Mining Actinomycetes for Novel Antibiotics in the Omics Era: Are We Ready to Exploit This New Paradigm?
Abstract
1. Introduction
2. Exploiting Diversity of Cultured Actinomycetes
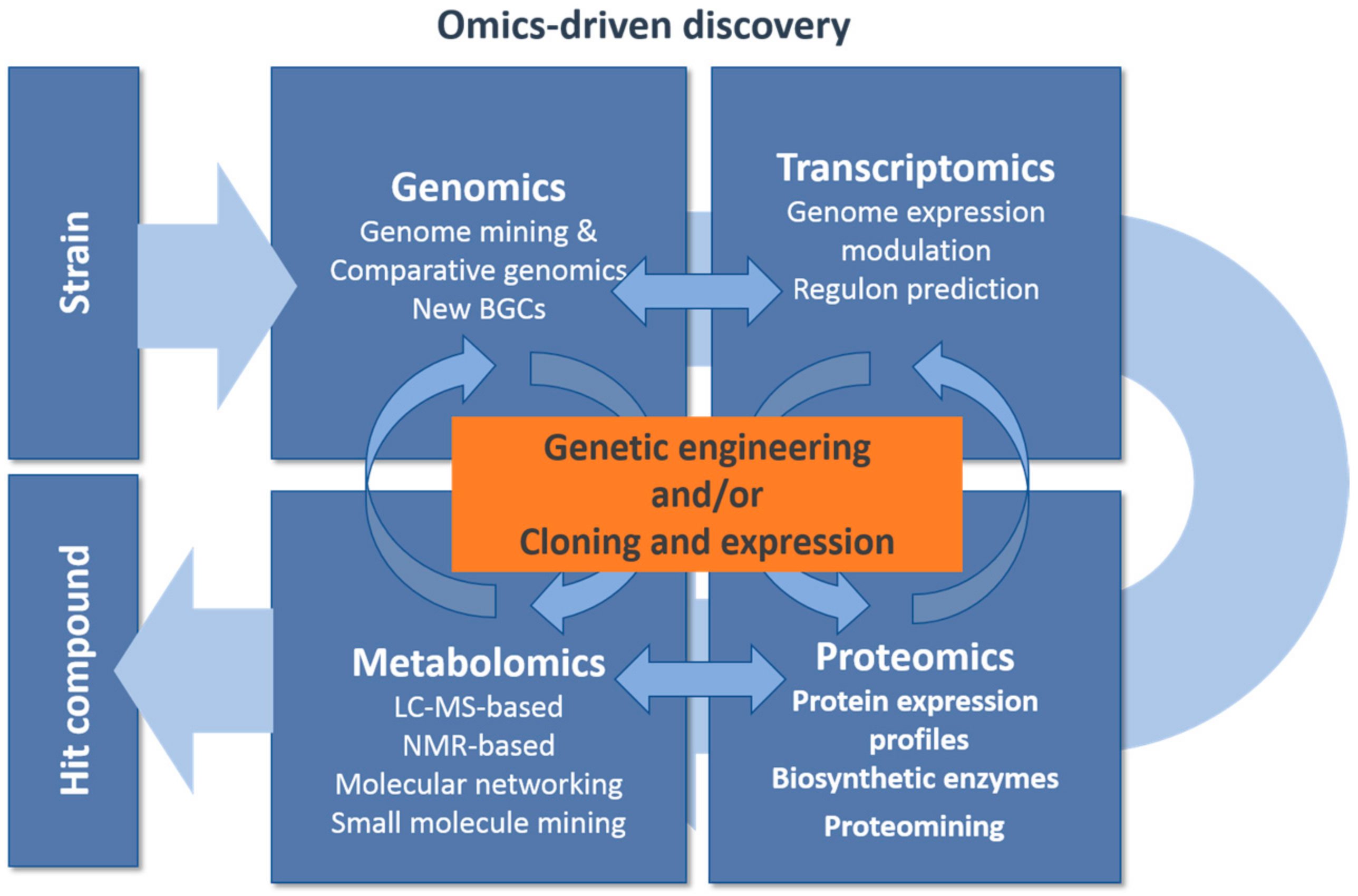
3. Genomics Driven Discovery
4. Eliciting Production from Silent Pathways
5. Harnessing Regulation of Primary and Secondary Metabolisms
6. Conclusions and Future Prospects
Funding
Conflicts of Interest
References
- Berdy, J.J. Thoughts and facts about antibiotics: Where we are now and where we are heading. J. Antibiot. 2012, 65, 385–395. [Google Scholar] [CrossRef] [PubMed]
- Landwehr, W.; Wolf, C.; Wink, J. Actinobacteria and myxobacteria-two of the most important bacterial resources for novel antibiotics. Curr. Top. Microbiol. Immunol. 2016, 398, 273–302. [Google Scholar] [PubMed]
- Fischbach, M.A.; Walsh, C.T. Antibiotics for emerging pathogens. Science 2009, 325, 1089–1093. [Google Scholar] [CrossRef] [PubMed]
- Silver, L.L. Challenges of antibacterial discovery. Clin. Microbiol. Rev. 2011, 24, 71–109. [Google Scholar] [CrossRef] [PubMed]
- Walsh, C.T.; Wencewicz, T.A. Prospects for new antibiotics: A molecule-centered perspective. J. Antibiot. 2014, 67, 7–22. [Google Scholar] [CrossRef] [PubMed]
- Rex, J.H.; Outterson, K. Antibiotic reimbursement in a sales-delinked model: Context and a benchmark-based global approach. Lancet Infect. Dis. 2016, 16, 500–505. [Google Scholar] [CrossRef]
- Editorial. Wanted: A reward for antibiotic development. Nat. Biotechnol. 2018, 36, 555. [Google Scholar] [CrossRef] [PubMed]
- Rex, J.H.; Goldberger, M.; Eisenstein, B.; Harney, C. The evolution of the regulatory framework for antibacterial agents. Ann. N. Y. Acad. Sci. 2014, 1323, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Simpkin, V.L.; Renwick, M.J.; Kelly, R.; Mossialos, E. Incentivising innovation in antibiotic drug discovery and development: Progress, challenges and next steps. J. Antibiot. 2017, 70, 1087–1096. [Google Scholar] [CrossRef] [PubMed]
- Antibiotics Currently in Global Clinical Development. Available online: http://www.pewtrusts.org/-/media/assets/2018/03/antibiotics_clinical_dev_table_february2018.pdf (accessed on 1 February 2018).
- Brown, E.D.; Wright, G.D. Antibacterial drug discovery in the resistance era. Nature 2016, 529, 336–343. [Google Scholar] [CrossRef] [PubMed]
- Payne, D.J.; Gwynn, M.N.; Holmes, D.J.; Pompliano, D.L. Drugs for bad bugs: Confronting the challenges of antibacterial discovery. Nat. Rev. Drug Discov. 2007, 6, 29–40. [Google Scholar] [CrossRef] [PubMed]
- Silver, L.L. Are natural products still the best source for antibacterial discovery? The bacterial entry factor. Expert Opin. Drug Discov. 2008, 3, 487–500. [Google Scholar] [CrossRef] [PubMed]
- Wright, G.D. Something old, something new: Revisiting natural products in antibiotic drug discovery. Can. J. Microbiol. 2014, 60, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Lewis, K. New approaches to antimicrobial discovery. Biochem. Pharmacol. 2017, 134, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Baltz, R.H. Gifted microbes for genome mining and natural product discovery. J. Ind. Microbiol. Biotechnol. 2017, 44, 573–588. [Google Scholar] [CrossRef] [PubMed]
- Nett, M.; Ikeda, H.; Moore, B.S. Genomic basis for natural product biosynthetic diversity in the actinomycetes. Nat. Prod. Rep. 2009, 26, 1362–1384. [Google Scholar] [CrossRef] [PubMed]
- Rutledge, P.J.; Challis, G.L. Discovery of microbial natural products by activation of silent biosynthetic gene clusters. Nat. Rev. Microbiol. 2015, 13, 509–523. [Google Scholar] [CrossRef] [PubMed]
- Genilloud, O. Actinomycetes: Still a source of novel antibiotics. Nat. Prod. Rep. 2017, 34, 1203–1232. [Google Scholar] [CrossRef] [PubMed]
- Katz, L.; Baltz, R.H. Natural product discovery: Past, present, and future. J. Ind. Microbiol. Biotechnol. 2016, 43, 155–176. [Google Scholar] [CrossRef] [PubMed]
- Kealey, C.; Creaven, C.A.; Murphy, C.D.; Brady, C.B. New approaches to antibiotic discovery. Biotechnol. Lett. 2017, 39, 805–817. [Google Scholar] [CrossRef] [PubMed]
- Wencewicz, T.A. New antibiotics from nature’s chemical inventory. Bioorg. Med. Chem. 2016, 24, 6227–6252. [Google Scholar] [CrossRef] [PubMed]
- Bull, A.T.; Asenjo, J.A.; Goodfellow, M.; Gomez-Silva, B. The atacama desert: Technical resources and the growing importance of novel microbial diversity. Ann. Rev. Microbiol. 2016, 70, 215–234. [Google Scholar] [CrossRef] [PubMed]
- Mohammadipanah, F.; Wink, J. Actinobacteria from arid and desert habitats: Diversity and biological activity. Front. Microbiol. 2016, 6, 1541. [Google Scholar] [CrossRef] [PubMed]
- Rateb, M.E.; Ebel, R.; Jaspars, M. Natural product diversity of actinobacteria in the atacama desert. Antonie Van Leeuwenhoek 2018, 111, 1467–1477. [Google Scholar] [CrossRef] [PubMed]
- Abdelkader, M.S.A.; Philippon, T.; Asenjo, J.A.; Bull, A.T.; Goodfellow, M.; Ebel, R.; Jaspars, M.; Rateb, M.E. Asenjonamides A-C, antibacterial metabolites isolated from Streptomyces asenjonii strain KNN 42.f from an extreme-hyper arid Atacama Desert soil. J. Antibiot. 2018, 71, 425–431. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Kuisiene, N.; Cheeptham, N. The cave microbiome as a source for drug discovery: Reality or pipe dream? Biochem. Pharmacol. 2017, 134, 18–34. [Google Scholar] [CrossRef] [PubMed]
- Adam, D.; Maciejewska, M.; Naômé, A.; Martinet, L.; Coppieters, W.; Karim, L.; Baurain, D.; Rigali, S. Isolation, characterization, and antibacterial activity of hard-to-culture actinobacteria from cave moonmilk deposits. Antibiotics 2018, 7, 28–48. [Google Scholar] [CrossRef] [PubMed]
- Gavrish, E.; Sit, C.S.; Cao, S.; Kandror, O.; Spoering, A.; Peoples, A.; Ling, L.A.; Hughes, D.; Bissell, A.; Torrey, H.; et al. Lassomycin, a ribosomally synthesized cyclic peptide, kills mycobacterium tuberculosis by targeting the ATP-dependent protease ClpC1P1P2. Chem. Biol. 2014, 21, 509–518. [Google Scholar] [CrossRef] [PubMed]
- Jang, K.H.; Nam, S.-J.; Locke, J.B.; Kauffman, C.A.; Beatty, D.S.; Paul, L.A.; Fenical, W. Anthracimycin, a potent anthrax antibiotic from a marine-derived actinomycete. Chem. Int. Ed. 2013, 52, 7822–7824. [Google Scholar] [CrossRef] [PubMed]
- Hassan, H.M.; Degen, D.; Jang, K.H.; Ebright, R.H.; Fenical, W.; Salinamide, F. New depsipeptide antibiotic and inhibitor of bacterial RNA polymerase from a marine-derived Streptomyces sp. J. Antibiot. 2015, 68, 206–209. [Google Scholar] [CrossRef] [PubMed]
- Palomo, S.; González, I.; Martín, J.; de la Cruz, M.; Vicente, F.; Reyes, F.; Tormo, J.R.; Anderson, M.; Hill, R.T.O.; Genilloud, O. Sponge-derived Kocuria and Micrococcus spp. as sources of the new thiazolyl peptide antibiotic kocurin. Mar. Drugs 2013, 11, 1071–1086. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Adnani, N.; Braun, D.R.; Ellis, G.A.; Barns, K.J.; Parker-Nance, S.; Guzei, I.A.; Bugni, T.S. Micromonohalimanes A and B: Antibacterial halimane-type diterpenoids from a marine Micromonospora species. J. Nat. Prod. 2016, 79, 2968–2972. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Bonilla, M.; Oves-Costales, D.; de la Cruz, M.; Kokkini, M.; Martín, J.; Vicente, F.; Genilloud, O.; Reyes, F. Phocoenamicins B and C, New antibacterial spirotetronates isolated from a marine Micromonospora sp. Mar. Drugs 2018, 16. [Google Scholar] [CrossRef] [PubMed]
- Ju, K.-S.; Gao, J.; Doroghazi, J.R.; Wang, K.-K.A.; Thibodeaux, C.J.; Lia, S.; Metzger, E.; Fudala, J.; Su, J.; Zhang, J.K.; et al. Discovery of phosphonic acid natural products by mining the genomes of 10,000 actinomycetes. Proc. Natl. Acad. Sci. USA 2015, 112, 12175–12180. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Li, J.; Millán-Aguiñaga, N.; Zhang, J.J.; O’Neill, E.C.; Ugalde, J.A.; Jensen, P.R.; Mantovani, S.M.; Moore, B.S. Identification of thiotetronic acid antibiotic biosynthetic pathways by target-directed genome mining. ACS Chem. Biol. 2015, 10, 2841–2849. [Google Scholar] [CrossRef] [PubMed]
- Yamanaka, K.; Reynolds, K.A.; Kersten, R.D.; Ryan, K.S.; Gonzalez, D.J.; Nizet, V.; Dorrestein, P.C.; Moore, B.S. Direct cloning and refactoring of a silent lipopeptide biosynthetic gene cluster yields the antibiotic taromycin A. Proc. Natl. Acad. Sci. USA 2014, 111, 1957–1962. [Google Scholar] [CrossRef] [PubMed]
- Bonet, B.; Teufel, R.; Cruüsemann, M.; Ziemert, N.; Moore, B.S. Direct capture and heterologous expression of Salinispora natural product genes for the biosynthesis of enterocin. J. Nat. Prod. 2014, 78, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Huang, C.; Zhang, W.; Zhu, Y.; Zhang, C. Heterologous expression of fluostatin gene cluster leads to a bioactive heterodimer. Org. Lett. 2015, 17, 5324–5327. [Google Scholar] [CrossRef] [PubMed]
- Onaka, H. Novel antibiotic screening methods to awaken silent or cryptic secondary metabolic pathways in actinomycetes. J. Antibiot. 2017, 70, 865–870. [Google Scholar] [CrossRef] [PubMed]
- Derewacz, D.K.; Covington, B.C.; McLean, J.A.; Bachmann, B.O. Mapping microbial response metabolomes for induced natural product discovery. ACS Chem. Biol. 2015, 10, 1998–2006. [Google Scholar] [CrossRef] [PubMed]
- Fenical, W.; Jensen, P.R. Developing a new resource for drug discovery: Marine actinomycete bacteria. Nat. Chem. Biol. 2006, 2, 666–673. [Google Scholar] [CrossRef] [PubMed]
- Bull, A.T.; Stach, J.E.M. Marine actinobacteria: New opportunities for natural product search and discovery. Trends Microbiol. 2007, 15, 491–499. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, A.; Naughton, L.M.; Montánchez, I.; Dobson, A.D.W.; Rai, D.K. Current status and future prospects of marine natural products (MNPs) as antimicrobials. Mar. Drugs 2017, 15, 272. [Google Scholar] [CrossRef] [PubMed]
- Dhakal, D.; Pokhrel, A.R.; Shrestha, B.; Sohng, J.K. Marine rare actinobacteria: Isolation, characterization, and strategies for harnessing bioactive compounds. Front. Microbiol. 2017, 8, 1106. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Han, L.; Li, C.; Cao, Q.; Zhu, D.; Barrett, N.H.; Harmody, D.; Chen, J.; Zhu, H.; McCarthy, P.J.; et al. Bioprospecting deep-sea actinobacteria for novel anti-infective natural products. Front. Microbiol. 2018, 9, 787. [Google Scholar] [CrossRef] [PubMed]
- Jose, P.A.; Jha, B. Intertidal marine sediment harbours actinobacteria with promising bioactive and biosynthetic potential. Sci. Rep. 2017, 7, 10041. [Google Scholar] [CrossRef] [PubMed]
- Sarmiento-Vizcaíno, A.; González, V.; Braña, A.F.; Palacios, J.J.; Otero, L.; Fernández, J.; Molina, A.; Kulik, A.; Vázquez, F.; Acuña, J.L.; et al. Pharmacological potential of phylogenetically diverse actinobacteria isolated from deep-sea coral ecosystems of the submarine Avilés canyon in the cantabrian sea. Microb. Ecol. 2017, 73, 338–352. [Google Scholar] [CrossRef] [PubMed]
- Patin, N.V.; Schorn, M.; Aguinaldo, K.; Lincecum, T.; Moore, B.S.; Jensen, P.R. Effects of actinomycete secondary metabolites on sediment microbial communities. Appl. Environ. Microbiol. 2017, 83, e02676-16. [Google Scholar] [CrossRef] [PubMed]
- Medema, M.H.; Fischbach, M.A. Computational approaches to natural product discovery. Nat. Chem. Biol. 2015, 11, 639–648. [Google Scholar] [CrossRef] [PubMed]
- Weber, T.; Kim, H.U. The secondary metabolite bioinformatics portal: Computational tools to facilitate synthetic biology of secondary metabolite production. Synth. Syst. Biotechnol. 2016, 1, 69–79. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Medema, M.H.; Kottmann, R.; Lee, S.Y.; Weber, T. The antiSMASH database, a comprehensive database of microbial secondary metabolite biosynthetic gene clusters. Nucleic Acids Res. 2017, 45, D555–D559. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Hidalgo, M.; González, I.; Díaz-Muñoz, C.; Martínez, G.; Genilloud, O. Comparative Genomics and Biosynthetic Potential Analysis of Two Lichen-Isolated Amycolatopsis Strains. Front. Microbiol. 2018, 9, 369. [Google Scholar] [CrossRef] [PubMed]
- Adamek, M.; Alanjary, M.; Sales-Ortells, H.; Goodfellow, M.; Bull, A.T.; Winkler, A.; Wibberg, D.; Kalinowski, J.; Ziemert, N. Comparative genomics reveals phylogenetic distribution patterns of secondary metabolites in Amycolatopsis species. BMC Genomics 2018, 19, 426. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Gu, J.; Li, Y.-Q.; Wang, Y. Genome plasticity and systems evolution in Streptomyces. BMC Bioinform. 2012, 13, S8. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-N.; Kim, Y.; Jeong, Y.; Roe, J.-H.; Kim, B.-G.; Cho, B.-K. Comparative Genomics Reveals the Core and Accessory Genomes of Streptomyces Species. J. Microbiol. Biotechnol. 2015, 25, 1599–1605. [Google Scholar] [CrossRef] [PubMed]
- Choudoir, M.J.; Pepe-Ranney, C.; Buckley, D.H. Diversification of Secondary Metabolite Biosynthetic Gene Clusters Coincides with Lineage Divergence in Streptomyces. Antibiotics 2018, 7, 12. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.M.; Wang, Y.; Ang, E.L.; Zhao, H. Engineering microbial hosts for production of bacterial natural products. Nat. Prod. Rep. 2016, 33, 963–987. [Google Scholar] [CrossRef] [PubMed]
- Myronovskyia, M.; Luzhetskyy, A. Native and engineered promoters in natural product discovery. Nat. Prod. Rep. 2016, 33, 1006–1020. [Google Scholar] [CrossRef] [PubMed]
- Weber, T.; Charusanti, P.; Musiol-Kroll, E.M.; Jiang, X.; Tong, Y.; Hyun Uk Kim, H.U.; Lee, S.Y. Metabolic engineering of antibiotic factories: New tools for antibiotic production in actinomycetes. Trends Biotechnol. 2015, 33, 15–26. [Google Scholar] [CrossRef] [PubMed]
- Ren, H.; Wang, B.; Zhao, H. Breaking the silence: New strategies for discovering novel natural products. Curr. Opin. Biotechnol. 2017, 48, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zhenga, G.; Chena, J.; Gec, M.; Jianga, W.; Lu, Y. New strategies and approaches for engineering biosynthetic gene clusters of microbial natural products. Metab. Eng. 2017, 40, 80–92. [Google Scholar] [CrossRef] [PubMed]
- Tong, Y.; Robertsen, H.L.; Blin, K.; Weber, T.; Lee, S.Y. CRISPR-Cas9 Toolkit for Actinomycete Genome Editing. In Synthetic Metabolic Pathways; Humana Press: New York, NY, USA, 2018; pp. 163–184. [Google Scholar]
- Xu, M.; Wang, Y.; Zhao, Z.; Gao, G.; Huang, S.-X.; Kang, Q.; He, X.; Lin, S.; Pang, X.; Deng, Z.; et al. Functional genome mining for metabolites encoded by large gene clusters through heterologous expression of a whole-genome bacterial artificial chromosome library in Streptomyces spp. Appl. Environ. Microbiol. 2016, 82, 5795–5805. [Google Scholar] [CrossRef] [PubMed]
- Arakawa, K. Manipulation of metabolic pathways controlled by signaling molecules, inducers of antibiotic production, for genome mining in Streptomyces spp. Antonie van Leeuwenhoek 2018, 111, 743–751. [Google Scholar] [CrossRef] [PubMed]
- Baltz, R.H. Genetic manipulation of secondary metabolite biosynthesis for improved production in Streptomyces and other actinomycetes. J. Ind. Microbiol. Biotechnol. 2016, 43, 343–370. [Google Scholar] [CrossRef] [PubMed]
- Hwang, K.-S.; Kim, H.U.; Charusanti, P.; Palsson, B.O.; Lee, S.Y. Systems biology and biotechnology of Streptomyces species for the production of secondary metabolites. Biotechnol. Adv. 2014, 32, 255–268. [Google Scholar] [CrossRef] [PubMed]
- Horbal, L.; Siegl, T.; Luzhetskyy, A. A set of synthetic versatile genetic control elements for the efficient expression of genes in Actinobacteria. Sci. Rep. 2018, 8, 491. [Google Scholar] [CrossRef] [PubMed]
- Goh, E.B.; Yim, G.; Tsui, W.; McClure, J.; Surette, M.G.; Davies, J. Transcriptional modulation of bacterial gene expression by subinhibitory concentrations of antibiotics Proc. Natl. Acad. Sci. USA 2002, 99, 17025–17030. [Google Scholar] [CrossRef] [PubMed]
- Rosen, P.C.; Seyedsayamdost, M.R. Though much is taken, much abides: Finding new antibiotics using old ones. Biochemistry 2017, 56, 4925–4926. [Google Scholar] [CrossRef] [PubMed]
- Okada, B.K.; Seyedsayamdost, M.R. Antibiotic dialogues: Induction of silent biosynthetic gene clusters by exogenous small molecules. FEMS Microbiol. Rev. 2017, 41, 19–33. [Google Scholar] [CrossRef] [PubMed]
- Goodwin, C.R.; Covington, B.C.; Derewacz, D.K.; McNees, C.R.; Wikswo, J.P.; McLean, J.A.; Bachmann, B.O. Structuring microbial metabolic responses to multiplexed stimuli via self-organizing metabolomics maps. Chem. Biol. 2015, 22, 661–670. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with GNPS. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Victoria, I.; Martín, J.; Reyes, F. Combined LC/UV/MS and NMR strategies for the dereplication of marine natural products. Planta Med. 2016, 82, 857–871. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Du, C.; Ichinose, K.; Choi, Y.H.; van Wezel, G.P. Discovery of C-Glycosylpyranonaphthoquinones in Streptomyces sp. MBT76 by a Combined NMR-Based Metabolomics and Bioinformatics Workflow. J. Nat. Prod. 2017, 80, 269. [Google Scholar] [CrossRef] [PubMed]
- Gubbens, J.; Zhu, H.; Girard, G.; Song, L.; Florea, B.I.; Aston, P.; Ichinose, K.; Filippov, D.V.; Choi, Y.H.; Overkleeft, H.S.; et al. Natural product proteomining, a quantitative proteomics platform, allows rapid discovery of biosynthetic gene clusters for different classes of natural products. Chem. Biol. 2014, 21, 707. [Google Scholar] [CrossRef] [PubMed]
- Chao Du, C.; van Wezel, G.P. Mining for microbial gems: Integrating proteomics in the postgenomic natural product discovery pipeline. Proteomics 2018. [Google Scholar] [CrossRef]
- Hou, B.; Lin, Y.; Wu, H.; Guo, M.; Petkovic, H.; Tao, L.; Zhu, X.; Ye, J.; Zhang, H. The novel transcriptional regulator LmbU promotes lincomycin biosynthesis through regulating expression of its target genes in Streptomyces lincolnensis. J. Bacteriol. 2018, 200, e00447-17. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Chater, K.F.; Chandra, G.; Niu, G.; Tan, H. Molecular regulation of antibiotic biosynthesis in Streptomyces. Microbiol. Mol. Biol. Rev. 2013, 77, 112–143. [Google Scholar] [CrossRef] [PubMed]
- Van Wezel, G.P.; Mcdowall, K.J. The regulation of the secondary metabolism of Streptomyces: New links and experimental advances. Nat. Prod. Rep. 2011, 28, 1311–1333. [Google Scholar] [CrossRef] [PubMed]
- Hoskisson, P.A.; Fernández-Martínez, L.T. Regulation of specialised metabolites in actinobacteria–expanding the paradigms. Environ. Microbiol. Rep. 2018, 10, 231–238. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.J.; Grant, C.E.; Noble, W.S. The MEME suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef] [PubMed]
- Rigali, S.; Anderssen, S.; Naômé, A.; van Wezel, G.P. Cracking the regulatory code of biosynthetic gene clusters as a strategy for natural product discovery. Biochem. Pharmacol. 2018, 153, 24–34. [Google Scholar] [CrossRef] [PubMed]
- Hiard, S.; Marée, R.; Colson, S.; Hoskisson, P.A.; Titgemeyer, F.; van Wezel, G.P.B.; Wehenkel, J.L.; Rigali, S. PREDetector: A new tool to identify regulatory elements in bacterial genomes. Biochem. Biophys. Res. Commun. 2007, 357, 861–864. [Google Scholar] [CrossRef] [PubMed]
- Rigali, S.; Titgemeyer, F.; Barends, S.; Mulder, S.; Thomae, A.W.; Hopwood, D.A.; van Wezel, G.P. Feast or famine: The global regulator DasR links nutrient stress to antibiotic production by Streptomyces. EMBO Rep. 2008, 9, 670–675. [Google Scholar] [CrossRef] [PubMed]
- Craig, M.; Lambert, S.; Jourdan, S.; Tenconi, E.; Colson, S.; Maciejewska, M.; Ongena, M.; Martin, J.F.; van Wezel, G.; Rigali, S. Unsuspected control of siderophore production by N-acetylglucosamine in streptomycetes. Environ. Microbiol. Rep. 2012, 4, 512–521. [Google Scholar] [CrossRef] [PubMed]
- Jourdan, S.; Francis, I.M.; Kim, M.J.; Salazar, J.J.C.; Planckaert, S.; Frère, J.-M.; Matagne, A.; Kerff, F.; Devreese, B.; Loria, R.; et al. The CebE/MsiK transporter is a doorway to the cello-oligosaccharide-mediated Induction of Streptomyces scabies pathogenicity. Sci. Rep. 2016, 6, 27144. [Google Scholar] [CrossRef] [PubMed]
- Schniete, J.K.; Cruz-Morales, P.; Selem-Mojica, N.; Fernández-Martínez, L.T.; Hunter, I.S.; Barona-Gómez, F.; Hoskisson, P.A. Expanding primary metabolism helps generatethe metabolic robustness to facilitate antibioticbiosynthesis in Streptomyces. mBio 2018, 9, e02283-17. [Google Scholar] [CrossRef] [PubMed]
- Craney, A.; Ozimok, C.; Pimentel-Elardo, S.M.; Capretta, A.; Nodwell, J.R. Chemical perturbation of secondary metabolism demonstrates important links to primary metabolism. Chem. Biol. 2012, 19, 1020–1027. [Google Scholar] [CrossRef] [PubMed]
- Seyedsayamdost, M.R. High-throughput platform for the discovery of elicitors of silent bacterial gene clusters. Proc. Natl. Acad. Sci. USA 2014, 111, 7266–7271. [Google Scholar] [CrossRef] [PubMed]
- Adnani, N.; Chevrette, M.G.; Adibhatla, S.N.; Zhang, F.; Yu, Q.; Braun, D.R.; Nelson, J.; Simpkins, S.W.; McDonald, B.R.; Myers, C.L.; et al. Coculture of marine invertebrate-associated bacteria and interdisciplinary technologies enable biosynthesis and discovery of a new antibiotic, keyicin. ACS Chem. Biol. 2017, 12, 3093–3102. [Google Scholar] [CrossRef] [PubMed]
- Sengesa, C.H.R.; Al-Dilaimib, A.; Marchbankc, D.H.; Wibbergb, D.; Winklerb, A.; Haltlic, B.; Nowrousiand, M.; Kalinowskib, J.; Kerrc, R.G.; Bandow, J.E. The secreted metabolome of Streptomyces chartreusis and implications for bacterial chemistry. Proc. Natl. Acad. Sci. USA 2018, 115, 2490–2495. [Google Scholar] [CrossRef] [PubMed]
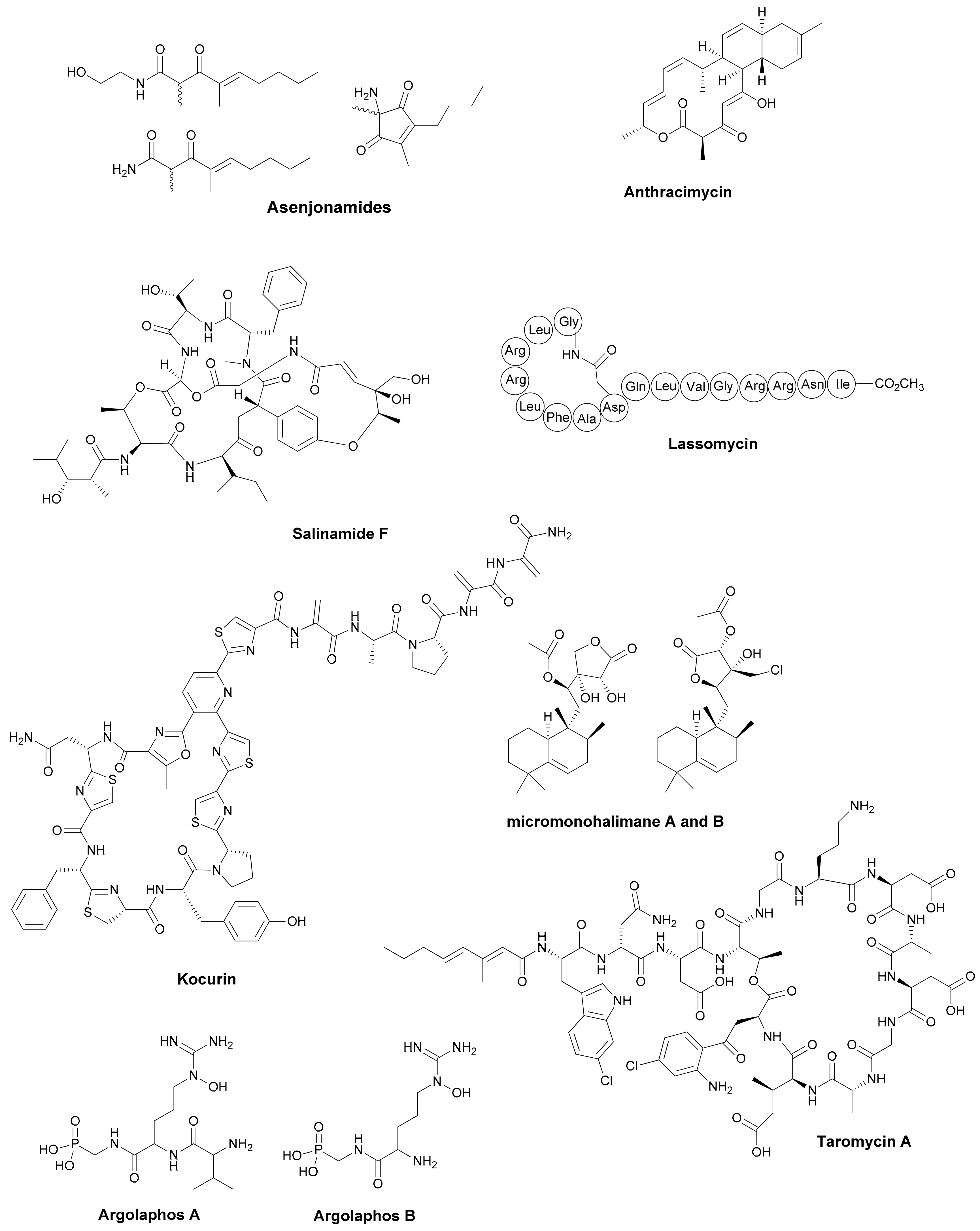
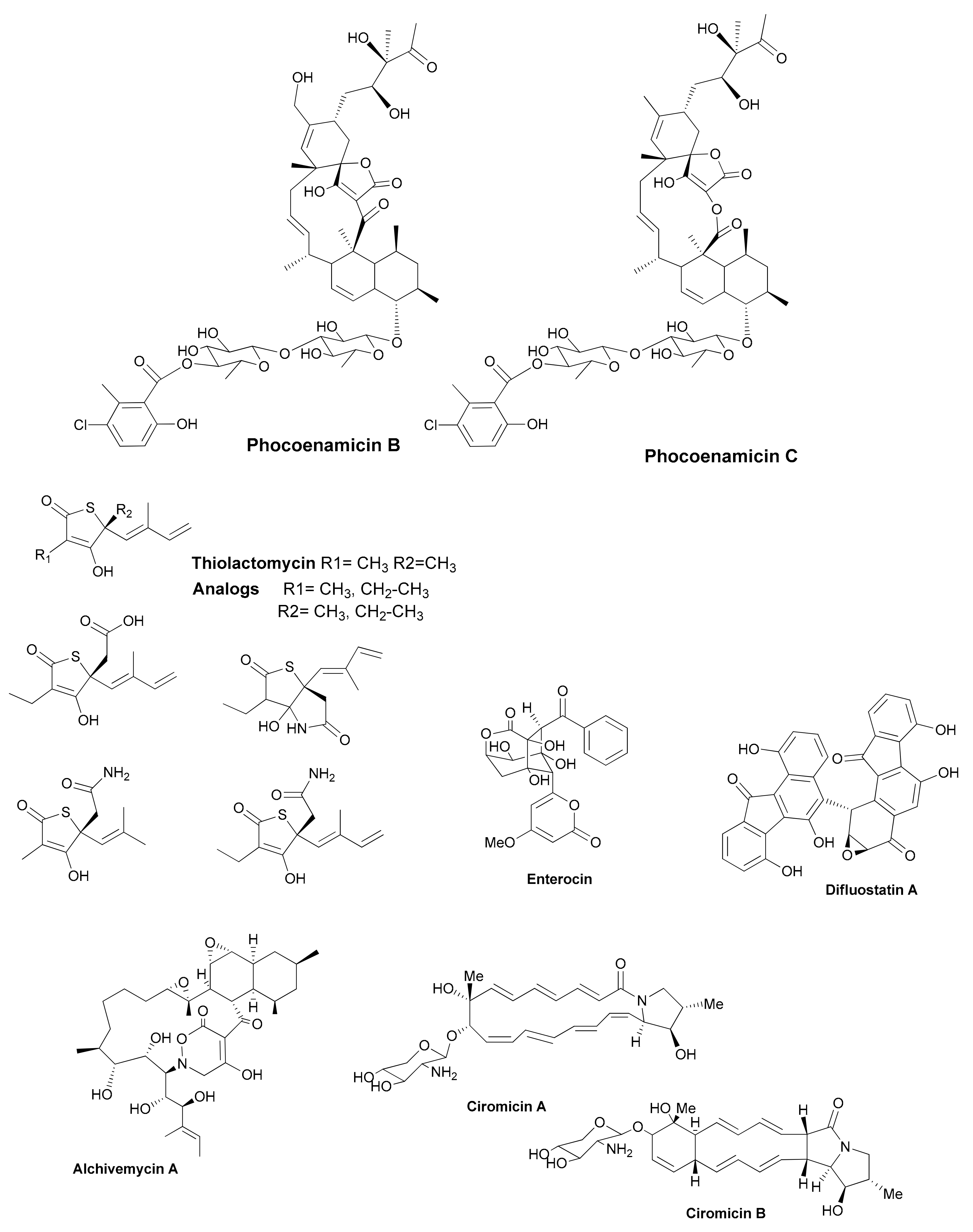
| Antibiotic | Structural Class | Producing Species | Antibiotic Spectrum | Discovery Approach | Reference |
|---|---|---|---|---|---|
| Asenjonamides A–C | di-ketone polyketides | S. asenjonii | Gram positive/ negative | Extreme environment | [26] |
| Lassomycin | cyclic peptide | Lentzea kentukyensis | M. tuberculosis | Diffusion chambers | [29] |
| Anthracimycin | tricarboxilic | Streptomyces sp. | Gram positive | Marine source | [30] |
| Salinamide F | depsipeptide | Streptomyces sp. | Gram positive/ negative | Marine source | [31] |
| Kocurin | thiazolylpeptide | Kocuria lacustris /Micrococcus sp. | Gram positive | Marine source | [32] |
| Micromonohalimanes A and B | diterpenoids | Micromonospora sp. | Gram positive | Marine source | [33] |
| Phocoenamicins B and C | spirotetronates | Micromonospora sp. | Gram positive | Marine source | [34] |
| Argolaphos A/B | phosphonopeptide | Streptomyces monomycini | Gram positive/negative | Genome-driven | [35] |
| Thiolactomycin and analogs | thiotetronic acids | Salinispora/S. afghaniensis | Gram positive | Genome-driven | [36] |
| taromycin A | lipopetide | Saccharomonospora sp. | Gram positive | Genome-driven | [37] |
| Enterocin | polyketide | Salinispora pacifica | Gram positive | Genome-driven | [38] |
| Difluostatin A | angucycline | Micromonospora rosaria | Gram positive | Genome-driven | [39] |
| Alchivemycin A and B | heterocyclic | S. endus + Tsukamurella pulmonis | Gram positive | Co-cultivation | [40] |
| Ciromicins | polyketide | Nocardiopsis sp. + Rhodococcus wratislaviensis | Not determined | Co-cultivation | [41] |
| Eliciting Production Approaches | Methods and Targets | References |
|---|---|---|
| Genetic engineering: | Genome expression modulation: Transcriptional repressors inactivation; Transcriptional activators overexpression Optimized ribosomal binding sequences Strong terminators Increase BGCs copies Alter metabolic fluxes | [63,65,66,67,68] |
| Culture-based approaches: | Small molecule signaling: Sub-inhibitory small molecule elicitors Co-cultivation: cell–cell signaling | [69,70][71] |
| Analytical mining: | Comparative metabolomics: LC-MS-based metabolomics NMR-based metabolomics Proteomining | [72,73,74,75][75][76,77] |
| Regulation of primary and secondary metabolism | Pathway specific regulatory elements: Two-component systems Sigma factors Pathways specific regulators | [78,79,80] |
| Global regulatory metabolic networks Master regulators | [81] | |
| Comparative genomics: Identification of Transcription factor orthologs | [82] | |
| Regulon prediction: identification of regulatory networks Primary metabolism transcription factors Signaling cascades Pleiotropic regulators | [83,84][85,86,87] | |
| Primary metabolism gene expansion | [88] |
© 2018 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Genilloud, O. Mining Actinomycetes for Novel Antibiotics in the Omics Era: Are We Ready to Exploit This New Paradigm? Antibiotics 2018, 7, 85. https://doi.org/10.3390/antibiotics7040085
Genilloud O. Mining Actinomycetes for Novel Antibiotics in the Omics Era: Are We Ready to Exploit This New Paradigm? Antibiotics. 2018; 7(4):85. https://doi.org/10.3390/antibiotics7040085
Chicago/Turabian StyleGenilloud, Olga. 2018. "Mining Actinomycetes for Novel Antibiotics in the Omics Era: Are We Ready to Exploit This New Paradigm?" Antibiotics 7, no. 4: 85. https://doi.org/10.3390/antibiotics7040085
APA StyleGenilloud, O. (2018). Mining Actinomycetes for Novel Antibiotics in the Omics Era: Are We Ready to Exploit This New Paradigm? Antibiotics, 7(4), 85. https://doi.org/10.3390/antibiotics7040085





