Isolation, Characterization, and Antibacterial Activity of Hard-to-Culture Actinobacteria from Cave Moonmilk Deposits
Abstract
1. Introduction
2. Results and Discussion
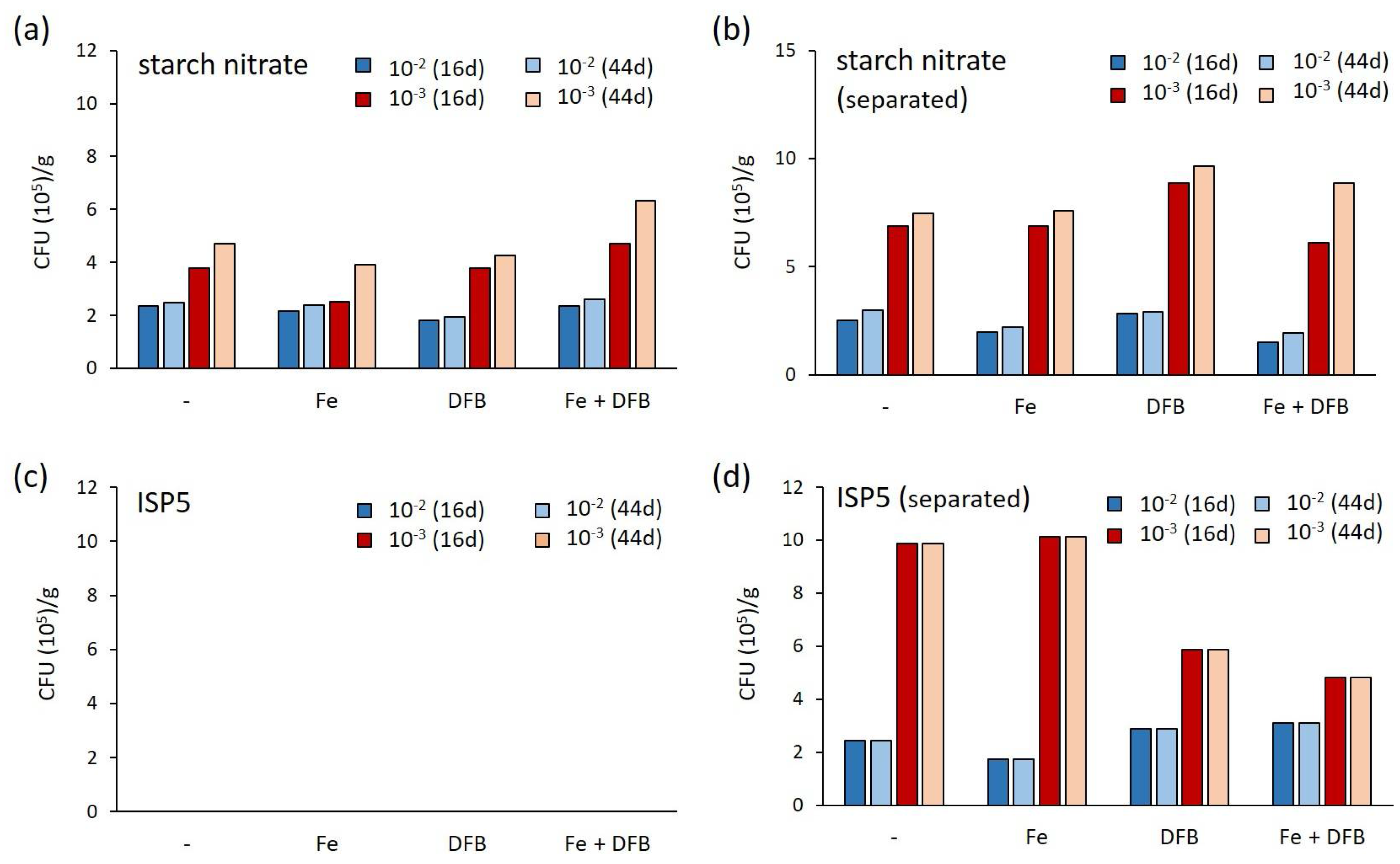
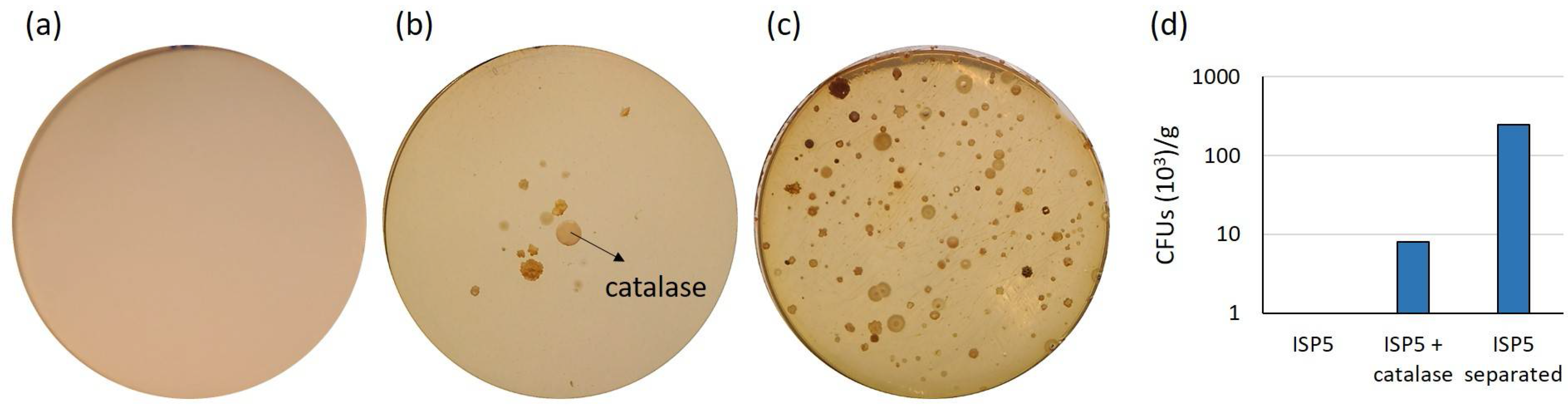
2.1. Assessment of Various Strategies for Isolating Moonmilk-Dwelling Rare Actinobacteria
2.2. Characterization of Culturable Moonmilk-Derived Actinobacterial Isolates
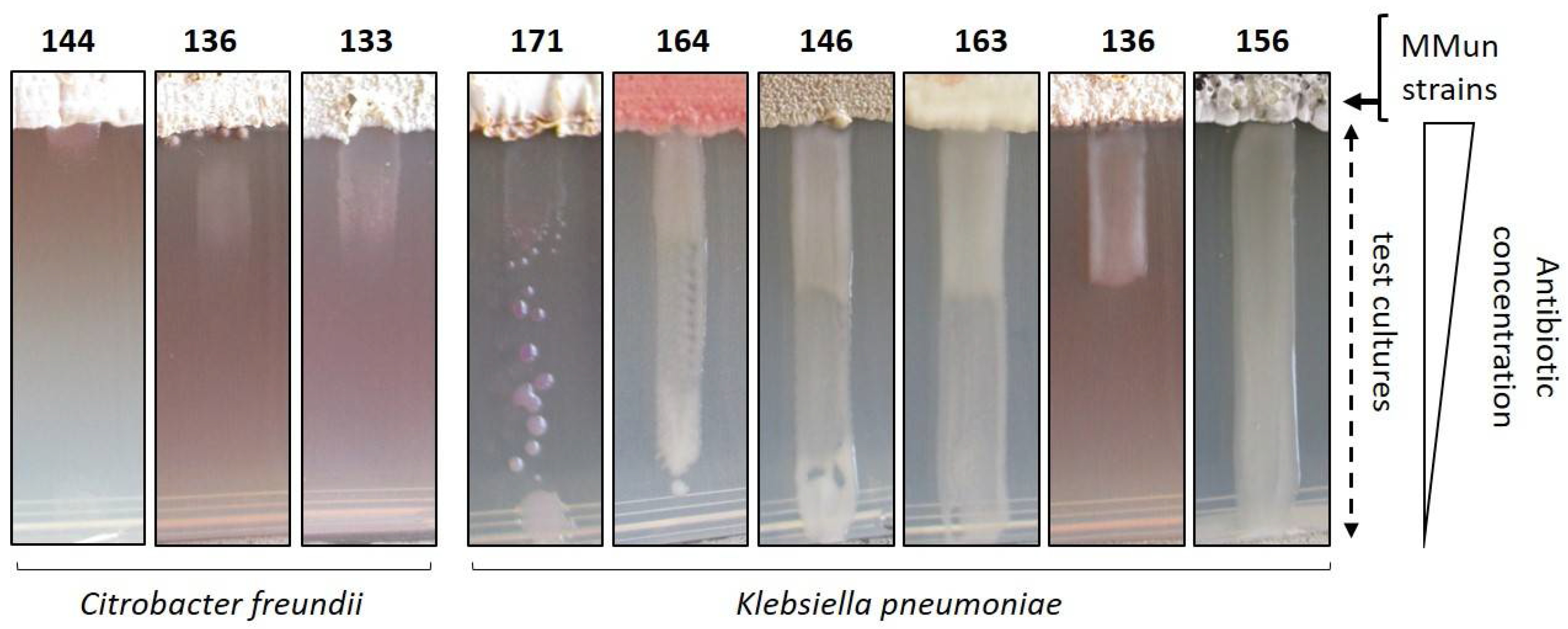
2.3. Evaluation of the Antibacterial Activity of the New Moonmilk Isolates
3. Materials and Methods
3.1. Preparation of Media and Moonmilk Suspension for Isolation of Rare Actinobacteria
3.2. Isolation and Sequencing of Genomic DNA
3.3. Phylogenetic Analyses of Actinobacterial Strains
3.4. Antimicrobial Activities of Rare Moonmilk Actinobacterial Strains
3.5. Genome Mining for Gene Clusters Involved in Secondary Metabolite Production.
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Hill, P.; Heberlig, G.W.; Boddy, C.N. Sampling Terrestrial Environments for Bacterial Polyketides. Molecules 2017, 22. [Google Scholar] [CrossRef] [PubMed]
- Goodfellow, M.; Fiedler, H.-P. A guide to successful bioprospecting: Informed by actinobacterial systematics. Antonie Leeuwenhoek 2010, 98, 119–142. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization (WHO). Antimicrobial Resistance: Global Report on Surveillance 2014; AMR Report; WHO: Geneva, Switzerland, 2014. [Google Scholar]
- Hutchinson, G.E. The Paradox of the Plankton. Am. Nat. 1961, 95, 137–145. [Google Scholar] [CrossRef]
- Bhullar, K.; Waglechner, N.; Pawlowski, A.; Koteva, K.; Banks, E.D.; Johnston, M.D.; Barton, H.A.; Wright, G.D. Antibiotic resistance is prevalent in an isolated cave microbiome. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Lavoie, K.; Ruhumbika, T.; Bawa, A.; Whitney, A.; de Ondarza, J. High levels of antibiotic resistance but no antibiotic production detected along a gypsum gradient in great Onyx Cave, KY, USA. Diversity 2017, 9. [Google Scholar] [CrossRef]
- Hopwood, D.A. Streptomyces in Nature and Medicine: The Antibiotic Makers; Oxford University Press: New York, NY, USA, 2007; ISBN 978-0-19-515066-7. [Google Scholar]
- Dhami, N.K.; Mukherjee, A.; Watkin, E.L.J. Microbial Diversity and Mineralogical-Mechanical Properties of Calcitic Cave Speleothems in Natural and in Vitro Biomineralization Conditions. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Portillo, M.C.; Gonzalez, J.M. Moonmilk Deposits Originate from Specific Bacterial Communities in Altamira Cave (Spain). Microb. Ecol. 2011, 61, 182–189. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rooney, D.C.; Hutchens, E.; Clipson, N.; Baldini, J.; McDermott, F. Microbial Community Diversity of Moonmilk Deposits at Ballynamintra Cave, Co. Waterford, Ireland. Microb. Ecol. 2010, 60, 753–761. [Google Scholar] [CrossRef] [PubMed]
- Maciejewska, M.; Adam, D.; Naômé, A.; Martinet, L.; Tenconi, E.; Całusińska, M.; Delfosse, P.; Hanikenne, M.; Baurain, D.; Compère, P.; et al. Assessment of the Potential Role of Streptomyces in Cave Moonmilk Formation. Front. Microbiol. 2017, 8, 1181. [Google Scholar] [CrossRef] [PubMed]
- Maciejewska, M.; Adam, D.; Martinet, L.; Naômé, A.; Całusińska, M.; Delfosse, P.; Carnol, M.; Barton, H.A.; Hayette, M.-P.; Smargiasso, N.; et al. A Phenotypic and Genotypic Analysis of the Antimicrobial Potential of Cultivable Streptomyces Isolated from Cave Moonmilk Deposits. Front. Microbiol. 2016, 7, 1455. [Google Scholar] [CrossRef] [PubMed]
- Maciejewska, M.; Calusinska, M.; Cornet, L.; Adam, D.; Pessi, I.S.; Malchair, S.; Delfosse, P.; Baurain, D.; Barton, H.A.; Carnol, M.; et al. High-throughput sequencing analysis of the actinobacterial spatial diversity in moonmilk deposits. Antibiotics 2018. [Google Scholar] [CrossRef]
- Oliver, J.D. The viable but nonculturable state in bacteria. J. Microbiol. 2005, 43, 93–100. [Google Scholar] [PubMed]
- Staley, J.T.; Konopka, A. Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu. Rev. Microbiol. 1985, 39, 321–346. [Google Scholar] [CrossRef] [PubMed]
- Hug, L.; Baker, B.; Anantharaman, K.; Brown, C.; Probst, A.; Castelle, C.; Butterfield, C.; Hernsdorf, A.; Amano, Y.; Ise, K.; et al. A new view of the tree of life. Nat. Microbiol. 2016, 1, 16048. [Google Scholar] [CrossRef] [PubMed]
- Button, D.K.; Schut, F.; Quang, P.; Martin, R.; Robertson, B.R. Viability and isolation of marine bacteria by dilution culture: Theory, procedures, and initial results. Appl. Environ. Microbiol. 1993, 59, 881–891. [Google Scholar] [PubMed]
- D’Onofrio, A.; Crawford, J.M.; Stewart, E.J.; Witt, K.; Gavrish, E.; Epstein, S.; Clardy, J.; Lewis, K. Siderophores from Neighboring Organisms Promote the Growth of Uncultured Bacteria. Chem. Biol. 2010, 17, 254–264. [Google Scholar] [CrossRef] [PubMed]
- Bruns, A.; Cypionka, H.; Overmann, J. Cyclic AMP and acyl homoserine lactones increase the cultivation efficiency of heterotrophic bacteria from the central Baltic Sea. Appl. Environ. Microbiol. 2002, 68, 3978–3987. [Google Scholar] [CrossRef] [PubMed]
- Sait, M.; Hugenholtz, P.; Janssen, P.H. Cultivation of globally distributed soil bacteria from phylogenetic lineages previously only detected in cultivation-independent surveys. Environ. Microbiol. 2002, 4, 654–666. [Google Scholar] [CrossRef] [PubMed]
- Nichols, D.; Cahoon, N.; Trakhtenberg, E.M.; Pham, L.; Mehta, A.; Belanger, A.; Kanigan, T.; Lewis, K.; Epstein, S.S. Use of Ichip for High-Throughput In Situ Cultivation of “Uncultivable” Microbial Species. Appl. Environ. Microbiol. 2010, 76, 2445–2450. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Kawasaki, K.; Daimon, S.; Kitagawa, W.; Yamamoto, K.; Tamaki, H.; Tanaka, M.; Nakatsu, C.H.; Kamagata, Y. A hidden pitfall in the preparation of agar media undermines microorganism cultivability. Appl. Environ. Microbiol. 2014, 80, 7659–7666. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, K.; Kamagata, Y. Phosphate-Catalyzed Hydrogen Peroxide Formation from Agar, Gellan, and κ-Carrageenan and Recovery of Microbial Cultivability via Catalase and Pyruvate. Appl. Environ. Microbiol. 2017, 83. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.-Y.; Dong, L.; Zhao, J.-K.; Hu, X.; Shen, C.; Qiao, Y.; Zhang, X.; Wang, Y.; Ismagilov, R.F.; Liu, S.-J.; et al. High-Throughput Single-Cell Cultivation on Microfluidic Streak Plates. Appl. Environ. Microbiol. 2016, 82, 2210–2218. [Google Scholar] [CrossRef] [PubMed]
- Hop, D.V.; Sakiyama, Y.; Binh, C.T.T.; Otoguro, M.; Hang, D.T.; Miyadoh, S.; Luong, D.T.; Ando, K. Taxonomic and ecological studies of actinomycetes from Vietnam: Isolation and genus-level diversity. J. Antibiot. 2011, 64, 599–606. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Otoguro, M.; Hayakawa, M.; Yamazaki, T.; Iimura, Y. An integrated method for the enrichment and selective isolation of Actinokineospora spp. in soil and plant litter. J. Appl. Microbiol. 2001, 91, 118–130. [Google Scholar] [PubMed]
- Hayakawa, M.; Otoguro, M.; Takeuchi, T.; Yamazaki, T.; Iimura, Y. Application of a method incorporating differential centrifugation for selective isolation of motile actinomycetes in soil and plant litter. Antonie Leeuwenhoek 2000, 78, 171–185. [Google Scholar] [CrossRef] [PubMed]
- Arias, A.A.; Lambert, S.; Martinet, L.; Adam, D.; Tenconi, E.; Hayette, M.-P.; Ongena, M.; Rigali, S. Growth of desferrioxamine-deficient Streptomyces mutants through xenosiderophore piracy of airborne fungal contaminations. FEMS Microbiol. Ecol. 2015, 91. [Google Scholar] [CrossRef] [PubMed]
- Maciejewska, M.; Pessi, I.S.; Arguelles-Arias, A.; Noirfalise, P.; Luis, G.; Ongena, M.; Barton, H.; Carnol, M.; Rigali, S. Streptomyces lunaelactis sp. nov., a novel ferroverdin A-producing Streptomyces species isolated from a moonmilk speleothem. Antonie Leeuwenhoek 2015, 107, 519–531. [Google Scholar] [CrossRef] [PubMed]
- Kis, Á.E.; Laczi, K.; Zsíros, S.; Kós, P.; Tengölics, R.; Bounedjoum, N.; Kovács, T.; Rákhely, G.; Perei, K. Characterization of the Rhodococcus sp. MK1 strain and its pilot application for bioremediation of diesel oil-contaminated soil. Acta Microbiol. Immunol. Hung. 2017, 64, 463–482. [Google Scholar] [CrossRef] [PubMed]
- Eagle, H.; Musselman, A.D. The rate of bactericidal action of penicillin in vitro as a function of its concentrations, and its paradoxically reduced activity at high concentrations against certain organisms. J. Exp. Med. 1948, 88, 99–131. [Google Scholar] [CrossRef] [PubMed]
- Waksman, S.; Lechevalier, H. The Actinomycetales, Classification, Identification and Description of Genera and Species; Williams & Wilkins Co.: Baltimore, MD, USA, 1961. [Google Scholar]
- Shirling, E.B.; Gottlieb, D. Methods for characterization of Streptomyces species. Int. J. Syst. Bacteriol. 1966, 16, 313–340. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef] [PubMed]
- Onstott, T.C.; McGown, D.J.; Bakermans, C.; Ruskeeniemi, T.; Ahonen, L.; Telling, J.; Soffientino, B.; Pfiffner, S.M.; Sherwood-Lollar, B.; Frape, S.; et al. Microbial communities in subpermafrost saline fracture water at the Lupin Au mine, Nunavut, Canada. Microb. Ecol. 2009, 58, 786–807. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- Philippe, H. MUST, a computer package of Management Utilities for Sequences and Trees. Nucleic Acids Res. 1993, 21, 5264–5272. [Google Scholar] [CrossRef] [PubMed]
- Gouy, M.; Guindon, S.; Gascuel, O. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 2010, 27, 221–224. [Google Scholar] [CrossRef] [PubMed]
- Rigali, S.; Anderssen, S.; Naômé, A.; van Wezel, G.P. Cracking the Regulatory Code of Biosynthetic Gene Clusters as a Strategy for Natural Product Discovery. Biochem. Pharmacol. 2018. [Google Scholar] [CrossRef] [PubMed]
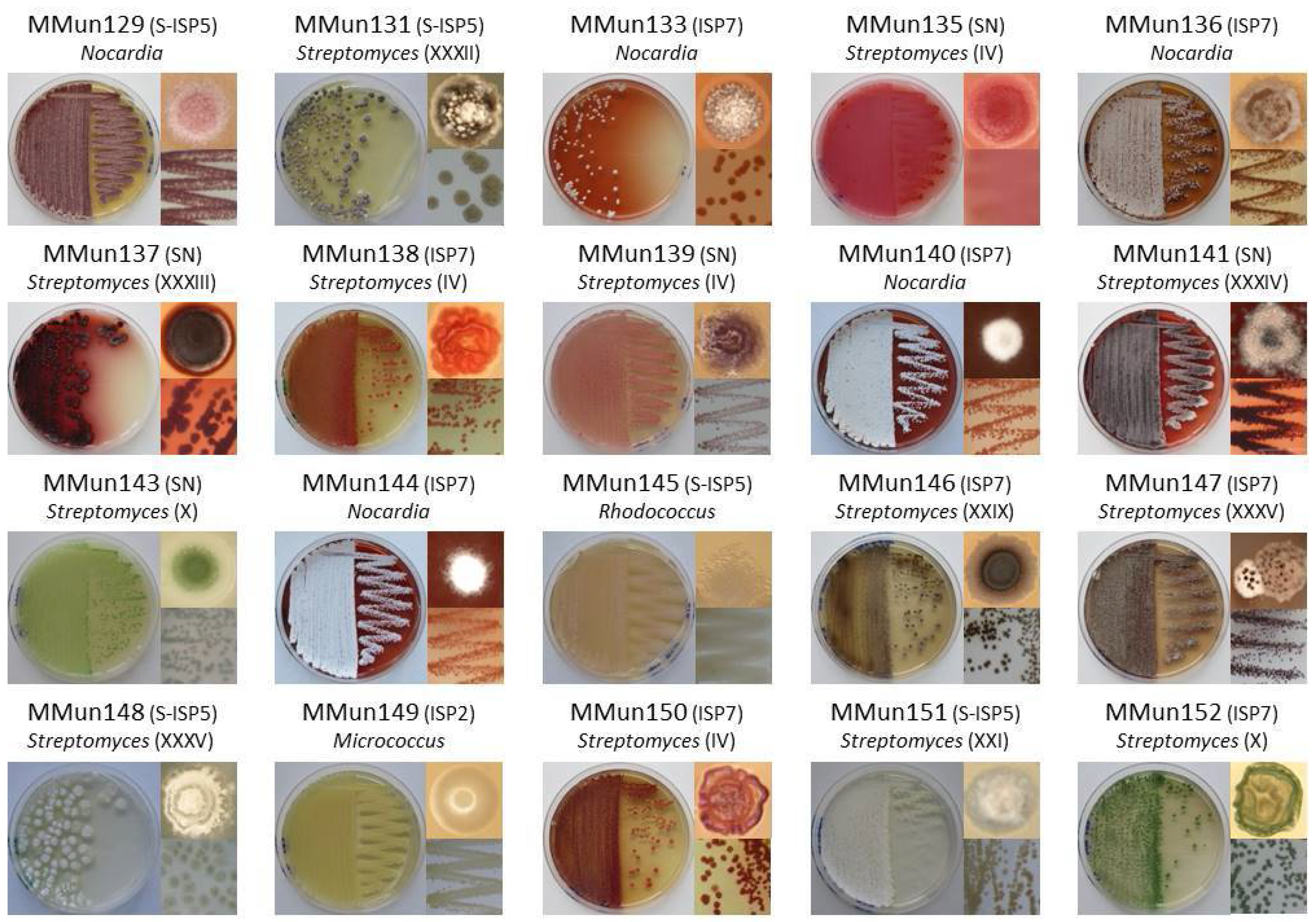
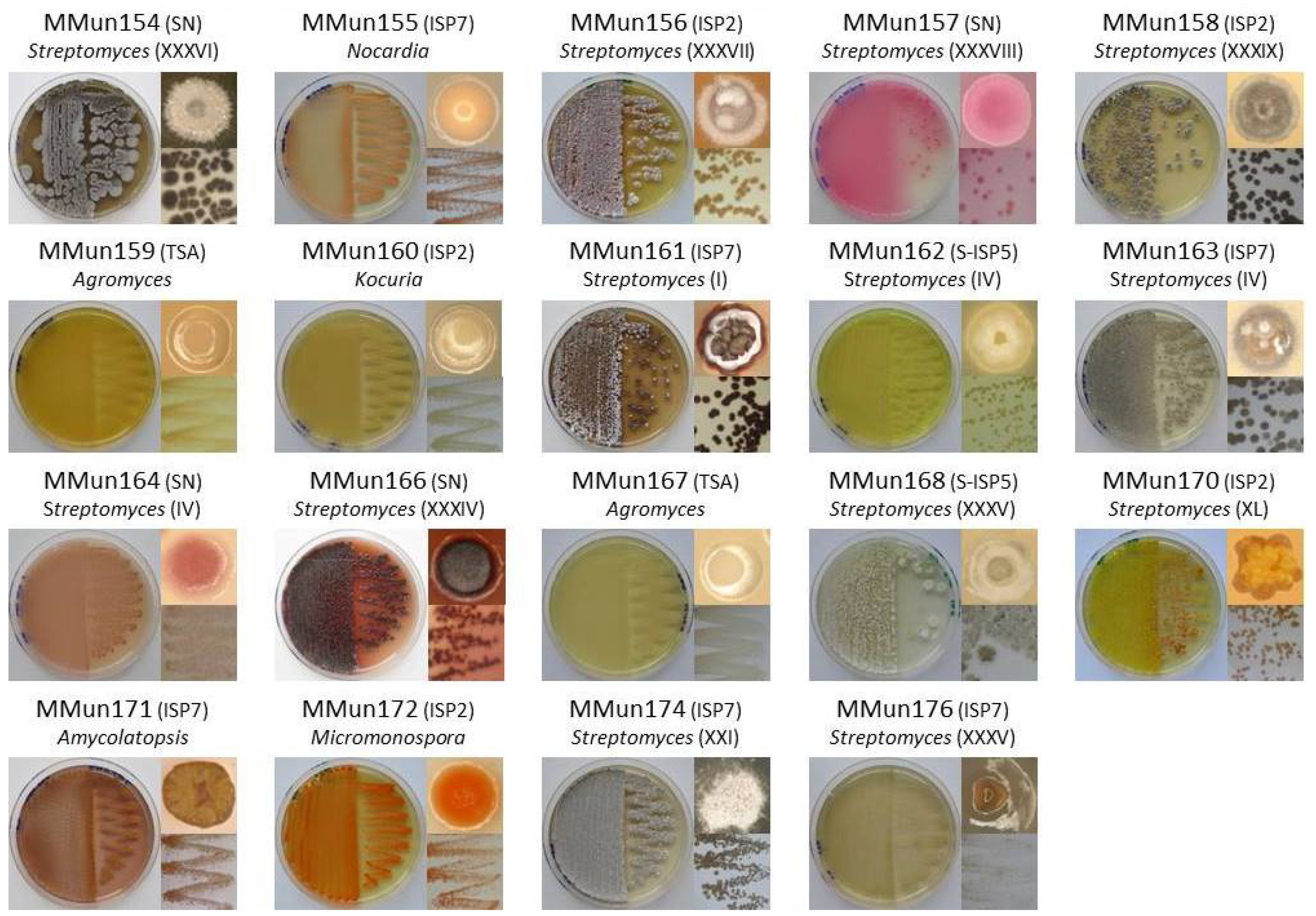
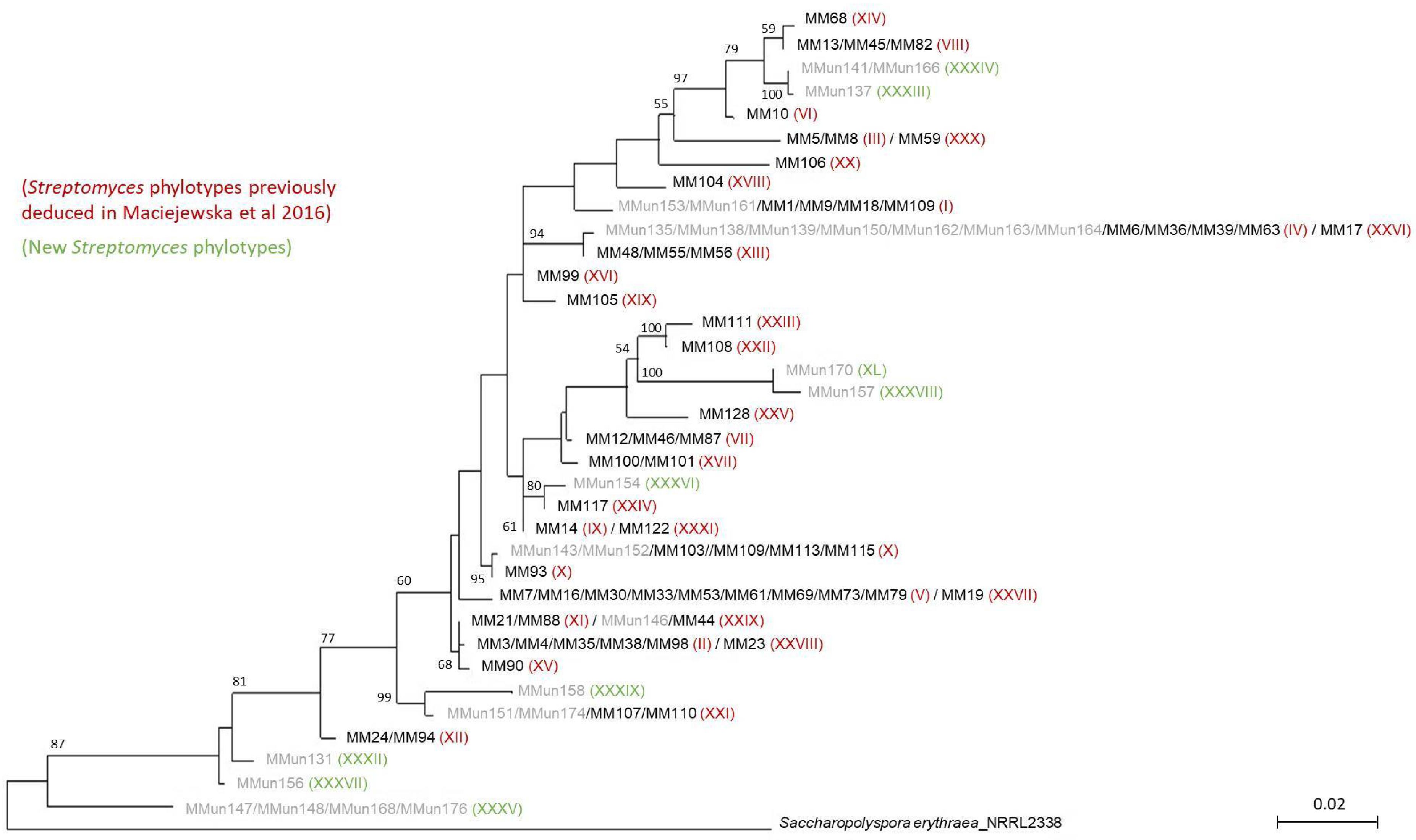
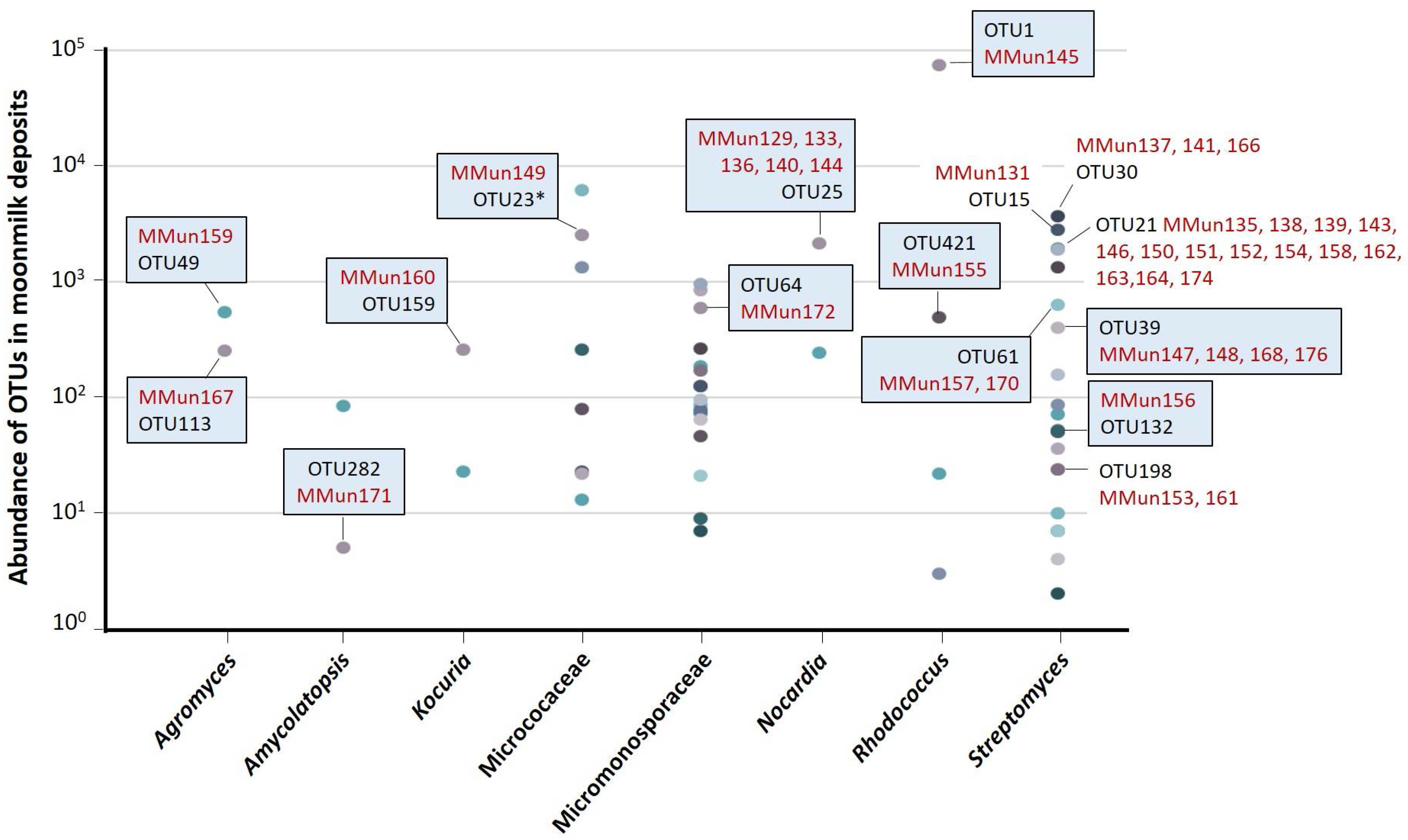
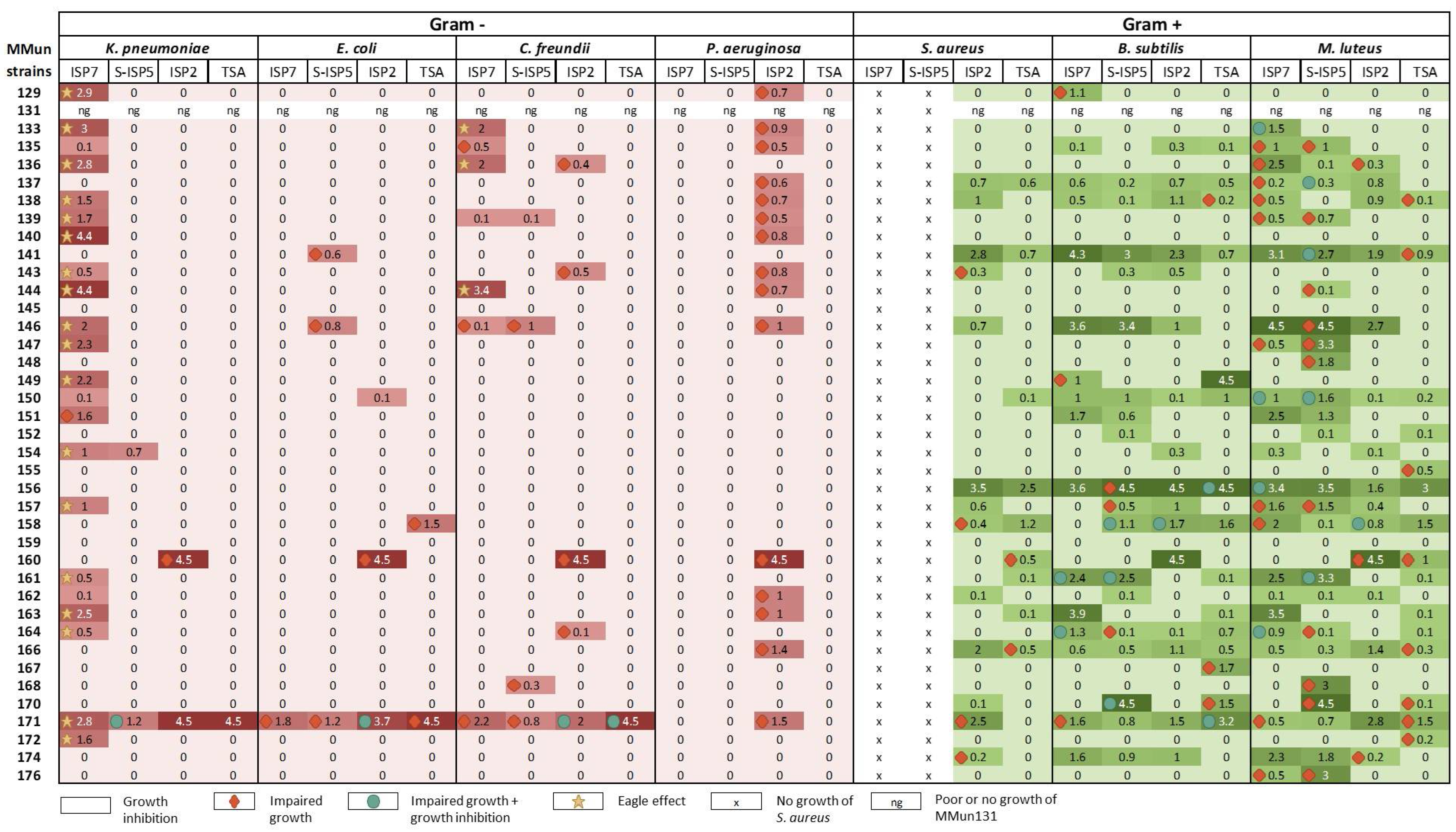
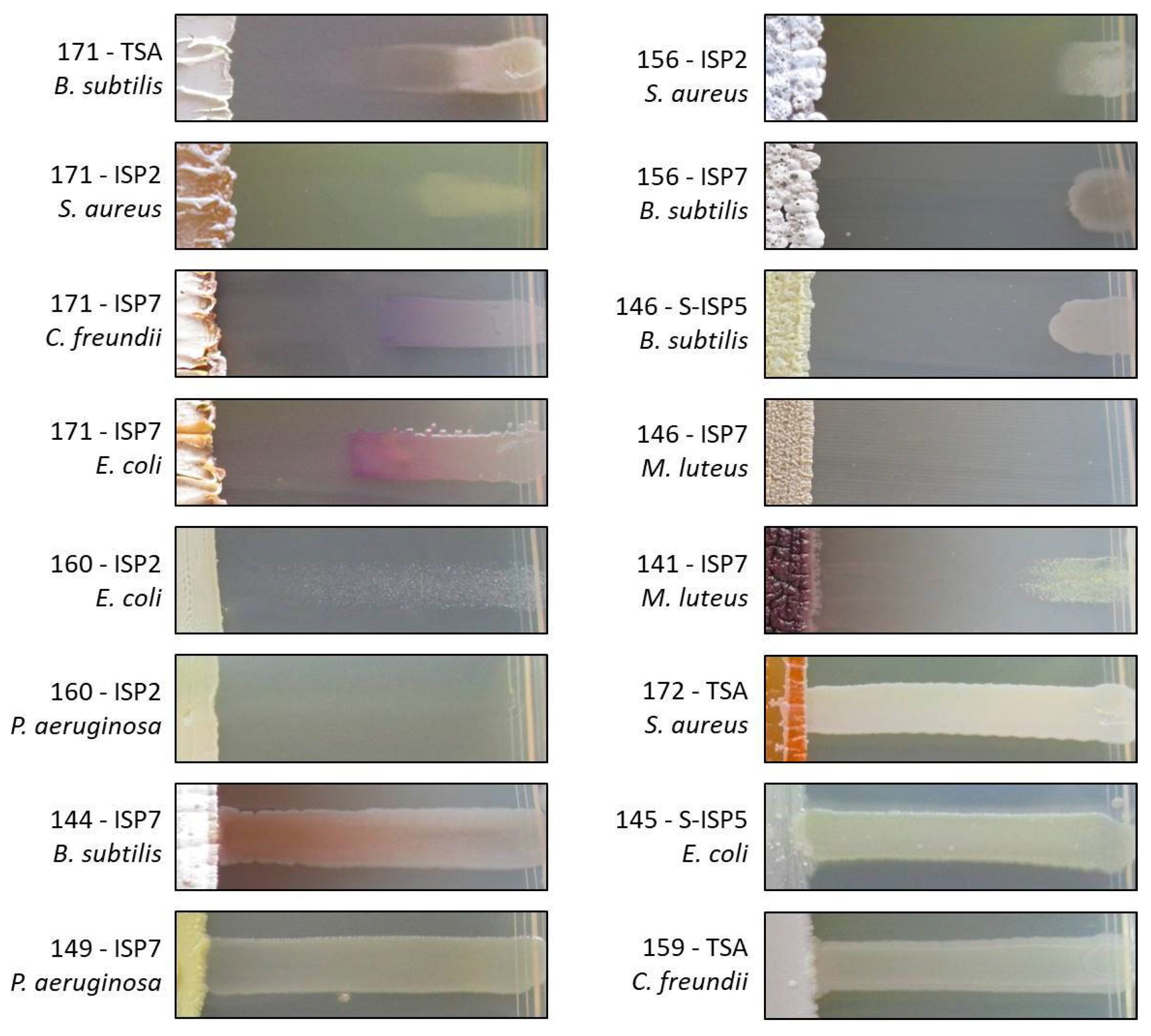

| MMun Strains | Family | Closest Match (BLAST) | 16S rRNA Ident. %; gaps | Isolation Medium | Phylotype | OTU |
|---|---|---|---|---|---|---|
| 129 ♣ | Nocardiaceae | Nocardia sp. strain 2M-SSA4 | 99.40; 1 | S-SN + Fe + DFB | - | 25 |
| 130 ♣ | Paenibacillaceae | Paenibacillus sp. DSL09-3 | 98.96; 1 | S-SN + Fe + DFB | - | na |
| 131 ♣ | Streptomycetaceae | Streptomyces sp. 2323.1 | 99.61; 0 | S-SN + Fe + DFB | XXXII | 15 |
| 133 ♣ | Nocardiaceae | Nocardia sp. strain 2M-SSA4 | 99.14; 3 | S-SN + Fe + DFB | - | 25 |
| 135 ♣ | Streptomycetaceae | Streptomyces sp. MM63 | 100; 0 | SN + Fe | IV * | 21 |
| 136 ♣ | Nocardiaceae | Nocardia soli strain DSM 44488 | 100; 0 | SN + Fe | - | 25 |
| 137 ♣ | Streptomycetaceae | Streptomyces sp. MM82 | 99.08; 2 | SN + DFB | XXXIII | 30 |
| 138 ♣ | Streptomycetaceae | Streptomyces sp. MM63 | 100; 0 | SN | IV * | 21 |
| 139 ♣ | Streptomycetaceae | Streptomyces sp. MM63 | 100; 0 | SN | IV * | 21 |
| 140 ♣ | Nocardiaceae | Nocardia sp. strain 2M-SSA4 | 99.54; 1 | SN | - | 25 |
| 141 ♣ | Streptomycetaceae | Streptomyces sp. MM82 | 99.15; 2 | SN | XXXIV | 30 |
| 142 ♣ | Rhodobacteraceae | Uncult. Paracoccus sp. clone 54 | 99.93; 0 | SN + catalase | - | na |
| 143 ♣ | Streptomycetaceae | Streptomyces lunaelactis strain MM126 | 100; 0 | SN + catalase | X * | 21 |
| 144 ♣ | Nocardiaceae | Nocardia soli strain Y48 | 99.34; 0 | SN + catalase | - | 25 |
| 145 ♣ | Nocardiaceae | Rhodococcus sp. MTM3W5.2 | 99.80; 1 | SN + catalase | - | 1 |
| 146 ♣ | Streptomycetaceae | Streptomyces sp. MM44 | 100; 0 | S-SN + DFB | XXIX * | 21 |
| 147 ♣ | Streptomycetaceae | Streptomyces xiamenensis strain 318 | 98.69; 8 | S-SN + DFB | XXXV | 39 |
| 148 ♣ | Streptomycetaceae | Streptomyces xiamenensis strain 318 | 98.69; 8 | S-SN + DFB | XXXV | 39 |
| 149 | Micrococcaceae | Micrococcus sp. 3455 | 100; 0 | SN | - | 23 |
| 150 ♣ | Streptomycetaceae | Streptomyces sp. MM63 | 100; 0 | SN | IV * | 21 |
| 151 ♣ | Streptomycetaceae | Streptomyces sp. MM110 | 100; 0 | SN | XXI * | 21 |
| 152 | Streptomycetaceae | Streptomyces lunaelactis strain MM126 | 100; 0 | SN + Fe | X * | 21 |
| 153 | Streptomycetaceae | Streptomyces sp. MM18 | 99.93; 1 | SN + DFB | I * (lost) | 198 |
| 154 ♣ | Streptomycetaceae | Streptomyces sp. ND04-1H | 99.93; 0 | SN | XXXVI | 21 |
| 155 | Nocardiaceae | Rhodococcus sp. strain MAK1 | 99.86; 2 | SN | - | 421 |
| 156 | Streptomycetaceae | Streptomyces sp. 1C-HV8 | 99.86; 2 | S-SN + Fe + DFB | XXXVII | 132 |
| 157 ♣ | Streptomycetaceae | Streptomyces sp. 2M-TWYE1 | 99.47; 0 | SN + catalase | XXXVIII | 61 |
| 158 | Streptomycetaceae | Streptomyces sp. SpC090624KE_06 | 99.51; 0 | S-SN + Fe + DFB | XXXIX | 21 |
| 159 | Microbacteriaceae | Agromyces sp. strain 4K403B | 97.88; 14 | S-SN + DFB | - | 49 |
| 160 | Micrococcaceae | Kocuria rhizophila strain R-42745 | 99.93; 0 | S-SN | - | 159 |
| 161 | Streptomycetaceae | Streptomyces sp. MM18 | 100; 0 | S-SN + Fe + DFB | I * | 198 |
| 162 | Streptomycetaceae | Streptomyces sp. MM63 | 99.93; 1 | S-SN + Fe + DFB | IV * | 21 |
| 163 | Streptomycetaceae | Streptomyces sp. MM63 | 100; 0 | S-SN | IV * | 21 |
| 164 | Streptomycetaceae | Streptomyces sp. MM63 | 100; 0 | S-SN | IV * | 21 |
| 166 | Streptomycetaceae | Streptomyces sp. strain A301 | 99.23; 3 | S-ISP5 + Fe + DFB | XXXIV | 30 |
| 167 | Microbacteriaceae | Agromyces cerinus strain DSM 8595 | 99.58; 0 | ISP5 + catalase | - | 113 |
| 168 | Streptomycetaceae | Streptomyces sp. 28a-5-1 | 98.60; 8 | ISP5 + catalase | XXXV | 39 |
| 170 | Streptomycetaceae | Streptomyces sp. 2C-SSA16-1 | 99.64; 0 | S-ISP5 + Fe + DFB | XL | 61 |
| 171 | Pseudonocardiaceae | Amycolatopsis sp. CA11 | 98.25; 16 | S-ISP5 + Fe + DFB | - | 282 |
| 172 | Micromonosporaceae | Micromonospora maoerensis strain NEAU-MES19 | 99.93; 0 | S-ISP5 + Fe + DFB | - | 64 |
| 174 | Streptomycetaceae | Streptomyces sp. MM110 | 100; 0 | ISP5 + catalase | XXI * | 21 |
| 176 | Streptomycetaceae | Streptomyces sp. 28a-5-1 | 98.67; 7 | S-ISP5 | XXXV | 39 |
| MMun | PKS-I | PKS-II | PKS-III | NRPS | Total | Genus |
|---|---|---|---|---|---|---|
| 129 | 2 | 2 | 1 | 34 | 39 | Nocardia |
| 131 | 2 | 6 | 1 | 27 | 36 | Streptomyces |
| 133 | 7 | 4 | 1 | 33 | 45 | Nocardia |
| 135 | 10 | 4 | 1 | 16 | 31 | Streptomyces |
| 136 | 6 | 2 | 1 | 31 | 40 | Nocardia |
| 137 | 9 | 6 | 0 | 6 | 21 | Streptomyces |
| 138 | 11 | 4 | 1 | 16 | 32 | Streptomyces |
| 139 | 4 | 4 | 0 | 13 | 21 | Streptomyces |
| 140 | 2 | 2 | 1 | 32 | 37 | Nocardia |
| 141 | 9 | 6 | 0 | 6 | 21 | Streptomyces |
| 143 | 6 | 4 | 1 | 9 | 20 | Streptomyces |
| 144 | 4 | 2 | 2 | 31 | 39 | Nocardia |
| 145 | 5 | 0 | 0 | 12 | 17 | Rhodococcus |
| 146 | 21 | 4 | 0 | 9 | 34 | Streptomyces |
| 147 | 10 | 2 | 1 | 4 | 17 | Streptomyces |
| 148 | 10 | 2 | 1 | 4 | 17 | Streptomyces |
| 150 | 12 | 4 | 1 | 14 | 31 | Streptomyces |
| 151 | 4 | 4 | 0 | 22 | 30 | Streptomyces |
| 154 | 0 | 4 | 1 | 10 | 15 | Streptomyces |
| 157 | 12 | 4 | 0 | 9 | 25 | Streptomyces |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adam, D.; Maciejewska, M.; Naômé, A.; Martinet, L.; Coppieters, W.; Karim, L.; Baurain, D.; Rigali, S. Isolation, Characterization, and Antibacterial Activity of Hard-to-Culture Actinobacteria from Cave Moonmilk Deposits. Antibiotics 2018, 7, 28. https://doi.org/10.3390/antibiotics7020028
Adam D, Maciejewska M, Naômé A, Martinet L, Coppieters W, Karim L, Baurain D, Rigali S. Isolation, Characterization, and Antibacterial Activity of Hard-to-Culture Actinobacteria from Cave Moonmilk Deposits. Antibiotics. 2018; 7(2):28. https://doi.org/10.3390/antibiotics7020028
Chicago/Turabian StyleAdam, Delphine, Marta Maciejewska, Aymeric Naômé, Loïc Martinet, Wouter Coppieters, Latifa Karim, Denis Baurain, and Sébastien Rigali. 2018. "Isolation, Characterization, and Antibacterial Activity of Hard-to-Culture Actinobacteria from Cave Moonmilk Deposits" Antibiotics 7, no. 2: 28. https://doi.org/10.3390/antibiotics7020028
APA StyleAdam, D., Maciejewska, M., Naômé, A., Martinet, L., Coppieters, W., Karim, L., Baurain, D., & Rigali, S. (2018). Isolation, Characterization, and Antibacterial Activity of Hard-to-Culture Actinobacteria from Cave Moonmilk Deposits. Antibiotics, 7(2), 28. https://doi.org/10.3390/antibiotics7020028





