 67% | 0 | 17% | 9 | 6 |
| Primer Name | Primer Sequence 5’—3’ | Product Size (bp) | Amplification Efficiency (From Standard Curve) | Reference |
|---|---|---|---|---|
| accD2_for | CTAACAGGCTATGCAGGCGA | 168 | 109% | This study |
| accD2_rev | ACATTACTCCCACCCGCAAG | |||
| gapA2_for | GTTGACCTGACCGTTCGTCT | 172 | 111% | This study |
| gapA2_rev | CCGCTTTAGCATCGAACACG | |||
| idnT2_for | CGGCGTTAATGGCTAACACG | 139 | 105% | This study |
| idnT2_rev | TCACACGTAAACGACCCTGG | |||
| arnT4_for | TTGCACTGGATGATGCCCAA | 167 | 102% | This study |
| arnT4_rev | CGGCATTATCGTCCAGCTCA | |||
| kgtP3_for | GTGAAACCAGAAACGCCACC | 131 | 97% | This study |
| kgtP3_rev | ATATGCGGTCGCCAATGCTA | |||
| papA_for_S | GTGCCTGCAGAAAATGCAGAT | 88 | 103% | [74] |
| papA_rev_S | CCCGTTTTCCACTCGAATCA | |||
| papH2_for | TAATCTGCCAGGCGTCTTCC | 70 | 112% | This study |
| papH2_rev | AGGGCTGCTTTTCATGGTGA |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ligowska-Marzęta, M.; Hancock, V.; Ingmer, H.; M. Aarestrup, F. Comparison of Gene Expression Profiles of Uropathogenic Escherichia Coli CFT073 after Prolonged Exposure to Subinhibitory Concentrations of Different Biocides. Antibiotics 2019, 8, 167. https://doi.org/10.3390/antibiotics8040167
Ligowska-Marzęta M, Hancock V, Ingmer H, M. Aarestrup F. Comparison of Gene Expression Profiles of Uropathogenic Escherichia Coli CFT073 after Prolonged Exposure to Subinhibitory Concentrations of Different Biocides. Antibiotics. 2019; 8(4):167. https://doi.org/10.3390/antibiotics8040167
Chicago/Turabian StyleLigowska-Marzęta, Małgorzata, Viktoria Hancock, Hanne Ingmer, and Frank M. Aarestrup. 2019. "Comparison of Gene Expression Profiles of Uropathogenic Escherichia Coli CFT073 after Prolonged Exposure to Subinhibitory Concentrations of Different Biocides" Antibiotics 8, no. 4: 167. https://doi.org/10.3390/antibiotics8040167
APA StyleLigowska-Marzęta, M., Hancock, V., Ingmer, H., & M. Aarestrup, F. (2019). Comparison of Gene Expression Profiles of Uropathogenic Escherichia Coli CFT073 after Prolonged Exposure to Subinhibitory Concentrations of Different Biocides. Antibiotics, 8(4), 167. https://doi.org/10.3390/antibiotics8040167





