Antibiotic Activity of Actinobacteria from the Digestive Tract of Millipede Nedyopus dawydoffiae (Diplopoda)
Abstract
1. Introduction
2. Results and Discussion
3. Materials and Methods
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- O’Neill, J. The Review on Antimicrobial Resistance. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations. 2016. Available online: http://amr-review.org/sites/default/files/160518_Final%20paper_with%20cover.pdf (accessed on 25 October 2018).
- Bérdy, J. Bioactive microbial metabolites. J. Antibiot. 2005, 58, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Bérdy, J. Thoughts and facts about antibiotics: Where we are now and where we are heading. J. Antibiot. 2012, 65, 385–395. [Google Scholar] [CrossRef] [PubMed]
- Hopkin, S.P.; Read, H.J. The Biology of Millipedes; Oxford University Press: Oxford, UK, 1992; ISBN 9780198576990. [Google Scholar]
- Zhang, Z.-Q. Animal biodiversity: An outline of higher-level classification and survey of taxonomic richness (Addenda 2013). Zootaxa 2013, 3703, 1–82. [Google Scholar] [CrossRef] [PubMed]
- Dhivya, A.; Alagesan, P. Millipedes as Host for Microbes (Review). Int. J. Microbial. Res. 2017, 8, 19–24. [Google Scholar]
- Byzov, B.A.; Zenova, G.M.; Babkina, N.I.; Dobrovolskaya, T.G.; Tretjakova, E.B.; Zvyagintsev, D.G. Actinomycetes in the food, gut and faeces of soil millipede, Pachyiulus flavipes C.L. Koch. Microbiology 1993, 62, 916–927. [Google Scholar]
- Byzov, B.A. Intestinal Microbiota of Millipedes. In Intestinal Microorganisms of Termites and Other Invertebrates; König, H., Varma, A., Eds.; Springer: Berlin/Heidelberg, Germany, 2006; Volume 6, pp. 89–114. [Google Scholar]
- Chu, T.L.; Szabo, I.M.; Szabo, I. Nocardioform gut actinomycetes of Glomeris hexasticha Brandt (Diplopoda). Biol. Fert. Soils 1987, 3, 113–116. [Google Scholar] [CrossRef]
- Jager, K.; Marialigeti, K.; Hauck, M.; Barabas, G. Promicromonospora enterophila sp. nov., a New Species of Monospore Actinomycetes. IJSB 1983, 33, 525–531. [Google Scholar] [CrossRef]
- Knapp, B.A.; Seeber, J.; Rief, A.; Meyer, E.; Insam, H. Bacterial community composition of the gut microbiota of Cylindroiulus fulviceps (diplopoda) as revealed by molecular fingerprinting and cloning. Folia Microbiol. 2010, 55, 489–496. [Google Scholar] [CrossRef] [PubMed]
- Byzov, B.A.; Thanh, V.N.; Bab’eva, I.P.; Tretyakova, E.B.; Dyvak, I.A.; Rabinovich, Y.M. Killing and hydrolytic activities of the gut fluid of the millipede Pachyiulus flavipes C.L. Koch on yeast cells. Soil Biol. Biochem. 1998, 30, 1137–1145. [Google Scholar] [CrossRef]
- Jarosz, J.; Kania, G. The question of whether gut microflora of the millipede Ommatoiulus sabulosus could function as a threshold to food infections. Pedobiologia 2000, 44, 705–708. [Google Scholar] [CrossRef]
- Marialigeti, K.; Contreras, E.; Barabas, G.; Heydrich, M.; Szabo, I.M. True intestinal actinomycetes of Millipedes (Diplopoda). J. Invertebr. Pathol. 1985, 45, 120–121. [Google Scholar] [CrossRef]
- Nguyen Duc, T.L.; Byzov, B.A.; Zenova, G.M.; Zveagintsev, D.G. Antagonistic properties of actinomycetes associated with intestinal tract of soil invertebrates. Vestnik Mosk. Un-ta 1996, 3, 70–77. (In Russian) [Google Scholar]
- Gause, G.F.; Preobrazhenskaya, T.P.; Sveshnikova, M.A.; Terekhova, L.P.; Maksimova, T.S. The Guide for Identification of Actinomycetes; Publishing House “Nauka”: Moscow, Russia, 1983. (In Russian) [Google Scholar]
- Shirling, E.B.; Gottlieb, D. Methods for Characterization of Streptomyces Species. IJSB 1966, 16, 317–327. [Google Scholar] [CrossRef]
- Miyajima, K.; Tanaka, F.; Takeuchi, T.; Kuninaga, S. Streptomyces turgidiscabies sp. nov. IJSEM 1998, 48, 495–502. [Google Scholar]
- Lambert, D.H.; Loria, R. Streptomyces acidiscabies sp. nov. IJSEM 1989, 39, 393–396. [Google Scholar] [CrossRef]
- Oka, H.; Yoshinari, T.; Murai, T.; Kawamura, K.; Satoh, F.; Funaishi, K.; Okura, A.; Suda, H.; Okanishi, M.; Shizuri, Y. A new topoisomerase-II inhibitor, BE-10988, produced by a streptomycete. I. Taxonomy, fermentation, isolation and characterization. J. Antibiot. 1991, 44, 486–491. [Google Scholar] [CrossRef] [PubMed]
- Dornberger, K.; Berger, U.; Knöll, H. Griseorubins, a new family of antibiotics with antimicrobial and antitumor activity. I. Taxonomy of the producing strain, fermentation, isolation and chemical characterization. J. Antibiot. 1980, 33, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Dornberger, K.; Berger, U.; Gutsche, W.; Jungstand, W.; Wohlrabe, K.; Härtl, A.; Knöll, H. Griseorubins, a new family of antibiotics with antimicrobial and antitumor activity. II. Biological properties and antitumor activity of the antibiotic complex griseorubin. J. Antibiot. 1980, 33, 9–12. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Lei, X.; Zhang, C.; Jiang, Z.; Shi, Y.; Wang, S.; Wang, L.; Hong, B. Complete genome sequence of Streptomyces globisporus C-1027, the producer of an enediyne antibiotic lidamycin. J. Biotechnol. 2016, 222, 9–10. [Google Scholar] [CrossRef] [PubMed]
- Shao, R.; Zhen, Y. Enediyne Anticancer Antibiotic Lidamycin: Chemistry, Biology and Pharmacology. Anti-Cancer Agents Med. Chem. 2008, 8, 123–131. [Google Scholar] [CrossRef]
- Gromyko, O.; Rebets, Y.; Ostash, B.; Luzhetskyy, A.; Fukuhara, M.; Bechthold, A.; Nakamura, T.; Fedorenko, V. Generation of Streptomyces globisporus SMY622 Strain with Increased Landomycin E Production and It’s Initial Characterization. J. Antibiot. 2004, 57, 383–389. [Google Scholar] [CrossRef] [PubMed]
- Krohk, K.; Rohr, J. Angucyclines: Total syntheses, new structures and biosynthetic studies of an emerging new class of antibiotics. Top. Curr. Chem. 1997, 188, 127–195. [Google Scholar]
- Khokhlov, A.S.; Cherches, B.Z.; Reshetov, P.D.; Smirnova, G.M.; Sorokina, I.B.; Prokoptzeva, T.A.; Koloditskaya, T.A.; Smirnov, W.; Navashin, S.M.; Fomina, I.P. Physico-chemical and biological studies on actinoxanthin, an antibiotic from Actinomyces globisporus 1131. J. Antibiot. 1969, 22, 541–544. [Google Scholar] [CrossRef] [PubMed]
- Khokhlov, A.S.; Reshetov, P.D.; Chupova, L.A.; Cherches, B.Z.; Zhigis, L.S.; Stoyachenko, I.A. Chemical studies on actinoxanthin. J. Antibiot. 1976, 29, 1026–1034. [Google Scholar] [CrossRef] [PubMed]
- Patterson, E.L.; Johnson, B.L.; DeVoe, S.E.; Bohonos, N. Structure of the antitumor antibiotic alazopeptin. Antimicrob. Agents Chemother. 1965, 5, 115–118. [Google Scholar] [PubMed]
- Ishiyama, A.; Otoguro, K.; Namatame, M.; Nishihara, A.; Furusawa, T.; Masuma, R.; Shiomi, K.; Takahashi, Y.; Ichimura, M.; Yamada, H.; et al. In Vitro and in Vivo Antitrypanosomal Activity of Two Microbial Metabolites, KS-505a and Alazopeptin. J. Antibiot. 2008, 61, 627–632. [Google Scholar] [CrossRef] [PubMed]
- Hata, T.; Umezawa, I.; Iwai, Y.; Katagiri, M.; Awaya, J.; Komiyama, K.; Oiwa, R.; Atsumi, K. Studies on the antitumor activity of an alazopeptin isolated from a new strain of Streptomyces. J. Antibiot. 1973, 26, 181–183. [Google Scholar] [CrossRef] [PubMed]
- Neuss, N.; Molloy, B.B.; Shah, R.; DeLaHiguera, N. The structure of anticapsin, a new biologically active metabolite of Streptomyces griseoplanus. Biochem. J. 1970, 118, 571–575. [Google Scholar] [CrossRef] [PubMed]
- Boeck, L.D.; Christy, K.L.; Shah, R. Production of anticapsin by Streptomyces griseoplanus. Appl. Microbiol. 1971, 21, 1075–1079. [Google Scholar] [PubMed]
- Thompson, R.M.; Strong, F.M. Identification of erythromycin A in cultures of Streptomyces griseoplanus. Biochem. Biophys. Res. Commun. 1971, 43, 213–216. [Google Scholar] [CrossRef]
- Kaur, T.; Manhas, R.K. Antifungal, insecticidal, and plant growth promoting potential of Streptomyces hydrogenans DH16. J. Basic Microbiol. 2014, 54, 1175–1185. [Google Scholar] [CrossRef] [PubMed]
- Kaur, T.; Vasudev, A.; Sohal, S.K.; Manhas, R.K. Insecticidal and growth inhibitory potential of Streptomyces hydrogenans DH16 on major pest of India, Spodoptera litura (Fab.) (Lepidoptera: Noctuidae). BMC Microbiol. 2014, 14, 227. [Google Scholar] [CrossRef] [PubMed]
- Manhas, R.K.; Talwinder, K. Biocontrol Potential of Streptomyces hydrogenans Strain DH16 toward Alternaria brassicicola to Control Damping Off and Black Leaf Spot of Raphanus sativus. Front. Plant. Sci. 2016, 7, 1869. [Google Scholar] [CrossRef] [PubMed]
- Kaur, T.; Kaur, A.; Sharma, V.; Manhas, R.K. Purification and Characterization of a New Antifungal Compound 10-(2,2-dimethyl-cyclohexyl)-6,9-dihydroxy-4,9-dimethyl-dec-2-enoic Acid Methyl Ester from Streptomyces hydrogenans Strain DH16. Front. Microbiol. 2016, 7, 1004. [Google Scholar] [CrossRef] [PubMed]
- Kaur, T.; Jasrotia, S.; Manhas, K. Evaluation of in vitro and in vivo nematicidal potential of a multifunctional streptomycete, Streptomyces hydrogenans strain DH16 against Meloidogyne incognita. Microbiol. Res. 2016, 192, 247–252. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, M.; Gorthi, S.; Chattopadhyay, P.; Banerjee, G. Production, characterization and optimization of actinomycin D from Streptomyces hydrogenans IB310, an antagonistic bacterium against phytopathogens. Biocatal. Agricult. Biotechnol. 2017, 10, 69–74. [Google Scholar] [CrossRef]
- Barghouthi, S.A.; Ayyad, I.; Ayesh, M.; Abu-Lafi, S. Isolation, Identification, and Characterization of the Novel Antibacterial Agent Methoxyphenyl-Oxime from Streptomyces pratensis QUBC97 Isolate. J. Antibiot. Res. 2017, 1, 105. [Google Scholar]
- Iwamoto, I.; Tsujii, E.; Ezaki, M.; Fujie, A.; Hashimoto, S.; Okuhara, M.; Kohsaka, M.; Imanaka, H.; Kawabata, K.; Inamoto, Y.; et al. FR109615, a new antifungal antibiotic from Streptomyces setonii. Taxonomy, fermentation, isolation, physico-chemical properties and biological activity. J. Antibiot. 1990, 43, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Larsen, S.H.; Boeck, L.D.; Mertz, F.P.; Paschal, J.W.; Occolowitz, J.L. 16-Deethylindanomycin (A83094A), a novel pyrrole-ether antibiotic produced by a strain of Streptomyces setonii. Taxonomy, fermentation, isolation and characterization. J. Antibiot. 1988, 41, 1170–1177. [Google Scholar] [CrossRef] [PubMed]
- Al-Askar, A.A.; Abdul Khair, W.M.; Rashad, Y.M. In vitro antifungal activity of Streptomyces spororaveus RDS28 against some phytopathogenic fungi. Afr. J. Agric. Res. 2011, 6, 2835–2842. [Google Scholar]
- Kotiaho, M.; Aittamaa, M.; Andersson, M.A.; Mikkola, R.; Valkonen, J.P.T.; Salkinoja-Salonen, M. Antimycin A-producing nonphytopathogenic Streptomyces turgidiscabies from potato. J. Appl. Microbiol. 2008, 104, 1332–1340. [Google Scholar] [CrossRef] [PubMed]
- Blanc, L.; Maury-Lechon, G.; Pascal, J.-P. Structure, floristic composition and natural regeneration in the forests of Cat Tien National Park, Vietnam: An analysis of the successional trends. J. Biogeogr. 2000, 27, 141–157. [Google Scholar] [CrossRef]

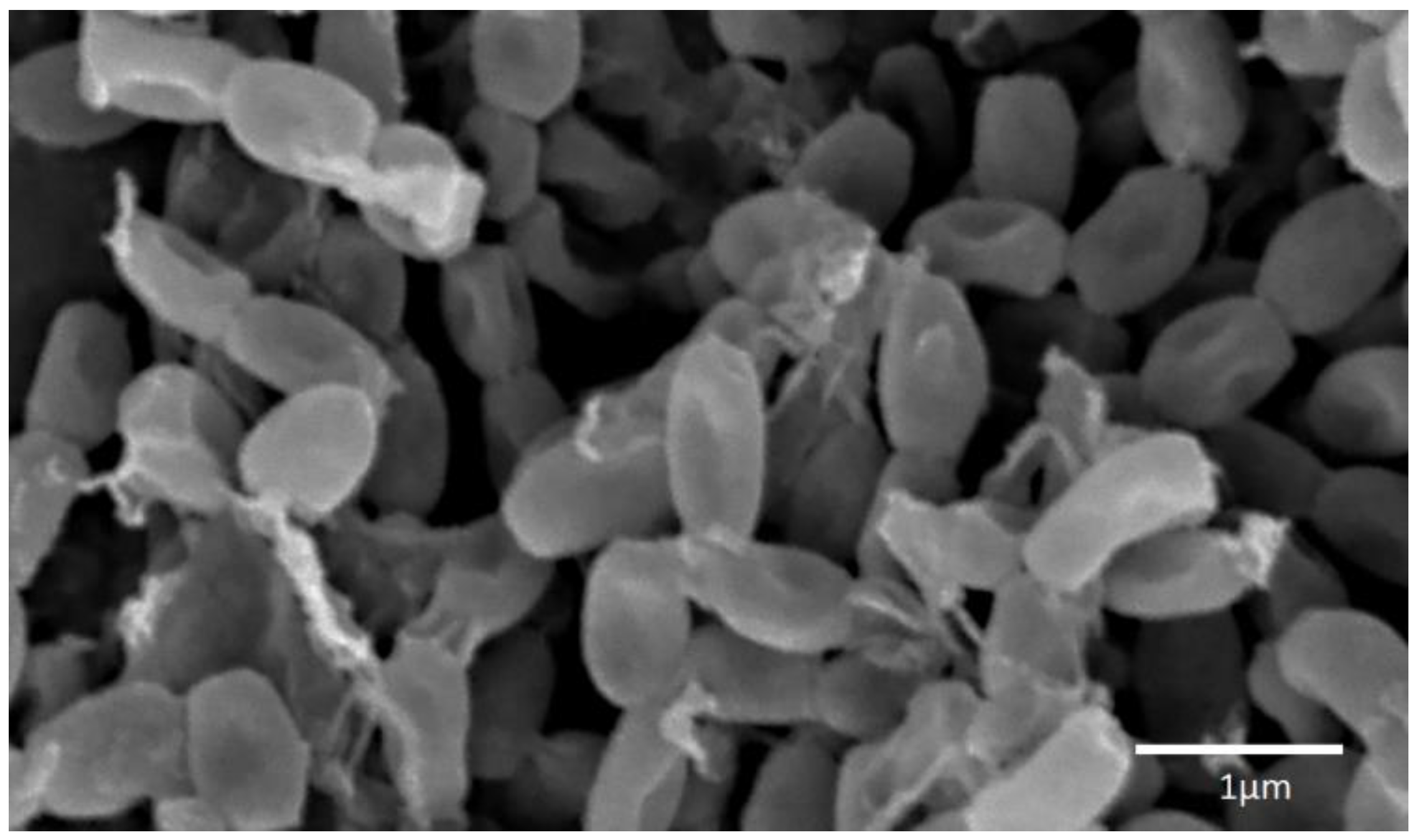

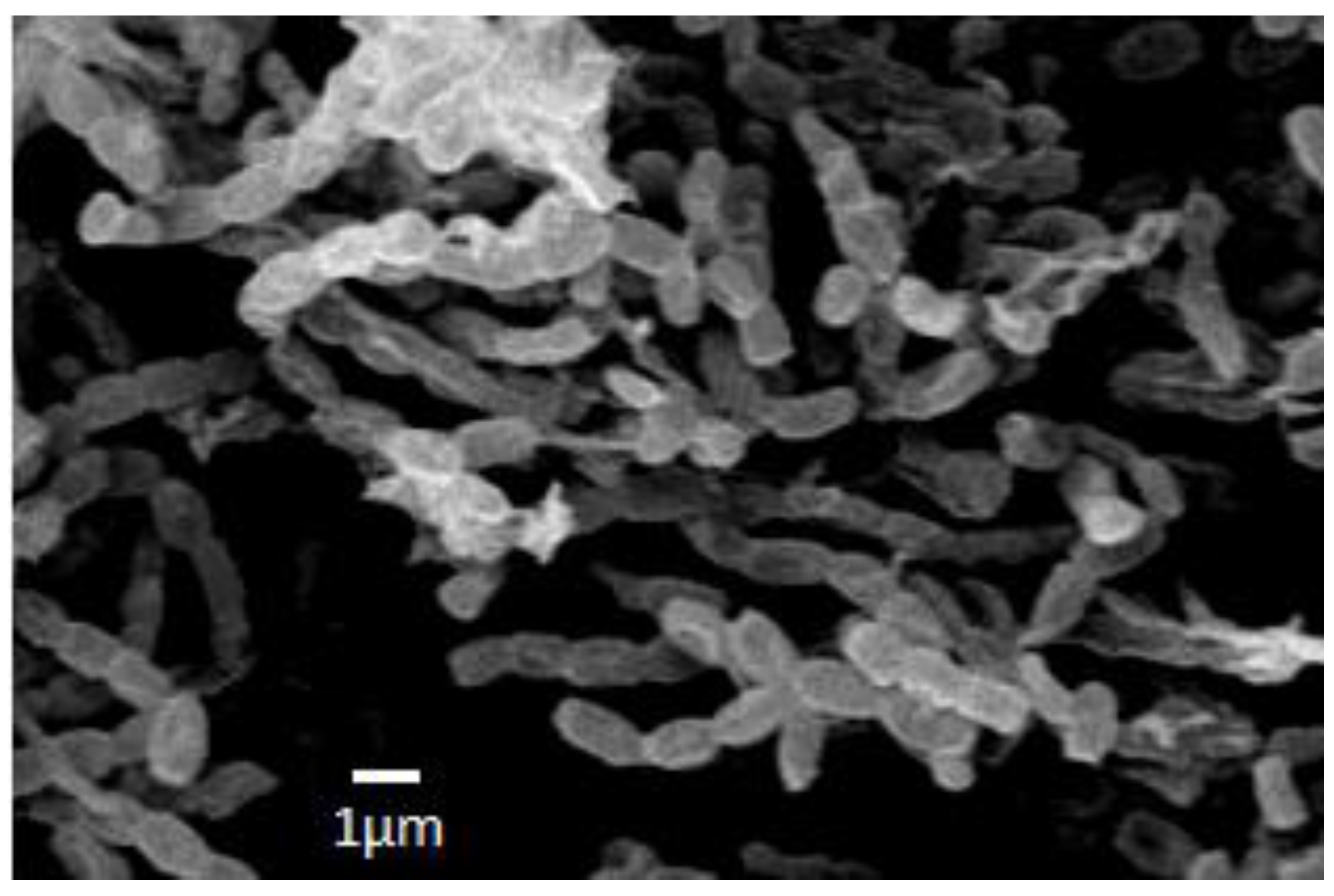
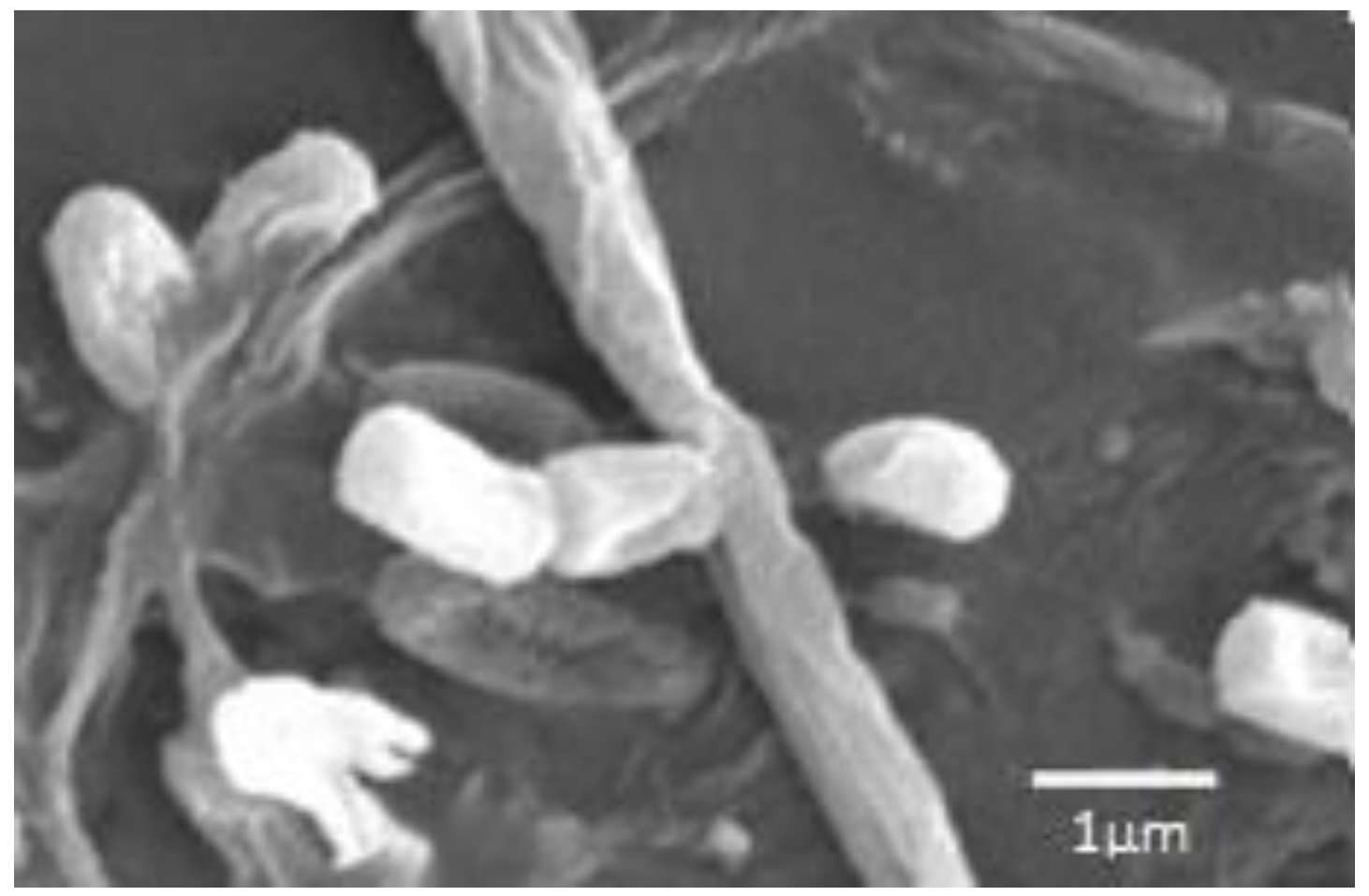

| Species, Strains | DNA (bp) | Alignment (%) * | ID of the Deposited Sequences (GenBank) |
|---|---|---|---|
| S. fimicarius INA 01179 | 1436 | 99.4 | MH635263 |
| S. griseoplanus INA 01177 | 1485 | 99.2 | MH635265 |
| S. hydrogenans INA 01175 ** | 1387 | 100 | - |
| S. hydrogenans INA 01173 ** | 1116 | 100 | - |
| S. sp. INA 01174 *** | 551 | 100 | - |
| S. sp. INA 01176 *** | 569 | 100 | - |
| S. pratensis INA 01182 | 1426 | 98.4 | MH635266 |
| S. setonii INA 01181 | 1468 | 100 | MH635267 |
| S. spororaveus INA 01183 | 1451 | 100 | MH635268 |
| S. turgidiscabies INA 01184 ** | 1309 | 96.5 | - |
| Streptomyces sp. INA 01180 **** | 568 | 99.3 | MH635264 |
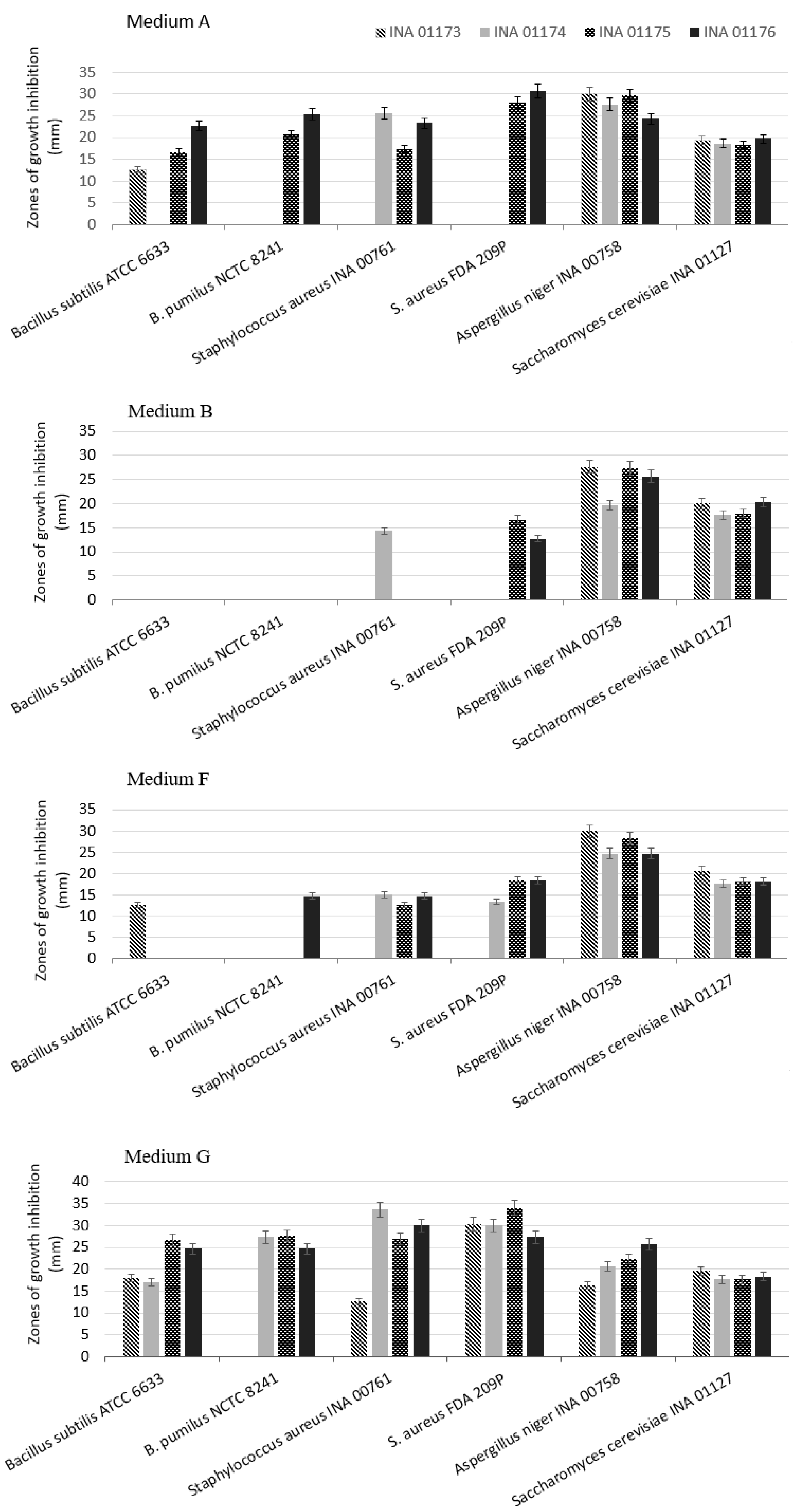
| Species, Strains | Media | Bacillus subtilis ATCC 6633 | Bacillus pumilus NCTC 8241 | Staphylococcus aureus INA 00761 (MRSA) | Staphylococcus aureus FDA 209P (MSSA) | Aspergillus niger INA 00760 | Saccharomyces cerevisiae INA 01129 |
|---|---|---|---|---|---|---|---|
| S. fimicarius INA 01179 | A, G | − | − | − | − | − | − |
| B, C, D, F | − | − | ++ | +++ | − | − | |
| E | − | − | − | − | − | ++ | |
| S. globisporus INA 01180 | A, B, G | − | − | − | − | − | − |
| C, D, F | − | − | +++ | +++ | − | − | |
| S. griseoplanus INA 01177 | A | − | − | − | − | − | − |
| B, F | − | − | +++ | ++++ | − | − | |
| C | ++ | − | +++ | ++ | − | − | |
| D | − | − | ++ | ++ | − | − | |
| E | − | − | ++ | ++ | − | ++ | |
| G | +++ | − | − | − | − | − | |
| S. hydrogenans INA 01175 | A | + | − | − | +++ | ++++ | +++ |
| B | − | − | − | − | ++++ | +++ | |
| C | + | + | ++ | +++ | ++++ | +++ | |
| D, E, F, G | ++++ | ++++ | ++++ | ++++ | ++++ | +++ | |
| S. pratensis INA01182 | A, E | ++ | − | − | − | − | − |
| B, D, G | − | − | − | − | − | − | |
| F | ++ | − | +++ | − | − | − | |
| S. setonii INA 01181 | A | ++ | ++ | − | + | +++ | nd |
| B | − | ++ | − | ++ | − | nd | |
| C | + | +++ | − | − | +++ | nd | |
| D | ++ | − | − | − | +++ | nd | |
| E | ++++ | ++++ | − | +++ | ++++ | nd | |
| F | ++ | ++++ | − | ++ | − | nd | |
| G | − | − | − | − | − | − | |
| S. spororaveus INA 01183 | A, F, G | ++ | ++ | − | − | +++ | ++ |
| B, D | − | − | − | − | ++ | − | |
| C, E | +++ | +++ | +++ | ++ | ++++ | ++ | |
| S. turgidiscabies INA 01184 | A, E, G | − | − | − | − | ++ | − |
| B, D, F | − | − | − | − | − | − | |
| C | − | − | − | − | +++ | ++ |
| Name | Organic Components (%) | Salts (%) | Water | pH |
|---|---|---|---|---|
| A (2663) | glycerin—3, soy flour—1.5, | NaCl—0.3, chalk—0.3, | tap | 7.0 |
| B (A4) | glucose—1, soy flour—1 | NaCl—0.5, chalk—0.25 | tap | 6.8 |
| C (Suc) | sucrose—2, soy flour—1 | NaCl—0.3, chalk—0.3 | tap | 6.8–7.0 |
| D (5339) | glycerin—2, soy flour—0.5 | (NH4)2SO4—0.15, NaCl—0.3, chalk—0.3 | tap | 6.8 |
| E (6613) | starch—2, corn extract—0.3 | KNO3—0.4, NaCl—0.5, chalk—0.5 | tap | 7.0–7.2 |
| F (330) | sucrose—2.1, starch—0.85, pea flour—1.5 | NaCl—0.5, NaNO3—0.5, chalk—0.5 | tap | 7.0 |
| G (Am) | sucrose—4, yeast extract—0.25 | K2HPO4—0.1, Na2SO4—0.1, NaCl—0.1, (NH4)2SO4—0.2, FeSO4 7H2O—0.0001, MnCl2 4H2O—0.0001, NaI—0.00005, chalk—0.2 | distilled | 6.5–6.7 |
| H (STR) | glucose—1, peptone—0.5, tryptone—0.3 | NaCl—0.5 | tap | 7.2–7.4 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Glukhova, A.A.; Karabanova, A.A.; Yakushev, A.V.; Semenyuk, I.I.; Boykova, Y.V.; Malkina, N.D.; Efimenko, T.A.; Ivankova, T.D.; Terekhova, L.P.; Efremenkova, O.V. Antibiotic Activity of Actinobacteria from the Digestive Tract of Millipede Nedyopus dawydoffiae (Diplopoda). Antibiotics 2018, 7, 94. https://doi.org/10.3390/antibiotics7040094
Glukhova AA, Karabanova AA, Yakushev AV, Semenyuk II, Boykova YV, Malkina ND, Efimenko TA, Ivankova TD, Terekhova LP, Efremenkova OV. Antibiotic Activity of Actinobacteria from the Digestive Tract of Millipede Nedyopus dawydoffiae (Diplopoda). Antibiotics. 2018; 7(4):94. https://doi.org/10.3390/antibiotics7040094
Chicago/Turabian StyleGlukhova, Alla A., Anna A. Karabanova, Andrey V. Yakushev, Irina I. Semenyuk, Yuliya V. Boykova, Natalia D. Malkina, Tatiana A. Efimenko, Tatiana D. Ivankova, Larissa P. Terekhova, and Olga V. Efremenkova. 2018. "Antibiotic Activity of Actinobacteria from the Digestive Tract of Millipede Nedyopus dawydoffiae (Diplopoda)" Antibiotics 7, no. 4: 94. https://doi.org/10.3390/antibiotics7040094
APA StyleGlukhova, A. A., Karabanova, A. A., Yakushev, A. V., Semenyuk, I. I., Boykova, Y. V., Malkina, N. D., Efimenko, T. A., Ivankova, T. D., Terekhova, L. P., & Efremenkova, O. V. (2018). Antibiotic Activity of Actinobacteria from the Digestive Tract of Millipede Nedyopus dawydoffiae (Diplopoda). Antibiotics, 7(4), 94. https://doi.org/10.3390/antibiotics7040094





