Characterization of Two New Multidrug-Resistant Strains of Mycobacterium smegmatis: Tools for Routine In Vitro Screening of Novel Anti-Mycobacterial Agents
Abstract
1. Introduction
2. Materials and Method
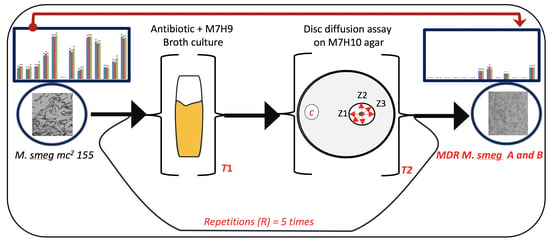
2.1. Bacterial Strains, Media Preparation, and Antibiotic Treatments
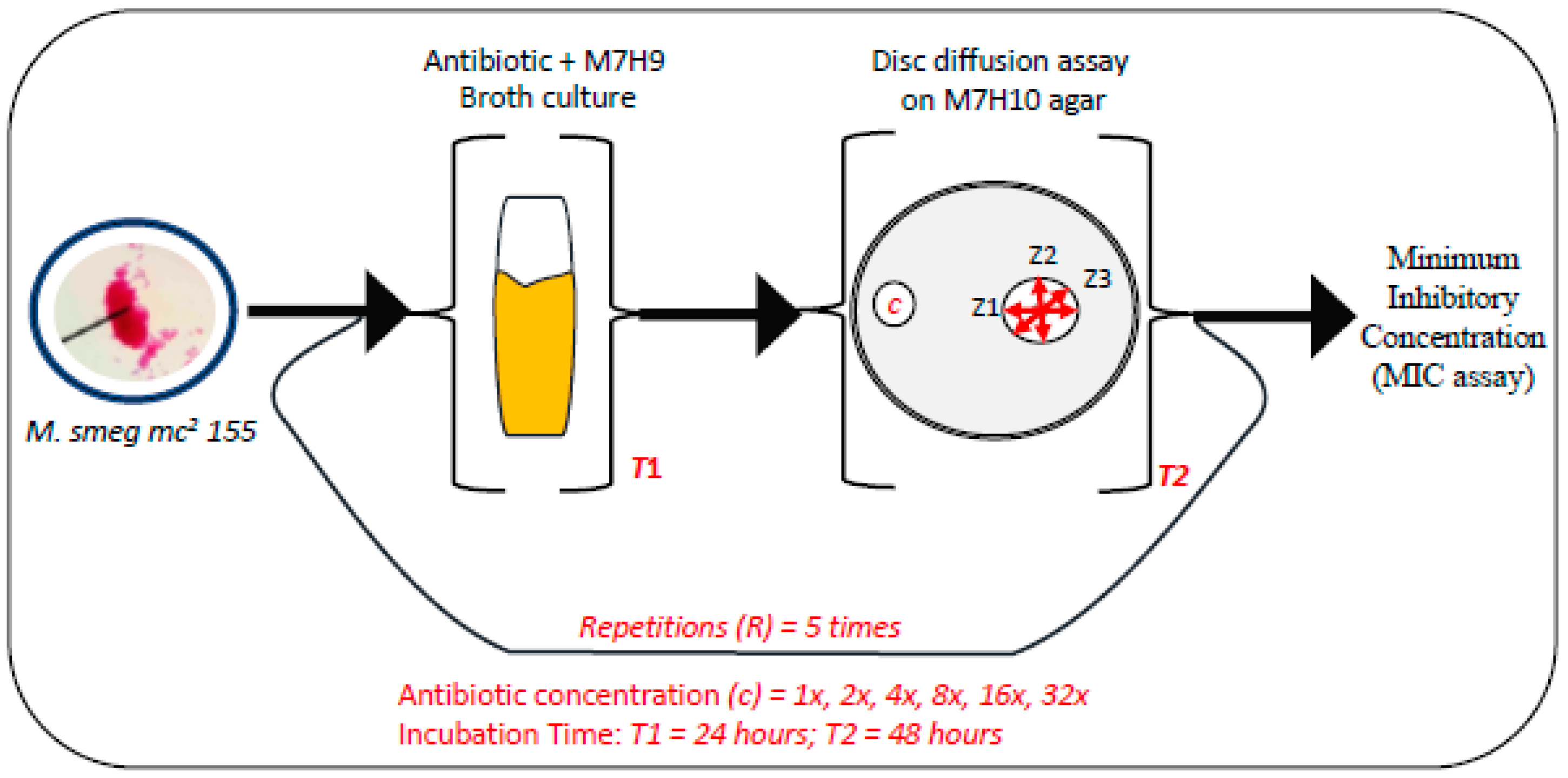
2.2. Drug Susceptibility Assay
2.3. Testing Cell Viability via Spot Test Assay
2.4. Growth Profile of Mycobacterial Cells
2.5. Colony Morphology Assay
2.6. Staining of Mycobacterial Cells
3. Results
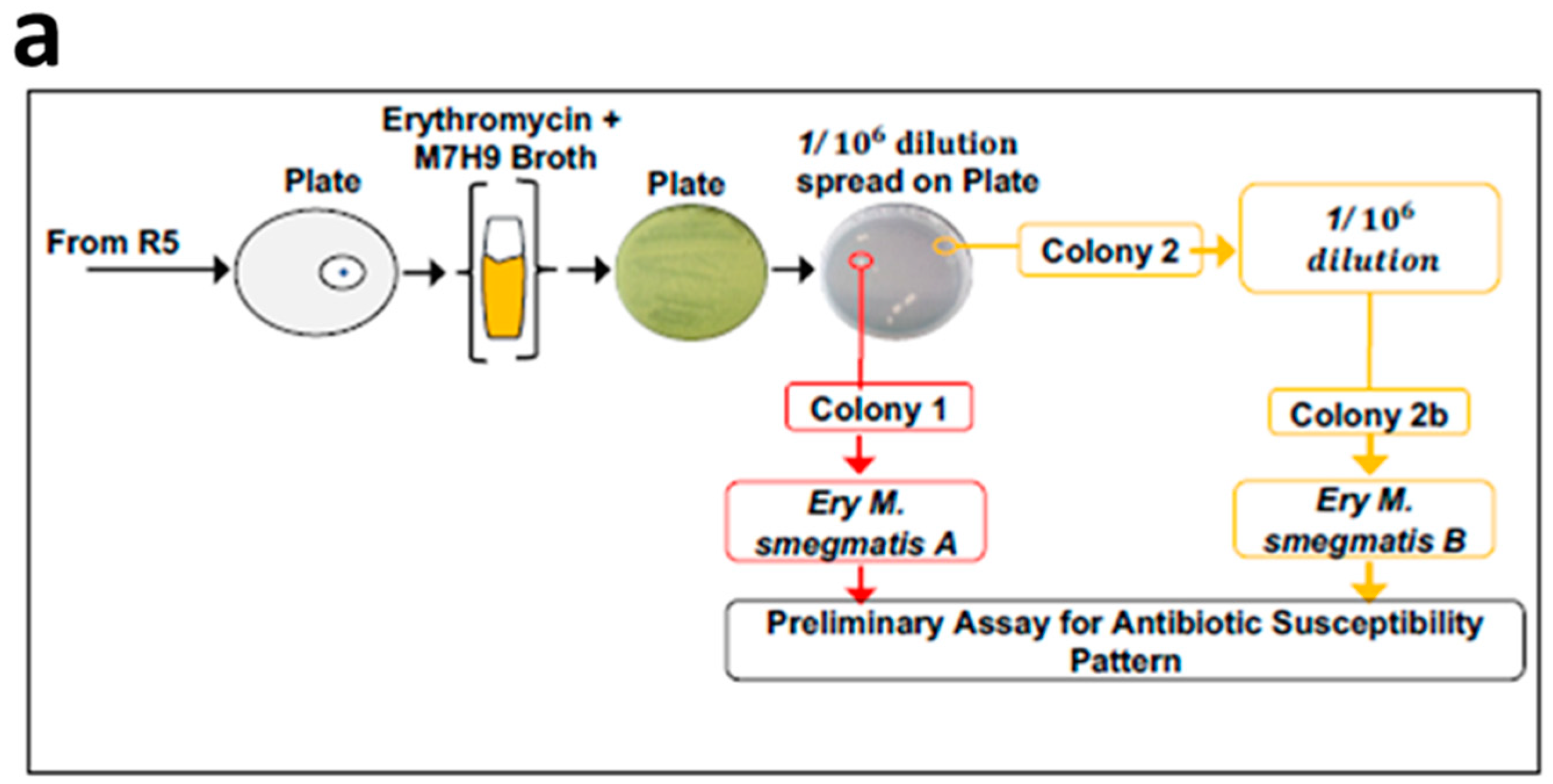
3.1. Exposure of Mycobacteria to Increasing Antibiotic Concentrations Drives Evolution of Drug Resistance
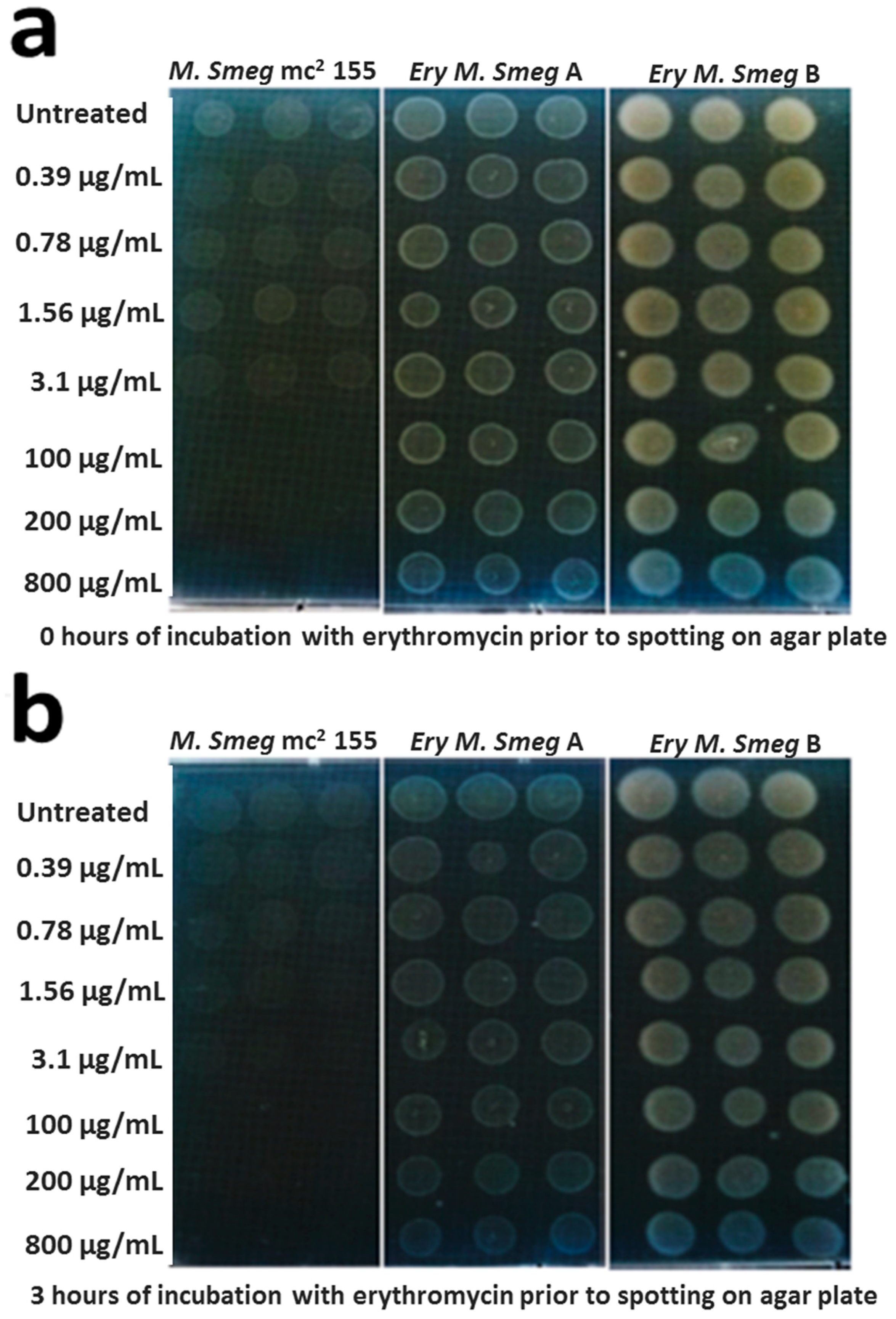
3.2. MDR Mycobacterium Smegmatis is Viable in the Presence of Higher Concentrations of Erythromycin
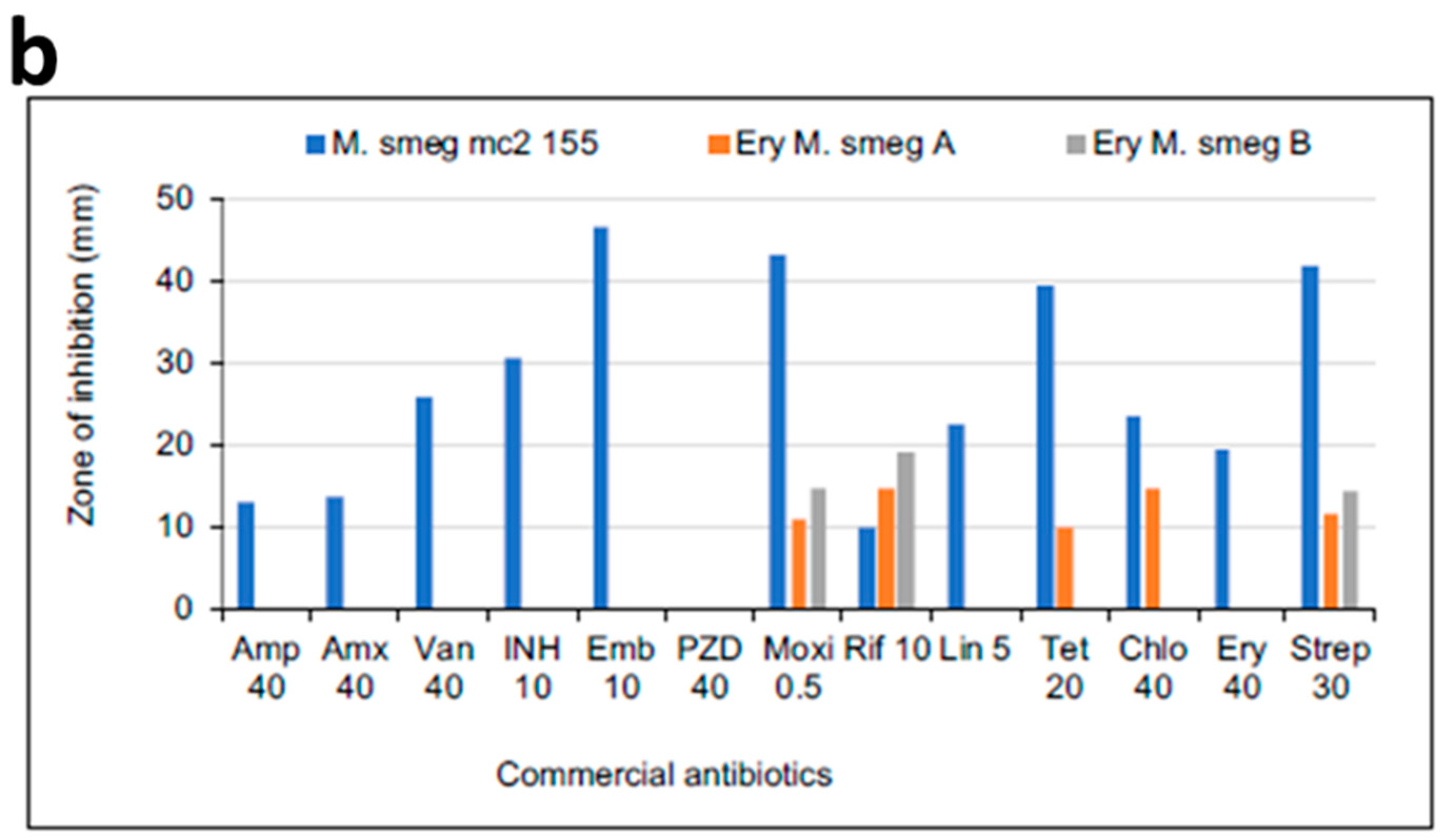
3.3. MICs of Selected Antibiotics against MDR Mycobacterium Smegmatis
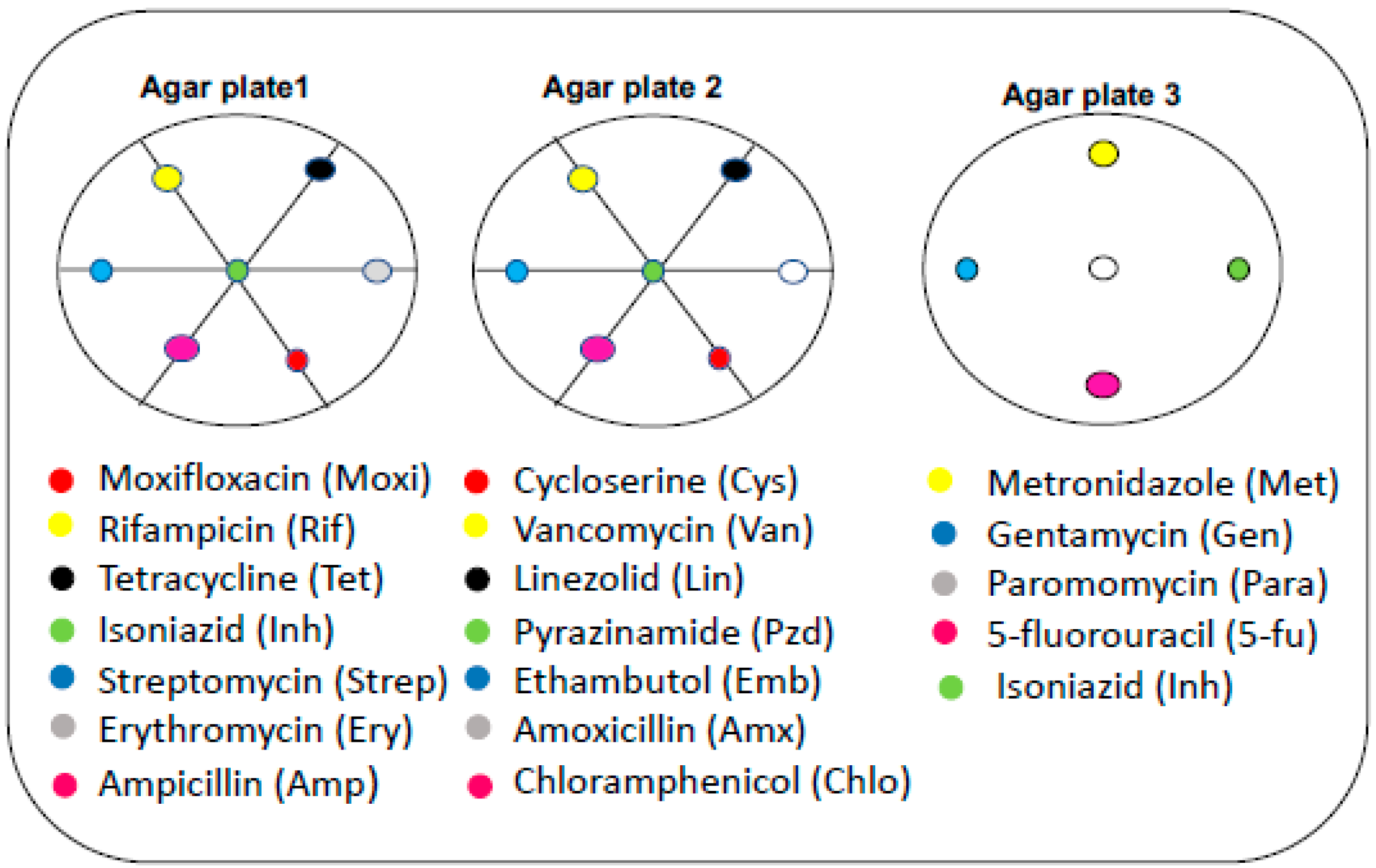
3.4. Effect of Neighboring Antibiotic Discs on the Antimycobacterial Activity of Selected Antibiotics
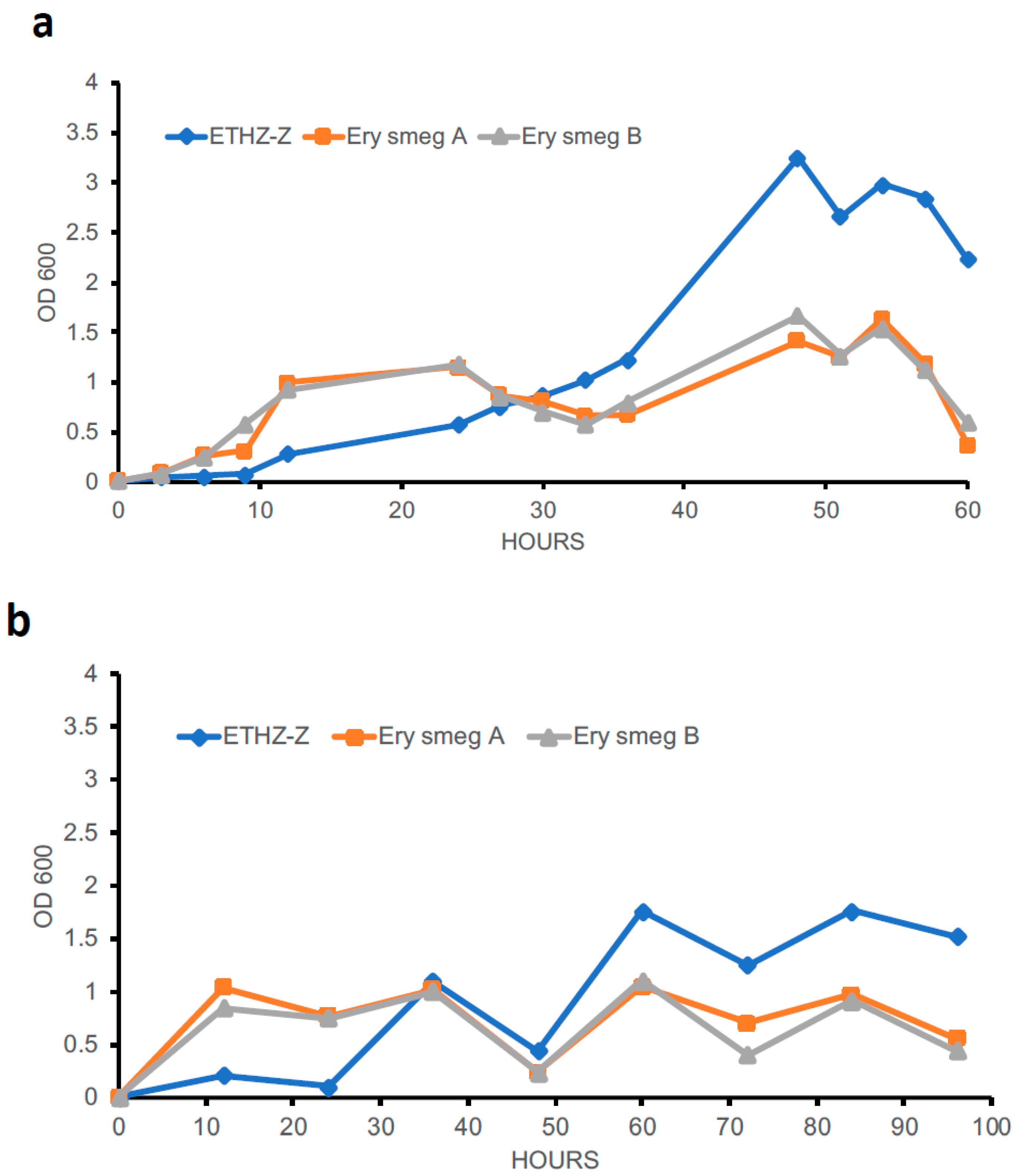
3.5. Evolution of the MDR Phenotype Affects the Growth Profile of Mycobacterium Smegmatis
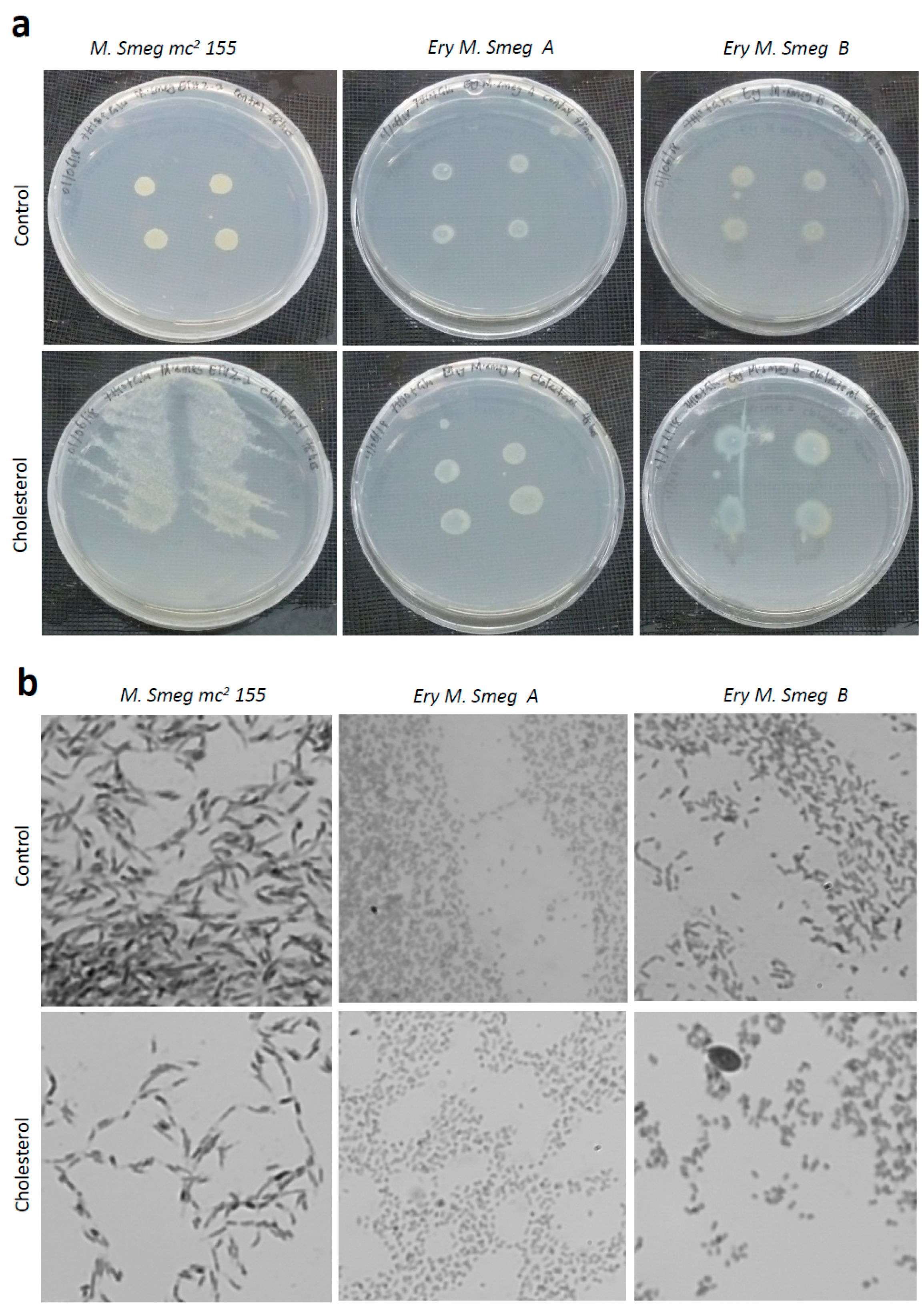
3.6. Cholesterol Enhances the Growth of Colonies of Mycobacterium Smegmatis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kirschner, D.E.; Young, D.; Flynn, J.L. Tuberculosis: Global approaches to a global disease. Curr. Opin. Biotechnol. 2010, 21, 524–531. [Google Scholar] [CrossRef] [PubMed]
- Forrellad, M.A.; Klepp, L.I.; Gioffré, A.; Sabio y Garcia, J.; Morbidoni, H.R.; Santangelo, M.d.L.P.; Cataldi, A.A.; Bigi, F. Virulence factors of the Mycobacterium tuberculosis complex. Virulence 2013, 4, 3–66. [Google Scholar] [CrossRef] [PubMed]
- Borkowska, D.I.; Napiorkowska, A.M.; Brzezińska, S.A.; Kozińska, M.; Zabost, A.T.; Augustynowicz-Kopeć, E. From Latent Tuberculosis Infection to Tuberculosis. News in Diagnostics (QuantiFERON-Plus). Pol. J. Microbiol. 2017, 66, 5–8. [Google Scholar] [CrossRef] [PubMed]
- Pai, M.; Nicol, M.P.; Boehme, C.C. Tuberculosis Diagnostics: State of the Art and Future Directions. Microbiol. Spectr. 2016, 4. [Google Scholar] [CrossRef]
- McGrath, M.; Gey van Pittius, N.C.; van Helden, P.D.; Warren, R.M.; Warner, D.F. Mutation rate and the emergence of drug resistance in Mycobacterium tuberculosis. J. Antimicrob. Chemother. 2014, 69, 292–302. [Google Scholar] [CrossRef] [PubMed]
- Tadesse, M.; Aragaw, D.; Dimah, B.; Efa, F.; Abdella, K.; Kebede, W.; Abdissa, K.; Abebe, G. Drug resistance-conferring mutations in Mycobacterium tuberculosis from pulmonary tuberculosis patients in Southwest Ethiopia. Int. J. Mycobacteriol. 2016, 5, 185–191. [Google Scholar] [CrossRef] [PubMed]
- Yew, W.W.; Lange, C.; Leung, C.C. Treatment of tuberculosis: Update 2010. Eur. Respir. J. 2011, 37, 441–462. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.K.; Mohan, A. Multidrug-resistant tuberculosis. Indian J. Med. Res. 2004, 120, 354–376. [Google Scholar] [CrossRef]
- World Health Organization (WHO). Global Tuberculosis Report 2013; WHO: Geneva, Switzerland, 2013. [Google Scholar]
- Forbes, L.; Ebsworth-Mojica, K.; DiDone, L.; Li, S.G.; Freundlich, J.S.; Connell, N.; Dunman, P.M.; Krysan, D.J. A High Throughput Screening Assay for Anti-Mycobacterial Small Molecules Based on Adenylate Kinase Release as a Reporter of Cell Lysis. PLoS ONE 2015, 10, e0129234. [Google Scholar] [CrossRef]
- Andries, K.; Verhasselt, P.; Guillemont, J.; Gohlmann, H.W.; Neefs, J.M.; Winkler, H.; Van Gestel, J.; Timmerman, P.; Zhu, M.; Lee, E.; et al. A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science 2005, 307, 223–227. [Google Scholar] [CrossRef]
- Canton, R.; Morosini, M.I. Emergence and spread of antibiotic resistance following exposure to antibiotics. FEMS Microbiol. Rev. 2011, 35, 977–991. [Google Scholar] [CrossRef] [PubMed]
- Levin-Reisman, I.; Ronin, I.; Gefen, O.; Braniss, I.; Shoresh, N.; Balaban, N.Q. Antibiotic tolerance facilitates the evolution of resistance. Science 2017, 355, 826–830. [Google Scholar] [CrossRef] [PubMed]
- Baquero, F.; Lemonnier, M. Generational coexistence and ancestor’s inhibition in bacterial populations. FEMS Microbiol. Rev. 2009, 33, 958–967. [Google Scholar] [CrossRef] [PubMed]
- Händel, N.; Schuurmans, J.M.; Brul, S.; ter Kuile, B.H. Compensation of the metabolic costs of antibiotic resistance by physiological adaptation in Escherichia coli. Antimicrob. Agents Chemother. 2013. [Google Scholar] [CrossRef] [PubMed]
- Zampieri, M.; Enke, T.; Chubukov, V.; Ricci, V.; Piddock, L.; Sauer, U. Metabolic constraints on the evolution of antibiotic resistance. Mol. Syst. Biol. 2017, 13, 917. [Google Scholar] [CrossRef] [PubMed]
- Obolski, U.; Stein, G.Y.; Hadany, L. Antibiotic restriction might facilitate the emergence of multi-drug resistance. PLoS Comput. Biol. 2015, 11, e1004340. [Google Scholar] [CrossRef] [PubMed]
- Imamovic, L.; Sommer, M.O. Use of collateral sensitivity networks to design drug cycling protocols that avoid resistance development. Sci. Transl. Med. 2013, 5, 204ra132. [Google Scholar] [CrossRef] [PubMed]
- Noens, E.E.; Williams, C.; Anandhakrishnan, M.; Poulsen, C.; Ehebauer, M.T.; Wilmanns, M. Improved mycobacterial protein production using a Mycobacterium smegmatis groEL1DeltaC expression strain. BMC Biotechnol. 2011, 11, 27. [Google Scholar] [CrossRef]
- Love, W.J.; Zawack, K.A.; Booth, J.G.; Grhn, Y.T.; Lanzas, C. Markov Networks of Collateral Resistance: National Antimicrobial Resistance Monitoring System Surveillance Results from Escherichia coli Isolates, 2004–2012. PLoS Comput. Biol. 2016, 12, e1005160. [Google Scholar] [CrossRef]
- Ouellet, H.; Johnston, J.B.; de Montellano, P.R. Cholesterol catabolism as a therapeutic target in Mycobacterium tuberculosis. Trends Microbiol. 2011, 19, 530–539. [Google Scholar] [CrossRef]
- Pandey, A.K.; Sassetti, C.M. Mycobacterial persistence requires the utilization of host cholesterol. Proc. Natl. Acad. Sci. USA 2008, 105, 4376–4380. [Google Scholar] [CrossRef] [PubMed]
- Ryan, A.; Polycarpou, E.; Lack, N.A.; Evangelopoulos, D.; Sieg, C.; Halman, A.; Bhakta, S.; Eleftheriadou, O.; McHugh, T.D.; Keany, S.; et al. Investigation of the mycobacterial enzyme HsaD as a potential novel target for anti-tubercular agents using a fragment-based drug design approach. Br. J. Pharmacol. 2017, 174, 2209–2224. [Google Scholar] [CrossRef] [PubMed]
- Evangelopoulos, D.; Gupta, A.; Lack, N.A.; Maitra, A.; ten Bokum, A.M.; Kendall, S.; Sim, E.; Bhakta, S. Characterisation of a putative AraC transcriptional regulator from Mycobacterium smegmatis. Tuberculosis 2014, 94, 664–671. [Google Scholar] [CrossRef] [PubMed]
- Bhakta, S.; Besra, G.S.; Upton, A.M.; Parish, T.; Sholto-Douglas-Vernon, C.; Gibson, K.J.; Knutton, S.; Gordon, S.; DaSilva, R.P.; Anderton, M.C.; et al. Arylamine N-acetyltransferase is required for synthesis of mycolic acids and complex lipids in Mycobacterium bovis BCG and represents a novel drug target. J. Exp. Med. 2004, 199, 1191–1199. [Google Scholar] [CrossRef] [PubMed]
- Chaturvedi, V.; Dwivedi, N.; Tripathi, R.P.; Sinha, S. Evaluation of Mycobacterium smegmatis as a possible surrogate screen for selecting molecules active against multi-drug resistant Mycobacterium tuberculosis. J. Gen. Appl. Microbiol. 2007, 53, 333–337. [Google Scholar] [CrossRef] [PubMed]
- Kerantzas, C.A.; Jacobs, W.R., Jr. Origins of Combination Therapy for Tuberculosis: Lessons for Future Antimicrobial Development and Application. mBio 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Vogwill, T.; MacLean, R.C. The genetic basis of the fitness costs of antimicrobial resistance: A meta-analysis approach. Evol. Appl. 2015, 8, 284–295. [Google Scholar] [CrossRef] [PubMed]
- Blumberg, H.M.; Burman, W.J.; Chaisson, R.E.; Daley, C.L.; Etkind, S.C.; Friedman, L.N.; Fujiwara, P.; Grzemska, M.; Hopewell, P.C.; Iseman, M.D.; et al. American Thoracic Society/Centers for Disease Control and Prevention/Infectious Diseases Society of America: Treatment of tuberculosis. Am. J. Respir. Crit Care Med. 2003, 167, 603–662. [Google Scholar] [CrossRef] [PubMed]

| Zones of Inhibition (mm) | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Repetitions (R) | Streptomycin (µg/µL) | Erythromycin (µg/µL) | Tetracycline (µg/µL) | ||||||||||||
| 0.1 | 1 | 6 | 30 | 150 | 0.1 | 1 | 6 | 30 | 150 | 0.1 | 1 | 6 | 30 | 150 | |
| - | 0 | 6 | 12 | 15 | 22 | 0 | 0 | 0 | 11.5 | 19.5 | 8 | 9.5 | 18 | 22 | 27.5 |
| 1 | 0 | 0 | 10 | 12 | 17.5 | 0 | 0 | 0 | 6 | 13.5 | 0 | 8 | 11 | 15.5 | 19 |
| 2 | 0 | 0 | 9 | 8 | 13 | 0 | 0 | 0 | 6 | 10 | 0 | 0 | 8.5 | 11 | 14.5 |
| 3 | 0 | 0 | 0 | 0 | 18 | 0 | 0 | 0 | 16.5 | 24.5 | 10 | 6 | 10 | 13 | 15 |
| 4 | 0 | 0 | 0 | 0 | 17 | 0 | 0 | 0 | 10 | 17.5 | N/A | N/A | N/A | N/A | N/A |
| 5 | N/A | N/A | N/A | N/A | N/A | 0 | 0 | 0 | 6 | 13 | N/A | N/A | N/A | N/A | N/A |
| Antibiotic (×) | Antibiotic Concentration | Zones of Inhibition (mm) | ||
|---|---|---|---|---|
| M. smeg mc2 155 | Ery M. smeg A | Ery M. smeg B | ||
| Amp (40 µg) | 0.5× | 0 | 12 | 10 |
| 1× | 6 | 11 | 14 | |
| 2× | 9 | 17 | 14 | |
| 4× | 18 | 21 | 20 | |
| Amx (40 µg) | 0.5× | 6 | 12 | 11 |
| 1× | 9 | 18 | 17 | |
| 2× | 13 | 18 | 19 | |
| 4× | 10 | 18 | 20 | |
| Van (40 µg) | 0.5× | 13 | 0 | 0 |
| 1× | 20 | 0 | 0 | |
| 2× | 20 | 7 | 9 | |
| 4× | 21 | 9 | 10 | |
| Inh (10 µg) | 0.5× | 8 | 0 | 0 |
| 1× | 29 | 0 | 0 | |
| 2× | 37 | 0 | 0 | |
| 4× | 40 | 0 | 0 | |
| Emb (10 µg) | 0.5× | 15 | 0 | 0 |
| 1× | 36 | 0 | 0 | |
| 2× | 46 | 0 | 0 | |
| 4× | 51 | 0 | 0 | |
| Pzd (40 µg) | 0.5× | 0 | 0 | 0 |
| 1× | 0 | 0 | 0 | |
| 2× | 0 | 0 | 0 | |
| 4× | 0 | 0 | 0 | |
| Moxi (0.5 µg) | 0.5× | 30 | 10 | 10 |
| 1× | 36 | 18 | 12 | |
| 2× | 40 | 22 | 20 | |
| 4× | 47 | 29 | 24 | |
| Rif (10 µg) | 0.5× | 0 | 18 | 12 |
| 1× | 8 | 20 | 16 | |
| 2× | 8 | 23 | 19 | |
| 4× | 11 | 25 | 21 | |
| Lin (5 µg) | 0.5× | 0 | 0 | 0 |
| 1× | 7 | 0 | 0 | |
| 2× | 8 | 0 | 0 | |
| 4× | 10 | 0 | 0 | |
| Tet (30 µg) | 0.5× | 46 | 20 | 7 |
| 1× | 40 | 20 | 9 | |
| 2× | 42 | 23 | 14 | |
| 4× | 54 | 27 | 18 | |
| Chlo (30 µg) | 0.5× | 14 | 9 | 0 |
| 1× | 18 | 9 | 7 | |
| 2× | 22 | 15 | 10 | |
| 4× | 31 | 22 | 13 | |
| Ery (40 µg) | 0.5× | 8 | 0 | 0 |
| 1× | 10 | 0 | 0 | |
| 2× | 10 | 0 | 0 | |
| 4× | 15 | 0 | 0 | |
| Strep (20 µg) | 0.5× | 18 | 0 | 0 |
| 1× | 20 | 0 | 0 | |
| 2× | 31 | 6 | 6 | |
| 4× | 38 | 8 | 9 | |
| Selected Antibiotics | Selected Antibiotics in the Presence of Neighbors | Selected Antibiotics alone | Neighbors | ||
|---|---|---|---|---|---|
| M. smeg mc2 155 | Ery M. smeg A | M. smeg mc2 155 | Ery M. smeg A | ||
| Amp | N/A | 0 | N/A | 11 | Strep, Inh, Moxi |
| Amx | N/A | 0 | N/A | 18 | Lin, Pzd, Cys |
| Lin | 50 | N/A | 0 | N/A | Van, Pzd, Amx |
| Tet | N/A | 7 | N/A | 20 | Rif, Inh, Ery |
| Chlo | N/A | 0 | N/A | 9 | Emb, Pzd, Cys |
| Ery | 20 | N/A | 10 | N/A | Tet, Inh, Moxi |
| Strep | 48 | 14 | 20 | 0 | Rif, Inh, Amp |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arthur, P.K.; Amarh, V.; Cramer, P.; Arkaifie, G.B.; Blessie, E.J.S.; Fuseini, M.-S.; Carilo, I.; Yeboah, R.; Asare, L.; Robertson, B.D. Characterization of Two New Multidrug-Resistant Strains of Mycobacterium smegmatis: Tools for Routine In Vitro Screening of Novel Anti-Mycobacterial Agents. Antibiotics 2019, 8, 4. https://doi.org/10.3390/antibiotics8010004
Arthur PK, Amarh V, Cramer P, Arkaifie GB, Blessie EJS, Fuseini M-S, Carilo I, Yeboah R, Asare L, Robertson BD. Characterization of Two New Multidrug-Resistant Strains of Mycobacterium smegmatis: Tools for Routine In Vitro Screening of Novel Anti-Mycobacterial Agents. Antibiotics. 2019; 8(1):4. https://doi.org/10.3390/antibiotics8010004
Chicago/Turabian StyleArthur, Patrick K., Vincent Amarh, Precious Cramer, Gloria B. Arkaifie, Ethel J. S. Blessie, Mohammed-Sherrif Fuseini, Isaac Carilo, Rebecca Yeboah, Leonard Asare, and Brian D. Robertson. 2019. "Characterization of Two New Multidrug-Resistant Strains of Mycobacterium smegmatis: Tools for Routine In Vitro Screening of Novel Anti-Mycobacterial Agents" Antibiotics 8, no. 1: 4. https://doi.org/10.3390/antibiotics8010004
APA StyleArthur, P. K., Amarh, V., Cramer, P., Arkaifie, G. B., Blessie, E. J. S., Fuseini, M.-S., Carilo, I., Yeboah, R., Asare, L., & Robertson, B. D. (2019). Characterization of Two New Multidrug-Resistant Strains of Mycobacterium smegmatis: Tools for Routine In Vitro Screening of Novel Anti-Mycobacterial Agents. Antibiotics, 8(1), 4. https://doi.org/10.3390/antibiotics8010004