Where Did They Come from—Multi-Drug Resistant Pathogenic Escherichia coli in a Cemetery Environment?
Abstract
1. Introduction
2. Results
2.1. Enumeration of Escherichia coli
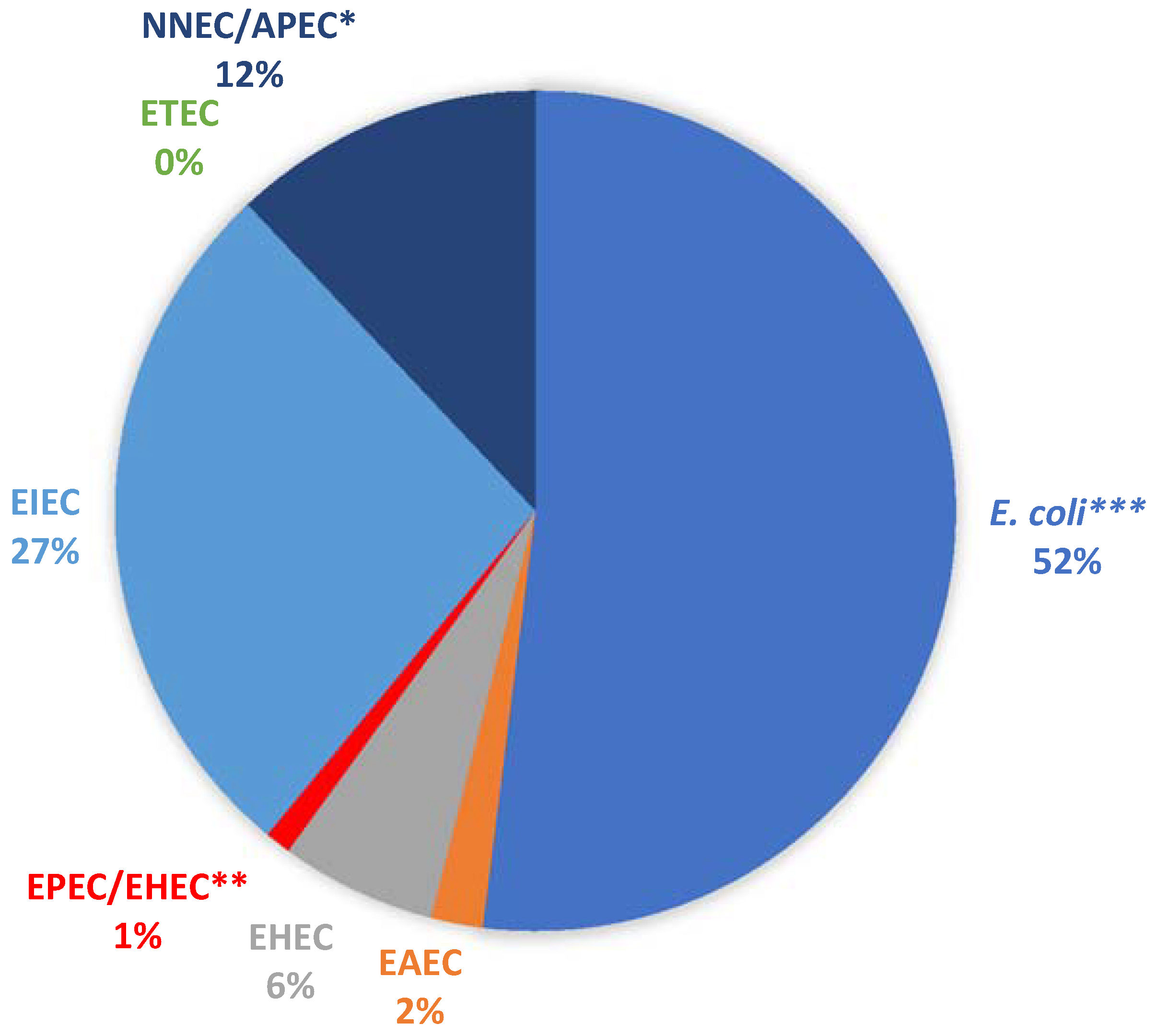
2.2. Virulence Potentials of Isolates
2.3. Antibiotic Resistance Profiles of Isolates
3. Discussion
3.1. Enumeration of Escherichia coli
3.2. Virulence Potentials of Isolates
3.3. Antibiotic Resistance Profiles of Isolates
4. Materials and Methods
4.1. Study Area
4.2. Sample Collection
4.3. Enumeration of Escherichia coli
4.4. Determination of Virulence Potentials of Isolates
4.5. Determination of Antibiotic Resistance Profiles
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Borel, N.; Leonard, C.; Slade, J.; Schoborg, R.V. Chlamydial antibiotic resistance and treatment failure in veterinary and human medicine. Curr. Clin. Microbiol. Rep. 2016, 3, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Mauldin, P.D.; Salgado, C.D.; Hansen, I.S.; Durup, D.T.; Bosso, J.A. Attributable hospital cost and length of stay associated with healthcare-associated infections caused by antibiotic-resistant gram-negative bacteria. Antimicrob. Agents Chemother. 2010, 54, 109–115. [Google Scholar] [CrossRef] [PubMed]
- De Kraker, M.E.A.; Stewardson, A.J.; Harbarth, S. Will 10 million people die a year due to antimicrobial resistance by 2050? PLoS Med. 2016, 13, e1002184. [Google Scholar] [CrossRef] [PubMed]
- Alanis, A.J. Resistance to antibiotics: Are we in the post-antibiotic era? Arch. Med. Res. 2005, 36, 697–705. [Google Scholar] [CrossRef] [PubMed]
- Adesoji, A.T.; Ogunjobi, A.A.; Olatoye, I.O.; Douglas, D.R. Prevalence of tetracycline resistance genes among multi-drug resistant bacteria from selected water distribution systems in southwestern Nigeria. Ann. Clin. Microbiol. Antimicrob. 2015, 14, 35. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, A.M.; de Andrade, D.; Rigotti, M.A.; de Almeida, M.T.G. Methicillin-resistant Staphylococcus aureus on surfaces of an intensive care unit. Acta Paul. Enferm. 2011, 24, 453–458. [Google Scholar] [CrossRef]
- Worthington, R.J.; Melander, C. Combination approaches to combat multi-drug resistant bacteria. Trends Biotechnol. 2014, 31, 177–184. [Google Scholar] [CrossRef] [PubMed]
- Fischbach, M.A. Combination therapies for combating antimicrobial resistance. Curr. Opin. Microbiol. 2011, 14, 519–523. [Google Scholar] [CrossRef] [PubMed]
- Stapleton, P.; Wu, P.J.; King, A.; Shannon, K.; French, G.; Phillips, I. Incidence and mechanisms of resistance to the combination of amoxicillin and clavulanic acid in Escherichia coli. Antimicrob. Agents Chemother. 1995, 39, 2478–2483. [Google Scholar] [CrossRef] [PubMed]
- Rahnama, M.S.; Wagenvoort, J.H.T.; van der Linden, C.J. Amoxicillin/Clavulanate (Augmentin®) resistant Escherichia coli in bacterial peritonitis after abdominal surgery—Clinical outcome in ICU patients. Neth. J. Med. 2009, 67, 173–176. [Google Scholar]
- Xu, J.; Xu, Y.; Wang, H.; Guo, C.; Qiu, H.; He, Y.; Zhang, Y.; Li, X.; Meng, W. Occurrence of antibiotics and antibiotic resistance genes in a sewage treatment plant and its effluent-receiving river. Chemosphere 2015, 119, 1379–1385. [Google Scholar] [CrossRef] [PubMed]
- Lau, C.H.F.; Li, B.; Zhang, T.; Tien, Y.C.; Scott, A.; Murray, R.; Sabourin, L.; Lapen, D.R.; Duenk, P.; Topp, E. Impact of pre-application treatment on municipal sludge composition, soil dynamics of antibiotic resistance genes, and abundance of antibiotic-resistance genes on vegetables at harvest. Sci. Total Environ. 2017, 587–588, 214–222. [Google Scholar] [CrossRef] [PubMed]
- Tien, Y.C.; Li, B.; Zhang, T.; Scott, A.; Murray, R.; Sabourin, L.; Marti, R.; Topp, E. Impact of dairy manure pre-application treatment on manure composition, soil dynamics of antibiotic resistance genes, and abundance of antibiotic-resistance genes on vegetables at harvest. Sci. Total Environ. 2017, 581–582, 32–39. [Google Scholar] [CrossRef] [PubMed]
- Sinthuchai, D.; Boontanon, S.K.; Boontanon, N.; Polprasert, C. Evaluation of removal efficiency of human antibiotics in wastewater treatment plants in Bangkok, Thailand. Water Sci. Technol. 2016, 73, 182–191. [Google Scholar] [CrossRef] [PubMed]
- Kohanski, M.A.; DePristo, M.A.; Collins, J.J. Sublethal antibiotic treatment leads to multidrug resistance via radical-induced mutagenesis. Mol. Cell 2010, 37, 311–320. [Google Scholar] [CrossRef] [PubMed]
- Jørgensen, K.M.; Wassermann, T.; Jensen, P.Ø.; Hengzuang, W.; Molin, S.; Høiby, N.; Ciofu, O. Sublethal ciprofloxacin treatment leads to rapid development of high-level ciprofloxacin resistance during long-term experimental evolution of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2013, 57, 4215–4221. [Google Scholar] [CrossRef] [PubMed]
- Abraham, W.-R. Megacities as sources for pathogenic bacteria in rivers and their fate downstream. Int. J. Microbiol. 2011, 2011, 798292. [Google Scholar] [CrossRef] [PubMed]
- Croxen, M.A.; Law, R.J.; Scholz, R.; Keeney, K.M.; Wlodarska, M.; Finlay, B.B. Recent advances in understanding enteric pathogenic Escherichia coli. Clin. Microbiol. Rev. 2013, 26, 822–880. [Google Scholar] [CrossRef] [PubMed]
- Luna, G.M.; Vignaroli, C.; Rinaldi, C.; Pusceddu, A.; Nicoletti, L.; Gabellini, M.; Danovaro, R.; Biavasco, F. Extraintestinal Escherichia coli carrying virulence genes in coastal marine sediments. Appl. Environ. Microbiol. 2010, 76, 5659–5668. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention (CDCP). Two Multistate Outbreaks of Shiga Toxin-Producing Escherichia coli Infections Linked to Beef from a Single Slaughter Facility—United States, 2008. Morb. Mortal. Wkly. Rep. 2010, 59, 557–560. [Google Scholar]
- Sartz, L.; De jong, B.; Hjertqvist, M.; Plym-forshell, L.; Alsterlund, R.; Löfdahl, S.; Osterman, B.; Ståhl, A.; Eriksson, E.; Hansson, H.-B.; et al. An outbreak of Escherichia coli O157:H7 infection in southern Sweden associated with consumption of fermented sausage; aspects of sausage production that increase the risk of contamination. Epidemiol. Infect. 2008, 136, 370–380. [Google Scholar] [CrossRef] [PubMed]
- Bamford, C.; Bonorchis, K.; Ryan, A.; Hoffmann, R.; Naicker, P.; Maloba, M.; Nana, T.; Zietsman, I.; Govind, C. Antimicrobial susceptibility patterns of Escherichia coli strains isolated from urine samples in South Africa from 2007–2011. S. Afr. J. Epidemiol. Infect. 2012, 27, 46–52. [Google Scholar] [CrossRef]
- Ibrahim, M.E.; Bilal, N.E.; Hamid, M.E. Increased multi-drug resistant Escherichia coli from hospitals in Khartoum state, Sudan. Afr. Health Sci. 2012, 12, 368–375. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, B.; Quinteiro, P.; Caetano, C.; Nadais, H.; Arroja, L.; Ferreira da Silva, E.; Senos Matias, M. Burial grounds’ impact on groundwater and public health: An overview. Water Environ. J. 2013, 27, 99–106. [Google Scholar] [CrossRef]
- Sofield, C.M. Living with the dead: Human burials in anglo-saxon settlement contexts. Archaeol. J. 2015, 172, 351–388. [Google Scholar] [CrossRef]
- Uslu, A.; Bari, E.; Erdo, E. Ecological concerns over cemeteries. Afr. J. Agric. Res. 2009, 4, 1505–1511. [Google Scholar]
- Całkosiński, I.; Płoneczka-Janeczko, K.; Ostapska, M.; Dudek, K.; Gamian, A.; Rypuła, K. Microbiological analysis of necrosols collected from urban cemeteries in Poland. BioMed Res. Int. 2015, 2015, 169573. [Google Scholar] [CrossRef] [PubMed]
- Rabe, L. Grave matters: Cemeteries as cultural landscapes—The German cemetery in Philippi. S. Afr. J. Cult. Hist. 2016, 30, 23–41. [Google Scholar]
- Van Der Hoven, C.; Ubomba-Jaswa, E.; van der Merwe, B.; Loubser, M.; Abia, A.L.K. The impact of various land uses on the microbial and physicochemical quality of surface water bodies in developing countries: Prioritisation of water resources management areas. Environ. Nanotechnol. Monit. Manag. 2017, 8, 280–289. [Google Scholar] [CrossRef]
- Hammerum, A.M.; Heuer, O.E. Human health hazards from antimicrobial-resistant Escherichia coli of animal origin. Clin. Infect. Dis. 2009, 48, 916–921. [Google Scholar] [CrossRef] [PubMed]
- Menrath, A.; Wieler, L.H.; Heidemanns, K.; Semmler, T.; Fruth, A.; Kemper, N. Shiga toxin producing Escherichia coli: Identification of non-O157:H7-Super-Shedding cows and related risk factors. Gut Pathog. 2010, 2, 7. [Google Scholar] [CrossRef] [PubMed]
- Morato, E.P.; Leomil, L.; Beutin, L.; Krause, G.; Moura, R.A.; Pestana de Castro, A.F. Domestic cats constitute a natural reservoir of human enteropathogenic Escherichia coli types. Zoonoses Public Health 2009, 56, 229–237. [Google Scholar] [CrossRef] [PubMed]
- Żychowski, J.; Bryndal, T. Impact of cemeteries on groundwater contamination by bacteria and viruses—A review. J. Water Health 2015, 13, 285–301. [Google Scholar] [CrossRef] [PubMed]
- Semadeni-Davies, A. Implications of climate and urban development on the design of sustainable urban drainage systems (SUDS). J. Water Clim. Chang. 2012, 3, 239–256. [Google Scholar] [CrossRef]
- Abayomi, A.; Odiri, E. Embalmment: A veritable source of human body. Anat. J. Afr. 2017, 6, 995–999. [Google Scholar]
- Marais-Werner, A.; Myburgh, J.; Meyer, A.; Nienaber, W.C.; Steyn, M. Decomposition patterns of buried remains at different intervals in the Central Highveld region of South Africa. Med. Sci. Law 2017, 57, 115–123. [Google Scholar] [CrossRef] [PubMed]
- DeBruyn, J.M.; Hauther, K.A. Postmortem succession of gut microbial communities in deceased human subjects. PeerJ 2017, 5, e3437. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.B.; Dupont, H.L. Enteroaggregative Escherichia coli: An emerging pathogen in children. Semin. Pediatr. Infect. Dis. 2004, 15, 266–271. [Google Scholar] [CrossRef] [PubMed]
- Kaur, P.; Chakraborti, A.; Asea, A. Enteroaggregative Escherichia coli: An emerging enteric foodborne pathogen. Interdiscip. Perspect. Infect. Dis. 2010, 2010, 254159. [Google Scholar] [CrossRef] [PubMed]
- Edge, T.A.; Khan, I.U.H.; Bouchard, R.; Guo, J.; Hill, S.; Locas, A.; Moore, L.; Neumann, N.; Nowak, E.; Payment, P.; et al. Occurrence of waterborne pathogens and Escherichia coli at offshore drinking water intakes in Lake Ontario. Appl. Environ. Microbiol. 2013, 79, 5799–5813. [Google Scholar] [CrossRef] [PubMed]
- Walters, S.P.; Thebo, A.L.; Boehm, A.B. Impact of urbanization and agriculture on the occurrence of bacterial pathogens and stx genes in coastal waterbodies of central California. Water Res. 2010, 45, 1752–1762. [Google Scholar] [CrossRef] [PubMed]
- O’Dwyer, J.; Hynds, P.; Pot, M.; Adley, C.C.; Ryan, M.P. Evaluation of levels of antibiotic resistance in groundwater-derived E. coli isolates in the Midwest of Ireland and elucidation of potential predictors of resistance. Hydrogeol. J. 2017, 25, 939–951. [Google Scholar] [CrossRef]
- De Giglio, O.; Caggiano, G.; Bagordo, F.; Barbuti, G.; Brigida, S.; Lugoli, F.; Grassi, T.; La Rosa, G.; Lucentini, L.; Uricchio, V.F.; et al. Enteric viruses and fecal bacteria indicators to assess groundwater quality and suitability for irrigation. Int. J. Environ. Res. Public Health 2017, 14, 558. [Google Scholar] [CrossRef] [PubMed]
- Bolin, I.; Wiklund, G.; Qadri, F.; Torres, O.; Bourgeois, A.L.; Savarino, S.; Svennerholm, A.-M. Enterotoxigenic Escherichia coli with STh and STp genotypes is associated with diarrhea both in children in areas of endemicity and in travelers. J. Clin. Microbiol. 2006, 44, 3872–3877. [Google Scholar] [CrossRef] [PubMed]
- Frye, J.G.; Jackson, C.R. Genetic mechanisms of antimicrobial resistance identified in Salmonella enterica, Escherichia coli, and Enteroccocus spp. isolated from U.S. food animals. Front. Microbiol. 2013, 4, 135. [Google Scholar] [CrossRef] [PubMed]
- Steinke, D.T.; Seaton, R.A.; Phillips, G.; MacDonald, T.M.; Davey, P.G. Prior trimethoprim use and trimethoprim-resistant urinary tract infection: A nested case-control study with multivariate analysis for other risk factors. J. Antimicrob. Chemother. 2001, 47, 781–787. [Google Scholar] [CrossRef] [PubMed]
- Johnson, T.J.; Wannemeuhler, Y.M.; Scaccianoce, J.A.; Johnson, S.J.; Nolan, L.K. Complete DNA sequence, comparative genomics, and prevalence of an IncHI2 plasmid occurring among extraintestinal pathogenic Escherichia coli isolates. Antimicrob. Agents Chemother. 2006, 50, 3929–3933. [Google Scholar] [CrossRef] [PubMed]
- Awad, A.; Arafat, N.; Elhadidy, M. Genetic elements associated with antimicrobial resistance among Avian Pathogenic Escherichia coli. Ann. Clin. Microbiol. Antimicrob. 2016, 15, 59. [Google Scholar] [CrossRef] [PubMed]
- Scott, L.J. Ceftolozane/Tazobactam: A review in complicated intra-abdominal and urinary tract infections. Drugs 2016, 76, 231–242. [Google Scholar] [CrossRef] [PubMed]
- Zhanel, G.G.; Chung, P.; Adam, H.; Zelenitsky, S.; Denisuik, A.; Schweizer, F.; Lagacé-Wiens, P.R.S.; Rubinstein, E.; Gin, A.S.; Walkty, A.; et al. A novel cephalosporin-β-lactamase inhibitor combination with activity against multidrug-resistant gram-negative bacilli. Drugs 2014, 74, 31–51. [Google Scholar] [CrossRef] [PubMed]
- Gangcuangco, L.M.; Clark, P.; Stewart, C.; Miljkovic, G.; Saul, Z.K. Persistent bacteremia from Pseudomonas aeruginosa with In vitro resistance to the novel antibiotics Ceftolozane-Tazobactam and Ceftazidime-Avibactam. Case Rep. Infect. Dis. 2016, 2016, 152404. [Google Scholar]
- Teleb, M.; Soto-Ruiz, E.; Dominguez, D.C.; Antony, S. The rapid development of ESBL E. coli resistance to Ceftolozane-Tazobactam in a patient with a liver abscess. The search for an omnipotent antibiotic goes on! Infect. Disord. Drug Targets 2016, 16. [Google Scholar] [CrossRef]
- Knapp, C.W.; Callan, A.C.; Aitken, B.; Shearn, R.; Koenders, A.; Hinwood, A. Relationship between antibiotic resistance genes and metals in residential soil samples from Western Australia. Environ. Sci. Pollut. Res. 2016, 24, 2484–2494. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Wang, J.; Han, Y.; Chen, J.; Liu, G.; Lu, H.; Yan, B.; Chen, S. Nutrients, heavy metals and microbial communities co-driven distribution of antibiotic resistance genes in adjacent environment of mariculture. Environ. Pollut. 2017, 220, 909–918. [Google Scholar] [CrossRef] [PubMed]
- Jonker, C.; Olivier, J. Mineral contamination from cemetery soils: Case study of Zandfontein Cemetery, South Africa. Int. J. Environ. Res. Public Health 2012, 9, 511–520. [Google Scholar] [CrossRef] [PubMed]
- Adelana, S.; Yongxin, X.; Vrbka, P. A conceptual model for the development and management. Water SA 2010, 36, 461–474. [Google Scholar] [CrossRef]
- Seyler, H.; Bollaert, M.; Witthüser, K. Regional Water Sensitive Design Scenario Planning for Cape Town Using an Urban (Geo) Hydrology Model; WRC Report No. TT 708/16; Water Research Commission: Pretoria, South Africa, 2016. [Google Scholar]
- WRC. Groundwater Sampling Manual; Water Research Commission (WRC) Report No. TT 733/17; WRC: Pretoria, South Africa, 2017. [Google Scholar]
- Abia, A.L.K.; Ubomba-Jaswa, E.; Momba, M.N.B. High prevalence of multiple-antibiotic-resistant (MAR) Escherichia coli in river bed sediments of the Apies River, South Africa. Environ. Monit. Assess. 2015, 187, 652. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; Mazumder, Y.; Dutta, B.; Shome, B.; Bujarbaruah, K.; Kumar, R. Molecular typing of Clostridium perfringens isolated from diarrhoeic cattle. J. Anim. Sci. Adv. 2012, 2, 226–229. [Google Scholar]
- Abia, A.L.K.; Ubomba-Jaswa, E.; Momba, M.N.B. Impact of seasonal variation on Escherichia coli concentrations in the riverbed sediments in the Apies River, South Africa. Sci. Total Environ. 2015, 537, 462–469. [Google Scholar] [CrossRef] [PubMed]
- Abia, A.; Schaefer, L.; Ubomba-Jaswa, E.; Le Roux, W. Abundance of Pathogenic Escherichia coli virulence-associated genes in well and borehole water used for domestic purposes in a peri-urban community of South Africa. Int. J. Environ. Res. Public Health 2017, 14, 320. [Google Scholar] [CrossRef] [PubMed]
- Bauer, A.; Kirby, W.; Sherris, J.; Turck, M. Antibiotic susceptibility testing by a standardised single disk method. Am. J. Clin. Pathol. 1966, 45, 413–496. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 27th ed.; Clinical and Laboratory Standards Institute: Wayne, NJ, USA, 2017. [Google Scholar]

| Cemetery | Sample Site * | Depth (m) ** | Replicate Samples (MPN/100 mL) | Geometric Mean (MPN/100 mL) | Standard Deviation | ||
|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | |||||
| Delft | D2 | 3.1 | <1 | <1 | <1 | 0.0 | 0.0 |
| D3 | na | <1 | <1 | <1 | 0.0 | 0.0 | |
| Maitland | MDam1 | na | 1 | 1 | 4.1 | 1.6 | 1.8 |
| MDam2 | na | 1 | 4.1 | 1 | 1.6 | 1.8 | |
| MRiver1 | na | 2419.6 | 2419.6 | 2419.6 | 2419.6 | 0.0 | |
| MRiver2 | na | 2419.6 | 2419.6 | 2419.6 | 2419.6 | 0.0 | |
| M1 | 1.6 | <1 | <1 | <1 | 0.0 | 0.0 | |
| M2 | 2.8 | <1 | <1 | <1 | 0.0 | 0.0 | |
| M4 | 4 | <1 | <1 | <1 | 0.0 | 0.0 | |
| M5 | 2.5 | <1 | <1 | <1 | 0.0 | 0.0 | |
| M6 | 2 | 9.7 | 18.7 | 8.4 | 11.5 | 5.6 | |
| M7 | 2.7 | <1 | <1 | <1 | 0.0 | 0.0 | |
| M8 | 2.3 | 8.6 | 3.1 | 5.2 | 5.2 | 2.8 | |
| Welmoed | Wetland | na | 65.7 | 86.5 | 42 | 62.0 | 22.3 |
| W2 | 1.8 | <1 | <1 | <1 | 0.0 | 0.0 | |
| W3 | 2.4 | <1 | <1 | <1 | 0.0 | 0.0 | |
| Profile | Number of Isolates | |||||||
|---|---|---|---|---|---|---|---|---|
| C (25 µg) | S (10 µg) | T (25 µg) | CFX (30 µg) | TM (5 µg) | CIP (5 µg) | NI (300 µg) | C/T (40 µg) | |
| Susceptible | 71 | 26 | 38 | 65 | 29 | 53 | 66 | 48 |
| Intermediate | 3 | 3 | 0 | 0 | 0 | 2 | 6 | 0 |
| Resistant | 26 | 71 | 62 | 35 | 71 | 45 | 28 | 52 |
| Number of Antibiotic Resistance | Antibiotic Resistance Profile | Number of Isolates |
|---|---|---|
| 1 | C/T | 1 |
| 2 | S-C/T | 3 |
| S-T | 5 | |
| TM-C/T | 1 | |
| TM-CIP | 1 | |
| 3 | S-CFX-C/T | 1 |
| S-T-C/T | 2 | |
| S-T-CFX | 1 | |
| S-TM-C/T | 4 | |
| T-TM-C/T | 1 | |
| T-TM-CIP | 3 | |
| 4 | C-CFX-TM-NI | 2 |
| C-S-T-C/T | 2 | |
| S-CFX-TM-C/T | 1 | |
| S-TM-CIP-C/T | 1 | |
| S-TM-CIP-NI | 1 | |
| S-T-TM-C/T | 1 | |
| S-T-TM-CIP | 13 | |
| T-CFX-TM-CIP | 1 | |
| 5 | C-CFX-TM-CIP-NI | 1 |
| C-S-CFX-NI-C/T | 1 | |
| C-S-T-TM-C/T | 1 | |
| C-T-CFX-CIP-C/T | 1 | |
| S-CFX-TM-NI-C/T | 1 | |
| S-T-CFX-TM-CIP | 2 | |
| S-T-CFX-TM-C/T | 2 | |
| S-T-TM-CIP-C/T | 8 | |
| 6 | C-S-CFX-TM-CIP-C/T | 1 |
| C-S-CFX-TM-NI-C/T | 1 | |
| C-S-T-CFX-TM-C/T | 1 | |
| C-S-T-TM-NI-C/T | 1 | |
| C-T-CFX-TM-NI-C/T | 1 | |
| S-T-CFX-TM-NI-C/T | 2 | |
| S-T-TM-CIP-NI-C/T | 1 | |
| T-CFX-TM-CIP-NI-C/T | 1 | |
| 7 | C-S-CFX-TM-CIP-NI-C/T | 1 |
| C-S-T-CFX-TM-CIP-C/T | 2 | |
| C-S-T-CFX-TM-CIP-NI | 4 | |
| C-S-T-CFX-TM-NI-C/T | 4 | |
| C-S-T-TM-CIP-NI-C/T | 1 | |
| 8 | C-S-T-CFX-TM-CIP-NI-C/T | 4 |
| Designation | E. coli Pathotype | Genes Targeted |
|---|---|---|
| EPEC | Enteropathogenic E. coli | eaeA |
| EHEC | Enterohaemorrhagic E. coli (or STEC) | eaeA, stx1, stx2 |
| EAEC | Enteroaggregative E. coli | eagg |
| EIEC | Enteroinvasive E. coli | ipaH |
| ETEC | Enterotoxigenic E. coli | ST |
| NMEC | Neonatal Meningitis E. coli | ibeA |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abia, A.L.K.; Ubomba-Jaswa, E.; Schmidt, C.; Dippenaar, M.A. Where Did They Come from—Multi-Drug Resistant Pathogenic Escherichia coli in a Cemetery Environment? Antibiotics 2018, 7, 73. https://doi.org/10.3390/antibiotics7030073
Abia ALK, Ubomba-Jaswa E, Schmidt C, Dippenaar MA. Where Did They Come from—Multi-Drug Resistant Pathogenic Escherichia coli in a Cemetery Environment? Antibiotics. 2018; 7(3):73. https://doi.org/10.3390/antibiotics7030073
Chicago/Turabian StyleAbia, Akebe Luther King, Eunice Ubomba-Jaswa, Chantelle Schmidt, and Matthys Alois Dippenaar. 2018. "Where Did They Come from—Multi-Drug Resistant Pathogenic Escherichia coli in a Cemetery Environment?" Antibiotics 7, no. 3: 73. https://doi.org/10.3390/antibiotics7030073
APA StyleAbia, A. L. K., Ubomba-Jaswa, E., Schmidt, C., & Dippenaar, M. A. (2018). Where Did They Come from—Multi-Drug Resistant Pathogenic Escherichia coli in a Cemetery Environment? Antibiotics, 7(3), 73. https://doi.org/10.3390/antibiotics7030073






