Explorative Study on Isolation and Characterization of a Microviridae G4 Bacteriophage, EMCL318, against Multi-Drug-resistant Escherichia coli 15-318
Abstract
1. Introduction
2. Results
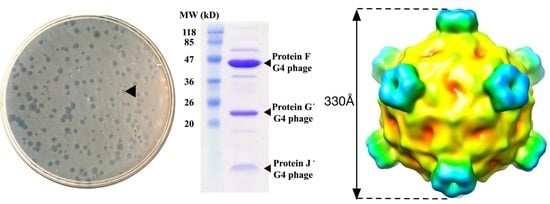
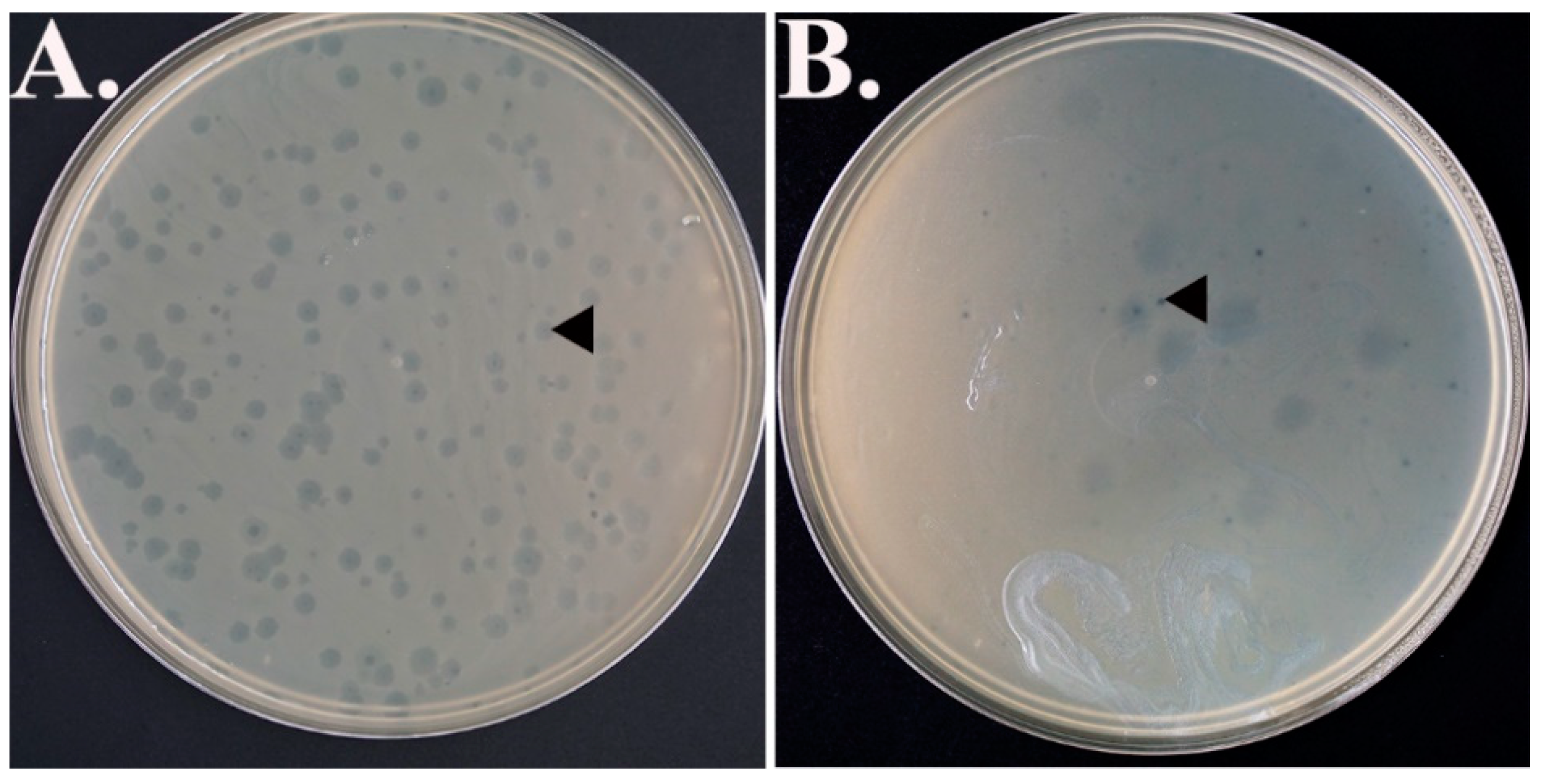
2.1. Bacteriophages Isolation
2.2. Bacteriophage Purification and Identification
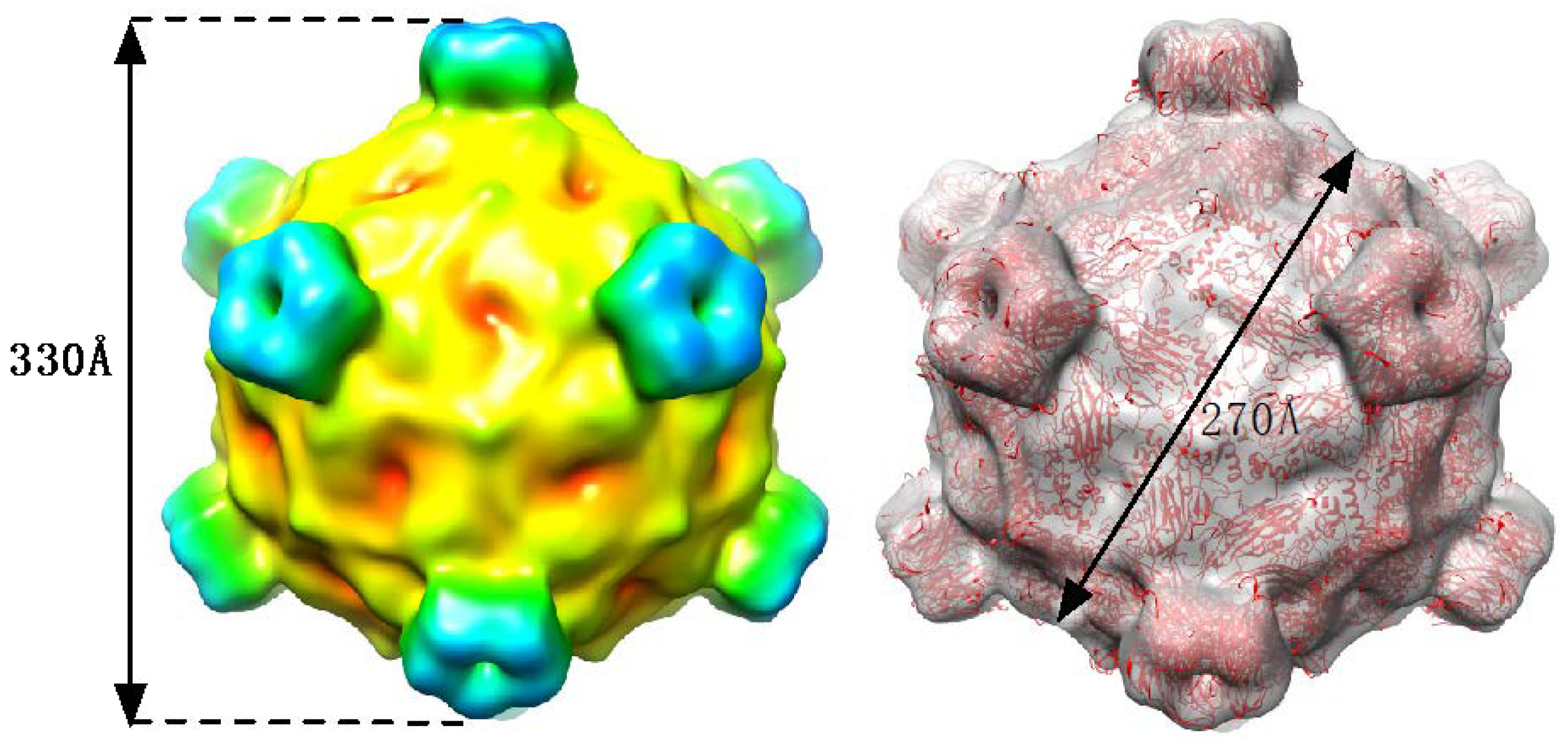
2.3. Cryo-EM Image Reconstruction
2.4. Sequencing Results
3. Discussion
4. Materials and Methods
4.1. Sample Collections
4.2. Processing of Water Samples
4.3. Host Microbes for Bacteriophage Screening
4.4. Isolation of Bacteriophages
4.5. Initial Purification and Identification of the Phage
4.6. Mass Spectroscopy
4.7. Electron Microscopy
4.8. Nucleotide Sequencing
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Milic, N.; Milanovic, M.; Letic, N.G.; Sekulic, M.T.; Radonic, J.; Mihajlovic, I.; Miloradov, M.V. Occurrence of antibiotics as emerging contaminant substances in aquatic environment. Int. J. Environ. Health Res. 2013, 23, 296–310. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, A.R.; Sures, B.; Schmidt, T.C. Cephalosporin antibiotics in the aquatic environment: A critical review of occurrence, fate, ecotoxicity and removal technologies. Environ. Pollut. 2018, 241, 1153–1166. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Tang, J.; Li, J.; Cheng, Z.; Chaemfa, C.; Liu, D.; Zheng, Q.; Song, M.; Luo, C.; Zhang, G. Occurrence and risks of antibiotics in the coastal aquatic environment of the yellow sea, north china. Sci. Total Environ. 2013, 450–451, 197–204. [Google Scholar] [CrossRef] [PubMed]
- Zuccato, E.; Castiglioni, S.; Bagnati, R.; Melis, M.; Fanelli, R. Source, occurrence and fate of antibiotics in the italian aquatic environment. J. Hazard. Mater. 2010, 179, 1042–1048. [Google Scholar] [CrossRef] [PubMed]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Levy, S.B.; Marshall, B. Antibacterial resistance worldwide: Causes, challenges and responses. Nat. Med. 2004, 10, S122–S129. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, L.; Manaia, C.; Merlin, C.; Schwartz, T.; Dagot, C.; Ploy, M.C.; Michael, I.; Fatta-Kassinos, D. Urban wastewater treatment plants as hotspots for antibiotic-resistant bacteria and genes spread into the environment: A review. Sci. Total Environ. 2013, 447, 345–360. [Google Scholar] [CrossRef] [PubMed]
- Levy, S.B. The challenge of antibiotic resistance. Sci. Am. 1998, 278, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Chan, B.K.; Sistrom, M.; Wertz, J.E.; Kortright, K.E.; Narayan, D.; Turner, P.E. Phage selection restores antibiotic sensitivity in MDR Pseudomonas aeruginosa. Sci. Rep. 2016, 6, 26717. [Google Scholar] [CrossRef] [PubMed]
- Queenan, A.M.; Bush, K. Carbapenemases: The versatile beta-lactamases. Clin. Microbiol. Rev. 2007, 20, 440–458. [Google Scholar] [CrossRef] [PubMed]
- Jimenez, A.; Abbo, L.; Gebrezgi, M.T. Importance of Carbapenemase production detection in Carbapenem-Resistant Enterobacteriaceae: Looking beyond epidemiological purposes. Clin. Infect. Dis. 2017, 15, 1424–1425. [Google Scholar] [CrossRef] [PubMed]
- De Kraker, M.E.; Jarlier, V.; Monen, J.C.; Heuer, O.E.; van de Sande, N.; Grundmann, H. The changing epidemiology of bacteraemias in europe: Trends from the european antimicrobial resistance surveillance system. Clin. Microbiol. Infect. 2013, 19, 860–868. [Google Scholar] [CrossRef] [PubMed]
- Alhashash, F.; Weston, V.; Diggle, M.; McNally, A. Multidrug-resistant Escherichia coli bacteremia. Emerg. Infect. Dis. 2013, 19, 1699–1701. [Google Scholar] [CrossRef] [PubMed]
- Hornsey, M.; Phee, L.; Wareham, D.W. A novel variant, ndm-5, of the new delhi metallo-beta-lactamase in a multidrug-resistant Escherichia coli ST648 isolate recovered from a patient in the United Kingdom. Antimicrob. Agents Chemother. 2011, 55, 5952–5954. [Google Scholar] [CrossRef] [PubMed]
- Abedon, S.T.; Kuhl, S.J.; Blasdel, B.G.; Kutter, E.M. Phage treatment of human infections. Bacteriophage 2011, 1, 66–85. [Google Scholar] [CrossRef] [PubMed]
- Carlton, R.M. Phage therapy: Past history and future prospects. Arch. Immunol. Ther. Exp. (Warsz) 1999, 47, 267–274. [Google Scholar] [PubMed]
- Sulakvelidze, A.; Alavidze, Z.; Morris, J.G. Bacteriophage therapy. Antimicrob. Agents Chemother. 2001, 45, 649–659. [Google Scholar] [CrossRef] [PubMed]
- McKenna, R.; Xia, D.; Willingmann, P.; Ilag, L.L.; Krishnaswamy, S.; Rossmann, M.G.; Olson, N.H.; Baker, T.S.; Incardona, N.L. Atomic structure of single-stranded DNA bacteriophage phi x174 and its functional implications. Nature 1992, 355, 137–143. [Google Scholar] [CrossRef] [PubMed]
- McKenna, R.; Bowman, B.; Ilag, L.L.; Rossmann, M.G.; Fane, B.A. The atomic structure of the degraded procapsid particle of the bacteriophage G4. J. Mol. Biol. 1996, 256, 736–750. [Google Scholar] [CrossRef] [PubMed]
- Ilag, L.L.; Olson, N.H.; Dokland, T.; Music, C.L.; Cheng, R.H.; Bowen, Z.; McKenna, R.; Rossmann, M.G.; Baker, T.S.; Incardona, N.L. DNA packaging intermediates of bacteriophage fx174. Structure 1995, 3, 353–363. [Google Scholar] [CrossRef]
- Rokyta, D.R.; Burch, C.L.; Caudle, S.B.; Wichman, H.A. Horizontal gene transfer and the evolution of microvirid coliphage genomes. J. Bacteriol. 2006, 188, 1134–1142. [Google Scholar] [CrossRef] [PubMed]
- Paterson, D.L. Resistance in gram-negative bacteria: Enterobacteriaceae. Am. J. Med. 2006, 119, S20–S28, discussion S62–S70. [Google Scholar] [CrossRef] [PubMed]
- Kaur, P.; Chakraborti, A.; Asea, A. Enteroaggregative Escherichia coli: An emerging enteric food borne pathogen. Interdiscip. Perspect. Infect. Dis. 2010, 2010, 254159. [Google Scholar] [CrossRef] [PubMed]
- Biswas, B.; Adhya, S.; Washart, P.; Paul, B.; Trostel, A.N.; Powell, B.; Carlton, R.; Merril, C.R. Bacteriophage therapy rescues mice bacteremic from a clinical isolate of vancomycin-resistant Enterococcus faecium. Infect. Immun. 2002, 70, 204–210. [Google Scholar] [CrossRef] [PubMed]
- Vinodkumar, C.S.; Kalsurmath, S.; Neelagund, Y.F. Utility of lytic bacteriophage in the treatment of multidrug-resistant Pseudomonas aeruginosa septicemia in mice. Indian J. Pathol. Microbiol. 2008, 51, 360–366. [Google Scholar] [CrossRef] [PubMed]
- Wright, A.; Hawkins, C.H.; Anggard, E.E.; Harper, D.R. A controlled clinical trial of a therapeutic bacteriophage preparation in chronic otitis due to antibiotic-resistant Pseudomonas aeruginosa; a preliminary report of efficacy. Clin. Otolaryngol. 2009, 34, 349–357. [Google Scholar] [CrossRef] [PubMed]
- Slanetz, L.W.; Bartley, C.H. Numbers of enterococci in water, sewage, and feces determined by the membrane filter technique with an improved medium. J. Bacteriol. 1957, 74, 591–595. [Google Scholar] [PubMed]
- Borsheim, K.Y. Native marine bacteriophages. FEMS Microbiol. Ecol. 1993, 102, 141–159. [Google Scholar] [CrossRef]
- Parhad, N.M.; Rao, N.U. Effect of pH on survival of Escherichia coli. J. Water Pollut. Control Fed. 1974, 46, 980–986. [Google Scholar]
- Roux, S.; Krupovic, M.; Poulet, A.; Debroas, D.; Enault, F. Evolution and diversity of the microviridae viral family through a collection of 81 new complete genomes assembled from virome reads. PLoS ONE 2012, 7, e40418. [Google Scholar] [CrossRef] [PubMed]
- Tan, G.H.; Nordin, M.; Napsiah, A.B. Isolation and characterization of lytic bacteriophages from sewage water. J. Trop. Agric. Food Sci. 2008, 36, 1–5. [Google Scholar]
- Wommack, K.E.; Colwell, R.R. Virioplankton: Viruses in aquatic ecosystems. Microbiol. Mol. Biol. Rev. 2000, 64, 69–114. [Google Scholar] [CrossRef] [PubMed]
- Hatfull, G.F.; Hendrix, R.W. Bacteriophages and their genomes. Curr. Opin. Virol. 2011, 1, 298–303. [Google Scholar] [CrossRef] [PubMed]
- Miller, R.V. Environmental bacteriophage-host interactions: Factors contribution to natural transduction. Antonie Van Leeuwenhoek 2001, 79, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Hendrix, R.W. Bacteriophages: Evolution of the majority. Theor. Popul. Biol. 2002, 61, 471–480. [Google Scholar] [CrossRef] [PubMed]
- Brentlinger, K.L.; Hafenstein, S.; Novak, C.R.; Fane, B.A.; Borgon, R.; McKenna, R.; Agbandje-McKenna, M. Microviridae, a family divided: Isolation, characterization, and genome sequence of phimh2k, a bacteriophage of the obligate intracellular parasitic bacterium Bdellovibrio bacteriovorus. J. Bacteriol. 2002, 184, 1089–1094. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Han, Y.; Ye, Y.; Liu, Y.; Jiang, Z.; Gui, Y.; Cai, Z. Chemical synthesis of bacteriophage G4. PLoS ONE 2011, 6, e27062. [Google Scholar] [CrossRef] [PubMed]
- Chan, B.K.; Abedon, S.T. Phage therapy pharmacology: Phage cocktails. Adv. Appl. Microbiol. 2012, 78, 1–23. [Google Scholar] [PubMed]
- Chan, B.K.; Abedon, S.T.; Loc-Carrillo, C. Phage cocktails and the future of phage therapy. Future Microbiol. 2013, 8, 769–783. [Google Scholar] [CrossRef] [PubMed]
- Chibani-Chennoufi, S.; Sidoti, J.; Bruttin, A.; Dillmann, M.L.; Kutter, E.; Qadri, F.; Sarker, S.A.; Brussow, H. Isolation of Escherichia coli bacteriophages from the stool of pediatric diarrhea patients in bangladesh. J. Bacteriol. 2004, 186, 8287–8294. [Google Scholar] [CrossRef] [PubMed]
- Sherman, M.B.; Guenther, R.H.; Tama, F.; Sit, T.L.; Brooks, C.L.; Mikhailov, A.M.; Orlova, E.V.; Baker, T.S.; Lommel, S.A. Removal of divalent cations induces structural transitions in red clover necrotic mosaic virus revealing a potential mechanism for rna release. J. Virol. 2006, 80, 10395–10406. [Google Scholar] [CrossRef] [PubMed]
- Tang, G.; Peng, L.; Baldwin, P.R.; Mann, D.S.; Jiang, W.; Rees, I.; Ludtke, S.J. Eman2: An extensible image processing suite for electron microscopy. J. Struc. Biol. 2007, 157, 38–46. [Google Scholar] [CrossRef] [PubMed]


© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ghosh, S.; Persad, E.; Shiue, T.-Y.; Lam, C.; Islam, A.; Mascibroda, L.G.; Sherman, M.B.; Smith, T.; Cheeptham, N. Explorative Study on Isolation and Characterization of a Microviridae G4 Bacteriophage, EMCL318, against Multi-Drug-resistant Escherichia coli 15-318. Antibiotics 2018, 7, 92. https://doi.org/10.3390/antibiotics7040092
Ghosh S, Persad E, Shiue T-Y, Lam C, Islam A, Mascibroda LG, Sherman MB, Smith T, Cheeptham N. Explorative Study on Isolation and Characterization of a Microviridae G4 Bacteriophage, EMCL318, against Multi-Drug-resistant Escherichia coli 15-318. Antibiotics. 2018; 7(4):92. https://doi.org/10.3390/antibiotics7040092
Chicago/Turabian StyleGhosh, Soumya, Emma Persad, Ting-Yun Shiue, Cindy Lam, Afsana Islam, Lauren G. Mascibroda, Michael B. Sherman, Thomas Smith, and Naowarat Cheeptham. 2018. "Explorative Study on Isolation and Characterization of a Microviridae G4 Bacteriophage, EMCL318, against Multi-Drug-resistant Escherichia coli 15-318" Antibiotics 7, no. 4: 92. https://doi.org/10.3390/antibiotics7040092
APA StyleGhosh, S., Persad, E., Shiue, T.-Y., Lam, C., Islam, A., Mascibroda, L. G., Sherman, M. B., Smith, T., & Cheeptham, N. (2018). Explorative Study on Isolation and Characterization of a Microviridae G4 Bacteriophage, EMCL318, against Multi-Drug-resistant Escherichia coli 15-318. Antibiotics, 7(4), 92. https://doi.org/10.3390/antibiotics7040092