Genetic Analysis of Virulence and β-Lactamase Determinants Related to β-Lactamase Inhibitors in Pseudomonas aeruginosa Strains from Nosocomial Infections
Abstract
1. Introduction
2. Results
2.1. Distribution of Virulence Genes with Respect to Strain Origin
2.2. Frequency of β-Lactamase Genotype and Resistance to β-Lactamase Inhibitor Antibiotics in the Strains
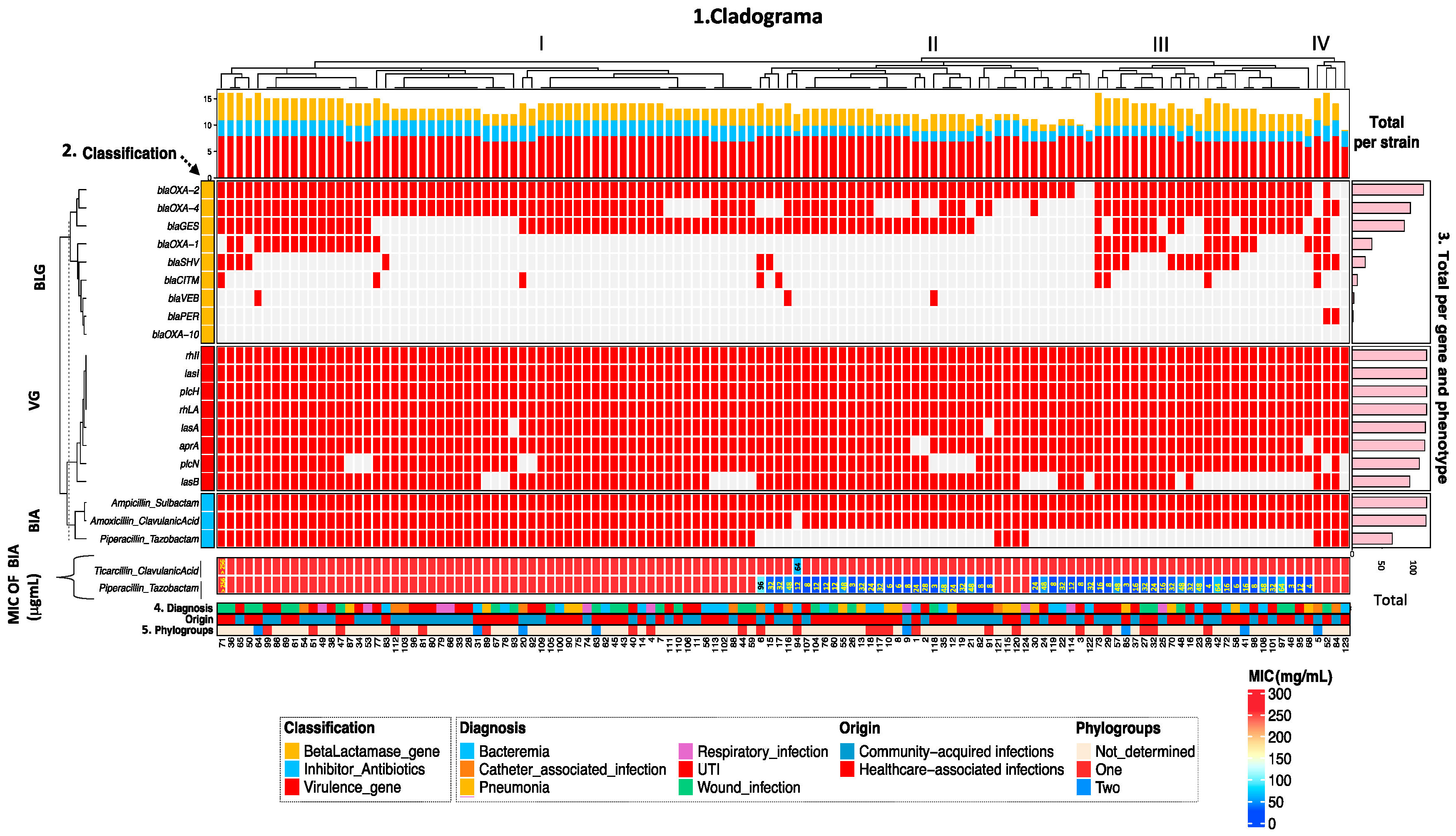
2.3. Frequency of Virulence Genes According to β-Lactamase and β-Lactamase Inhibitor Genotypes
2.4. Distribution of Phylogroups and MIC for β-Lactamase Inhibitor Antibiotics According to the Origin of the Strains
2.5. Analysis of the overall distribution of virulence and resistance to β-lactams
2.6. Patterns of Association of Virulence Genotype and β-Lactamases with the Phenotype of Resistance to β-Lactamase Inhibitor Antibiotics
3. Discussion
4. Methods
4.1. Origin of the Strains
4.2. Determination of Resistance to β-Lactam Antibiotics with β-Lactamase Inhibitors
4.3. Determination of the MIC of β-Lactamase Inhibitor Antibiotics
4.4. DNA Extraction
4.5. Strain Identification
4.6. Identification of Virulence Genes
4.7. Detection of BLES Genes
4.8. Identification of Phylogroups
4.9. Statistical Analysis
4.10. Unsupervised Hierarchical Clustering
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zupetic, J.; Peñaloza, H.F.; Bain, W.; Hulver, M.; Mettus, R.; Jorth, P.; Doi, Y.; Bomberger, J.; Pilewski, J.; Nouraie, M.; et al. Elastase Activity From Pseudomonas aeruginosa Respiratory Isolates and ICU Mortality. Chest 2021, 160, 1624–1633. [Google Scholar] [CrossRef] [PubMed]
- Vena, A.; Schenone, M.; Corcione, S.; Giannella, M.; Pascale, R.; Giacobbe, D.R.; Muccio, M.; Mornese Pinna, S.; Pari, B.; Giovannenze, F.; et al. Impact of adequate empirical combination therapy on mortality in septic shock due to Pseudomonas aeruginosa bloodstream infections: A multicentre retrospective cohort study. J. Antimicrob. Chemother. 2024, 79, 2846–2853. [Google Scholar] [CrossRef] [PubMed]
- Cotran-Lenrow, A.; Tefera, L.S.; Douglas-Vail, M.; Ayebare, A.; Kpokpah, L.N.; Davis, B.P. Community-Acquired Pseudomonas aeruginosa Meningitis in a Pediatric Patient. Cureus 2023, 15, e42376. [Google Scholar] [CrossRef] [PubMed]
- Recanatini, C.; van Werkhoven, C.H.; van der Schalk, T.E.; Paling, F.; Hazard, D.; Timbermont, L.; Torrens, G.; DiGiandomenico, A.; Esser, M.T.; Wolkewitz, M.; et al. Impact of Pseudomonas aeruginosa carriage on intensive care unit-acquired pneumonia: A European multicentre prospective cohort study. Clin. Microbiol. Infect. 2025, 31, 433–440. [Google Scholar] [CrossRef]
- Shah, S.; Kline, E.G.; Haidar, G.; Squires, K.M.; Pogue, J.M.; McCreary, E.K.; Ludwig, J.; Clarke, L.G.; Stellfox, M.; Van Tyne, D.; et al. Rates of Resistance to Ceftazidime-Avibactam and Ceftolozane-Tazobactam Among Patients Treated for Multidrug-Resistant Pseudomonas aeruginosa Bacteremia or Pneumonia. Clin. Infect. Dis. 2025, 80, 24–28. [Google Scholar] [CrossRef]
- Ghasemian, S.; Karami-Zarandi, M.; Heidari, H.; Khoshnood, S.; Kouhsari, E.; Ghafourian, S.; Maleki, A.; Kazemian, H.J. Molecular characterizations of antibiotic resistance, biofilm formation, and virulence determinants of Pseudomonas aeruginosa isolated from burn wound infection. J. Clin. Lab. Anal. 2023, 37, e24850. [Google Scholar] [CrossRef]
- El Husseini, N.; Carter, J.A.; Lee, V.T. Urinary tract infections and catheter-associated urinary tract infections caused by Pseudomonas aeruginosa. Microbiol. Mol. Biol. Rev. 2024, 88, e0006622. [Google Scholar] [CrossRef]
- Suetens, C.; Latour, K.; Kärki, T.; Ricchizzi, E.; Kinross, P.; Moro, M.L.; Jans, B.; Hopkins, S.; Hansen, S.; Lyytikäinen, O.; et al. Prevalence of healthcare-associated infections, estimated incidence and composite antimicrobial resistance index in acute care hospitals and long-term care facilities: Results from two European point prevalence surveys, 2016 to 2017. Eurosurveillance 2018, 23, 1800516. [Google Scholar] [CrossRef]
- Sastre-Femenia, M.À.; Fernández-Muñoz, A.; Gomis-Font, M.A.; Taltavull, B.; López-Causapé, C.; Arca-Suárez, J.; Martínez-Martínez, L.; Cantón, R.; Larrosa, N.; Oteo-Iglesias, J.; et al. Pseudomonas aeruginosa antibiotic susceptibility profiles, genomic epidemiology and resistance mechanisms: A nation-wide five-year time lapse analysis. Lancet Reg. Health Eur. 2023, 34, 100736. [Google Scholar] [CrossRef]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Sader, H.S.; Castanheira, M.; Duncan, L.R.; Flamm, R.K. Antimicrobial Susceptibility of Enterobacteriaceae and Pseudomonas aeruginosa Isolates from United States Medical Centers Stratified by Infection Type: Results from the International Network for Optimal Resistance Monitoring (INFORM) Surveillance Program, 2015–2016. Diagn. Microbiol. Infect. Dis. 2018, 92, 69–74. [Google Scholar] [CrossRef]
- CDC. COVID-19: U.S. Impact on Antimicrobial Resistance, Special Report 2022; U.S. Department of Health and Human Services, CDC: Atlanta, GA, USA, 2022.
- Jurado-Martín, I.; Sainz-Mejías, M.; McClean, S. Pseudomonas aeruginosa: An Audacious Pathogen with an Adaptable Arsenal of Virulence Factors. Int. J. Mol. Sci. 2021, 18, 3128. [Google Scholar] [CrossRef] [PubMed]
- Veetilvalappil, V.V.; Manuel, A.; Aranjani, J.M.; Tawale, R.; Koteshwara, A. Pathogenic arsenal of Pseudomonas aeruginosa: An update on virulence factors. Future Microbiol. 2022, 17, 465–481. [Google Scholar] [CrossRef] [PubMed]
- Ozer, E.A.; Nnah, E.; Didelot, X.; Whitaker, R.J.; Hauser, A.R. The population structure of Pseudomonas aeruginosa is characterized by genetic isolation of exoU+ and exoS+ lineages. Genome Biol. Evol. 2019, 11, 1780–1796. [Google Scholar] [CrossRef] [PubMed]
- Miranda, S.W.; Asfahl, K.L.; Dandekar, A.A.; Greenberg, E.P. Pseudomonas aeruginosa Quorum Sensing. Adv. Exp. Med. Biol. 2022, 1386, 95–115. [Google Scholar]
- De Sousa, T.; Hébraud, M.; Alves, O.; Costa, E.; Maltez, L.; Pereira, J.E.; Martins, Â.; Igrejas, G.; Poeta, P. Study of Antimicrobial Resistance, Biofilm Formation, and Motility of Pseudomonas aeruginosa Derived from Urine Samples. Microorganisms 2023, 11, 1345. [Google Scholar] [CrossRef]
- Thirumalmuthu, K.; Devarajan, B.; Prajna, L.; Mohankumar, V. Mechanisms of Fluoroquinolone and Aminoglycoside Resistance in Keratitis-Associated Pseudomonas aeruginosa. Microb. Drug Resist. 2019, 25, 813–823. [Google Scholar] [CrossRef]
- Tenover, F.C.; Nicolau, D.P.; Gill, C.M. Carbapenemase-producing Pseudomonas aeruginosa—An emerging challenge. Emerg. Microbes Infect. 2022, 11, 811–814. [Google Scholar] [CrossRef]
- Xiao, S.; Liang, X.; Han, L.; Zhao, S. Incidence, antimicrobial resistance and mortality of Pseudomonas aeruginosa bloodstream infections among hospitalized patients in China: A retrospective observational multicenter cohort study from 2017 to 2021. Front. Public Health 2024, 11, 1294141. [Google Scholar] [CrossRef]
- Mancini, S.; Garcia-Verellen, L.; Seth-Smith, H.M.B.; Keller, P.M.; Kolesnik-Goldmann, N.; Syed, M.A.; Ullah, I.; Hinic, V.; Roloff, T.; Egli, A.; et al. Diagnostic algorithm for the detection of carbapenemases and extended-spectrum β-lactamases in carbapenem-resistant Pseudomonas aeruginosa. Microbiol. Spectr. 2025, 13, e0319624. [Google Scholar] [CrossRef]
- Hosu, M.C.; Vasaikar, S.; Okuthe, G.E.; Apalata, T. Molecular Detection of Antibiotic-Resistant Genes in Pseudomonas aeruginosa from Nonclinical Environment: Public Health Implications in Mthatha, Eastern Cape Province, South Africa. Int. J. Microbiol. 2021, 2021, 8861074. [Google Scholar] [CrossRef]
- Valzano, F.; La Bella, G.; Lopizzo, T.; Curci, A.; Lupo, L.; Morelli, E.; Mosca, A.; Marangi, M.; Melfitano, R.; Rollo, T.; et al. Resistance to ceftazidime-avibactam and other new β-lactams in Pseudomonas aeruginosa clinical isolates: A multi-center surveillance study. Microbiol. Spectr. 2024, 12, e0426623. [Google Scholar] [CrossRef] [PubMed]
- Le Terrier, C.; Raro, O.H.F.; Saad, A.M.; Nordmann, P.; Poirel, L. In-vitro activity of newly-developed β-lactamase inhibitors avibactam, relebactam and vaborbactam in combination with anti-pseudomonal β-lactam antibiotics against AmpC-overproducing clinical Pseudomonas aeruginosa isolates. Eur. J. Clin. Microbiol. Infect. Dis. 2025, 44, 277–284. [Google Scholar] [CrossRef] [PubMed]
- Sargianou, M.; Stathopoulos, P.; Vrysis, C.; Tzvetanova, I.D.; Falagas, M.E. New β-Lactam/β-Lactamase Inhibitor Combination Antibiotics. Pathogens 2025, 14, 307. [Google Scholar] [CrossRef] [PubMed]
- Alonso-García, I.; Vázquez-Ucha, J.C.; Lasarte-Monterrubio, C.; González-Mayo, E.; Lada-Salvador, P.; Vela-Fernández, R.; Aja-Macaya, P.; Guijarro-Sánchez, P.; Rumbo-Feal, S.; Muíño-Andrade, M.; et al. Simultaneous and divergent evolution of resistance to cephalosporin/β-lactamase inhibitor combinations and imipenem/relebactam following ceftazidime/avibactam treatment of MDR Pseudomonas aeruginosa infections. J. Antimicrob. Chemother. 2023, 78, 1195–1200. [Google Scholar] [CrossRef]
- González-Olvera, E.M.; Pérez-Morales, R.; González-Zamora, A.; Castro-Escarpulli, G.; Palma-Martínez, I.; Alba-Romero, J.J. Antibiotic resistance, virulence factors and genotyping of Pseudomonas aeruginosa in public hospitals of northeastern Mexico. J. Infect. Dev. Ctries. 2019, 13, 374–383. [Google Scholar] [CrossRef]
- Tapia-Cornejo, A.S.; Ramírez-Castillo, F.Y.; Guerrero-Barrera, A.L.; Guillen-Padilla, D.E.; Arreola-Guerra, J.M.; González-Gámez, M.; Avelar-González, F.J.; Loera-Muro, A.; Hernández-Cuellar, E.; Ramos-Medellín, C.L.; et al. Occurrence of Plasmid-Mediated Quinolone Resistance and Carbapenemase-Encoding Genes in Pseudomonas aeruginosa Isolates from Nosocomial Patients in Aguascalientes, Mexico. Pathogens 2024, 13, 992. [Google Scholar] [CrossRef]
- Antochevis, L.C.; Wilhelm, C.M.; Arns, B.; Sganzerla, D.; Sudbrack, L.O.; Nogueira, T.C.R.L.; Guzman, R.D.; Martins, A.S.; Cappa, D.S.; Dos Santos, Â.C.; et al. World Health Organization priority antimicrobial resistance in Enterobacterales, Acinetobacter baumannii, Pseudomonas aeruginosa, Staphylococcus aureus and Enterococcus faecium healthcare-associated bloodstream infections in Brazil (ASCENSION): A prospective, multicentre, observational study. Lancet Reg. Health Am. 2025, 43, 101004. [Google Scholar]
- Tabak, Y.P.; Merchant, S.; Ye, G.; Vankeepuram, L.; Gupta, V.; Kurtz, S.G.; Puzniak, L.A. Incremental clinical and economic burden of suspected respiratory infections due to multi-drug-resistant Pseudomonas aeruginosa in the United States. J. Hosp. Infect. 2019, 103, 134–141. [Google Scholar] [CrossRef]
- Poovieng, J.; Sakboonyarat, B.; Nasomsong, W. Bacterial etiology and mortality rate in community-acquired pneumonia, healthcare-associated pneumonia and hospital-acquired pneumonia in Thai university hospital. Sci. Rep. 2022, 12, 9004. [Google Scholar] [CrossRef]
- Zhen, S.; Zhao, Y.; Chen, Z.; Zhang, T.; Wang, J.; Jiang, E.; Zhang, F.; Mi, Y.; Zhu, X.; Han, M.; et al. Assessment of mortality-related risk factors and effective antimicrobial regimens for treatment of bloodstream infections caused by carbapenem-resistant Pseudomonas aeruginosa in patients with hematological diseases. Front. Cell. Infect. Microbiol. 2023, 13, 1156651. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.Q.; Gong, Y.W.; Tian, J.Q.; Peng, C.; Niu, S.M.; Feng, H.Z.; You, C.G.; Wang, Z.P. Cross-talk between Pseudomonas aeruginosa and urinary tract infections. World J. Urol. 2025, 43, 380. [Google Scholar] [CrossRef] [PubMed]
- Liao, C.; Huang, X.; Wang, Q.; Yao, D.; Lu, W. Virulence Factors of Pseudomonas aeruginosa and Antivirulence Strategies to Combat Its Drug Resistance. Front. Cell. Infect. Microbiol. 2022, 12, 926758. [Google Scholar] [CrossRef] [PubMed]
- Park, M.H.; Kim, S.Y.; Roh, E.Y.; Lee, H.S. Difference of Type 3 secretion system (T3SS) effector gene genotypes (exoU and exoS) and its implication to antibiotics resistances in isolates of P. aeruginosa from chronic otitis media. Auris Nasus Larynx 2017, 44, 258–265. [Google Scholar] [CrossRef]
- Faraji, F.; Mahzounieh, M.; Ebrahimi, A.; Fallah, F.; Teymournejad, O.; Lajevardi, B. Molecular detection of virulence genes in Pseudomonas aeruginosa isolated from children with Cystic Fibrosis and burn wounds in Iran. Microb. Pathog. 2016, 99, 1–4. [Google Scholar] [CrossRef]
- Kaiser, S.J.; Mutters, N.T.; DeRosa, A.; Ewers, C.; Frank, U.; Günther, F. Determinants for persistence of Pseudomonas aeruginosa in hospitals: Interplay between resistance, virulence and biofilm formation. Eur. J. Clin. Microbiol. Infect. Dis. 2017, 36, 243–253. [Google Scholar] [CrossRef]
- Hemmati, J.; Nazari, M.; Abolhasani, F.S.; Ahmadi, A.; Asghari, B. In vitro investigation of relationship between quorum-sensing system genes, biofilm forming ability, and drug resistance in clinical isolates of Pseudomonas aeruginosa. BMC Microbiol. 2024, 24, 99. [Google Scholar] [CrossRef]
- de Sousa, T.; Silva, C.; Alves, O.; Costa, E.; Igrejas, G.; Poeta, P.; Hébraud, M. Determination of Antimicrobial Resistance and the Impact of Imipenem + Cilastatin Synergy with Tetracycline in Pseudomonas aeruginosa Isolates from Sepsis. Microorganisms 2023, 11, 2687. [Google Scholar] [CrossRef]
- Mateu-Borrás, M.; González-Alsina, A.; Doménech-Sánchez, A.; Querol-García, J.; Fernández, F.J.; Vega, M.C.; Albertí, S. Pseudomonas aeruginosa adaptation in cystic fibrosis patients increases C5a levels and promotes neutrophil recruitment. Virulence 2022, 13, 215–224. [Google Scholar] [CrossRef]
- Mackinder, J.R.; Hinkel, L.A.; Schutz, K.; Eckstrom, K.; Fisher, K.; Wargo, M.J. Sphingosine induction of the Pseudomonas aeruginosa hemolytic phospholipase C/sphingomyelinase (PlcH). J. Bacteriol. 2024, 206, e0038223. [Google Scholar] [CrossRef]
- Ali, N.M.; Chatta, S.; Liaqat, I.; Mazhar, S.A.; Mazhar, B.; Zahid, S. Pseudomonas aeruginosa associated pulmonary infections and in vitro amplification virulent rhamnolipid (rhlR) gene. Braz. J. Biol. 2021, 82, e228009. [Google Scholar] [CrossRef]
- Guo, L.; Ruan, Q.; Ma, D.; Wen, J. Revealing quorum-sensing networks in Pseudomonas aeruginosa infections through internal and external signals to prevent new resistance trends. Microbiol. Res. 2024, 289, 127915. [Google Scholar] [CrossRef] [PubMed]
- Frem, J.A.; Doumat, G.; Kazma, J.; Gharamti, A.; Kanj, S.S.; Abou Fayad, A.G.; Matar, G.M.; Kanafani, Z.A. Clinical predictors of mortality in patients with Pseudomonas aeruginosa infection. PLoS ONE 2023, 18, e02822. [Google Scholar] [CrossRef] [PubMed]
- Monroy-Pérez, E.; Herrera-Gabriel, J.P.; Olvera-Navarro, E.; Ugalde-Tecillo, L.; García-Cortés, L.R.; Moreno-Noguez, M.; Martínez-Gregorio, H.; Vaca-Paniagua, F.; Paniagua-Contreras, G.L. Molecular Properties of Virulence and Antibiotic Resistance of Pseudomonas aeruginosa Causing Clinically Critical Infections. Pathogens 2024, 13, 868. [Google Scholar] [CrossRef] [PubMed]
- Mack, A.R.; Hujer, A.M.; Mojica, M.F.; Taracila, M.A.; Feldgarden, M.; Haft, D.H.; Klimke, W.; Prasad, A.B.; Bonomo, R.A. β-Lactamase diversity in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2025, 69, e0078524. [Google Scholar] [CrossRef]
- Nasser, M.; Gayen, S.; Kharat, A.S. Prevalence of β-lactamase and antibiotic-resistant Pseudomonas aeruginosa in the Arab region. J. Glob. Antimicrob. Resist. 2020, 22, 152–160. [Google Scholar] [CrossRef]
- Santerre Henriksen, A.; Jeannot, K.; Oliver, A.; Perry, J.D.; Pletz, M.W.; Stefani, S.; Morrissey, I.; Longshaw, C. ARTEMIS Study Investigators. In vitro activity of cefiderocol against European Pseudomonas aeruginosa and Acinetobacter spp., including isolates resistant to meropenem and recent β-lactam/β-lactamase inhibitor combinations. Microbiol. Spectr. 2024, 12, e0383623. [Google Scholar] [CrossRef]
- Carcione, D.; Siracusa, C.; Sulejmani, A.; Leoni, V.; Intra, J. Old and New Beta-Lactamase Inhibitors: Molecular Structure, Mechanism of Action, and Clinical Use. Antibiotics 2021, 10, 995. [Google Scholar] [CrossRef]
- Zhao, W.H.; Hu, Z.Q. β-lactamases identified in clinical isolates of Pseudomonas aeruginosa. Crit. Rev. Microbiol. 2010, 36, 245–258. [Google Scholar] [CrossRef]
- Zhao, D.W.; Lohans, C.T. Combatting Pseudomonas aeruginosa with β-Lactam Antibiotics: A Revived Weapon? Antibiotics 2025, 14, 526. [Google Scholar] [CrossRef]
- Yahav, D.; Giske, C.G.; Grāmatniece, A.; Abodakpi, H.; Tam, V.H.; Leibovici, L. New β-Lactam–β-Lactamase Inhibitor Combinations. Clin. Microbiol. Rev. 2020, 34, e00115-20. [Google Scholar] [CrossRef] [PubMed]
- Asmare, Z.; Reta, M.A.; Gashaw, Y.; Getachew, E.; Sisay, A.; Gashaw, M.; Tamrat, E.; Kidie, A.A.; Abebe, W.; Misganaw, T.; et al. Antimicrobial resistance profile of Pseudomonas aeruginosa clinical isolates from healthcare-associated infections in Ethiopia: A systematic review and meta-analysis. PLoS ONE 2024, 19, e0308946. [Google Scholar] [CrossRef] [PubMed]
- Camargo, C.H.; Yamada, A.Y.; Souza, A.R.; Lima, M.J.C.; Cunha, M.P.V.; Ferraro, P.S.P.; Sacchi, C.T.; Santos, M.B.N.D.; Campos, K.R.; Tiba-Casas, M.R.; et al. Genomics and Antimicrobial Susceptibility of Clinical Pseudomonas aeruginosa Isolates from Hospitals in Brazil. Pathogens 2023, 12, 918. [Google Scholar] [CrossRef] [PubMed]
- Sader, H.S.; Flamm, R.K.; Carvalhaes, C.G.; Castanheira, M. Antimicrobial Susceptibility of Pseudomonas aeruginosa to Ceftazidime-Avibactam, Ceftolozane-Tazobactam, Piperacillin-Tazobactam, and Meropenem Stratified by U.S. Census Divisions: Results from the 2017 INFORM Program. Antimicrob. Agents Chemother. 2018, 62, e01587-18. [Google Scholar] [CrossRef]
- García-Betancur, J.C.; De La Cadena, E.; Mojica, M.F.; Hernández-Gómez, C.; Correa, A.; Radice, M.A.; Castañeda-Méndez, P.; Jaime-Villalon, D.A.; Gales, A.C.; Munita, J.M.; et al. Comparative in vitro activity of Ceftolozane/Tazobactam against clinical isolates of Pseudomonas aeruginosa and Enterobacterales from Five Latin American Countries. Antibiotics 2022, 11, 1101. [Google Scholar] [CrossRef]
- Camacho-Ortiz, A.; Flores-Treviño, S.; Bocanegra-Ibarias, P. Prevalence of difficult-to-treat resistance in ESKAPE pathogens in a third level hospital in Mexico. Infect. Prev. Pract. 2024, 7, 100426. [Google Scholar] [CrossRef]
- Cosentino, F.; Viale, P.; Giannella, M. MDR/XDR/PDR or DTR? Which definition best fits the resistance profile of Pseudomonas aeruginosa? Curr. Opi. Infect. Dis. 2023, 36, 564–571. [Google Scholar] [CrossRef]
- Tamma, P.D.; Aitken, S.L.; Bonomo, R.A.; Mathers, A.J.; van Duin, D.; Clancy, C.J. Infectious Diseases Society of America Guidance on the Treatment of Extended-Spectrum β-lactamase Producing Enterobacterales (ESBL-E), Carbapenem-Resistant Enterobacterales (CRE), and Pseudomonas aeruginosa with Difficult-to-Treat Resistance (DTR-P. aeruginosa). Clin. Infect. Dis. 2021, 72, e169–e183. [Google Scholar]
- Olana, M.D.; Asrat, D.; Swedberg, G. Molecular characterization of serotype and virulence genes of Pseudomonas aeruginosa isolated from patients admitted at two hospitals in Addis Ababa, Ethiopia. J. Med. Microbiol. 2025, 74, 002034. [Google Scholar] [CrossRef]
- Swain, J.; Askenasy, I.; Rudland Nazeer, R.; Ho, P.M.; Labrini, E.; Mancini, L.; Xu, Q.; Hollendung, F.; Sheldon, I.; Dickson, C.; et al. Pathogenicity and virulence of Pseudomonas aeruginosa: Recent advances and under-investigated topics. Virulence 2025, 16, 2503430. [Google Scholar] [CrossRef]
- Quiroz-Morales, S.E.; Piñón, J.; Salgado, N.; Medina, A.; Rojas-Ríos, R.; González-Valdez, A.; López-Jácome, E.; Franco-Cendejas, R.; Ponce-Soto, G.Y.; Servín-González, L.; et al. Phylogrouping of Pseudomonas aeruginosa from burn patients reveals distinct group 3 characteristics. J. Med. Microbiol. 2025, 74, 002078. [Google Scholar] [CrossRef] [PubMed]
- CLSI. Performance standards for antimicrobial susceptibility testing. In CLSI, Twenty-Third Informational Supplement M100-S23; Carpenter, D.E., Ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2013; Volume 74. [Google Scholar]
- Ooka, T.; Terajima, J.; Kusumoto, M.; Iguchi, A.; Kurokawa, K.; Ogura, Y.; Asadulghani, M.; Nakayama, K.; Murase, K.; Ohnishi, M.; et al. Development of a multiplex PCR-based rapid typing method for enterohemorrhagic Escherichia coli O157 strains. J. Clin. Microbiol. 2009, 47, 2888–2894. [Google Scholar] [CrossRef] [PubMed]
- Kaszab, E.; Szoboszlay, S.; Dobolyi, C.; Háhn, J.; Pék, N.; Kriszt, B. Antibiotic resistance profiles and virulence markers of Pseudomonas aeruginosa strains isolated from composts. Bioresour Technol. 2011, 102, 1543–1548. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.Q.; Li, T.T.; Li, Z.R.; Zhang, L.C.; Zhang, L.H.; Han, L.; Tang, P.F. Effect of Negative Pressure on Proliferation, Virulence Factor Secretion, Biofilm Formation, and Virulence-Regulated Gene Expression of Pseudomonas aeruginosa In Vitro. BioMed Res. Int. 2016, 2016, 7986234. [Google Scholar] [CrossRef]
- Momtaz, H.; Karimian, A.; Madani, M.; Safarpoor Dehkordi, F.; Ranjbar, R.; Sarshar, M.; Souod, N. Uropathogenic Escherichia coli in Iran: Serogroup distributions, virulence factors and antimicrobial resistance properties. Ann. Clin. Microbiol. Antimicrob. 2013, 12, 8. [Google Scholar] [CrossRef]
- Amirkamali, S.; Naserpour-Farivar, T.; Azarhoosh, K.; Peymani, A. Distribution of the blaOXA, blaVEB-1, and blaGES-1 genes and resistance patterns of ESBL-producing Pseudomonas aeruginosa isolated from hospitals in Tehran and Qazvin, Iran. Rev. Soc. Bras. Med. Trop. 2017, 50, 315–320. [Google Scholar] [CrossRef]
- Romero-Gonzalez, L.E.; Montelongo-Martinez, L.F.; González-Valdez, A.; Quiroz-Morales, S.E.; Cocotl-Yañez, M.; Franco-Cendejas, R.; Soberon-Chavez, G.; Pardo-Lopez, L.; Bustamante, V.H. Pseudomonas aeruginosa Isolates from Water Samples of the Gulf of Mexico Show Similar Virulence Properties but Different Antibiotic Susceptibility Profiles than Clinical Isolates. Int. J. Microbiol. 2024, 2024, 6959403. [Google Scholar] [CrossRef]
- Gower, J.C. A General Coefficient of Similarity and Some of Its Properties. Biometrics 1971, 27, 857–871. [Google Scholar] [CrossRef]
- Ward, J.H., Jr. Hierarchical Grouping to Optimize an Objective Function. J. Am. Stat. Assoc. 1963, 58, 236–244. [Google Scholar] [CrossRef]
| Strain Origin (n = 124) | Infection | Function (Gene) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Protease | Phospholipases | Elastases | Rhamnolipids | Quorum-Sensing | |||||
| aprA No. (%) | pIcH No. (%) | plcN No. (%) | lasA No. (%) | lasB No. (%) | rhlA No. (%) | lasI No. (%) | rhII No. (%) | ||
| Healthcare associated infections (n = 67) | Pneumonia (n = 16) | 15 (93.7) | 16 (100) | 14 (87.5) | 16 (100) | 12 (75) | 16 (100) | 16 (100) | 16 (100) |
| Bacteremia (n = 24) | 23 (95.8) | 24 (100) | 22 (91.6) | 24 (100) | 14 (58.3) | 24 (100) | 24 (100) | 24 (100) | |
| Wound infection (n = 27) | 27 (100) | 27 (100) | 25 (92.5) | 26 (96.2) | 21 (77.7) | 27 (100) | 27 (100) | 27 (100) | |
| Community-acquired infections (n = 57) | Respiratory infection (n = 10) | 10 (100) | 10 (100) | 9 (90) | 10 (100) | 9 (90) | 10 (100) | 10 (100) | 10 (100) |
| Catheter associated infection (n = 10) | 10 (100) | 10 (100) | 9 (90) | 10 (100) | 9 (90) | 10 (100) | 10 (100) | 10 (100) | |
| UTI (n = 37) | 36 (97.3) | 37 (100) | 33 (89.1) | 36 (97.3) | 31 (83.7) | 37 (100) | 37 (100) | 37 (100) | |
| p-value | 0.2903 | 1 | 0.995 | 1 | 0.5 | 1 | 1 | 1 | |
| Total, No. (%) | 121 (97.5) | 124 (100) | 112 (90.3) | 122 (98.3) | 96 (77.4) | 124 (100) | 124 (100) | 124 (100) | |
| Strain Origin (n = 124) | Infection | Phenotype of Resistance to β-Lactamase Inhibitor Antibiotics | β-Lactamase Genes | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Amoxicillin/ Clavulanic Acid | Ampicillin/ Sulbactam | Piperacillin /Tazobactam | blaSHV No. (%) | blaCITM No. (%) | blaOXA-1 No. (%) | blaOXA-2 No. (%) | blaOXA-4 No. (%) | blaOXA-10 No. (%) | blaGES No. (%) | blaPER No. (%) | blaVEB No. (%) | ||
| Healthcare associated infections (n = 67) | Pneumonia (n = 16) | 16 (100) | 16 (100) | 6 (37.5) | 3 (18.7) | 0 (0) | 4 (25) | 16 (100) | 12 (75) | 0 (0) | 10 (62.5) | 0 (0) | 0 (0) |
| Bacteremia (n = 24) | 23 (95.8) | 24 (100) | 10 (41.6) | 4 (16.6) | 0 (0) | 2 (8.3) | 22 (91.6) | 17 (70.8) | 0 (0) | 15 (62.5) | 0 (0) | 0 (0) | |
| Wound infection (n = 27) | 27 (100) | 27 (100) | 18 (66.6) | 5 (18.5) | 1 (3.7) | 8 (29.6) | 27 (100) | 25 (92.5) | 0 (0) | 25 (92.5) | 1 (3.7) | 2 (7.4) | |
| Community-acquired infections (n = 57) | Respiratory infection (n = 10) | 10 (100) | 10 (100) | 8 (80) | 0 (0) | 0 (0) | 3 (30) | 10 (100) | 7 (70) | 0 (0) | 5 (50) | 0 (0) | 0 (0) |
| Catheter associated infection (n = 10) | 10 (100) | 10 (100) | 7 (70) | 3 (30) | 3 (30) | 2 (20) | 8 (80) | 7 (70) | 0 (0) | 7 (70) | 1 (10) | 1 (10) | |
| UTI (n = 37) | 37 (100) | 37 (100) | 18 (48.6) | 7 (18.9) | 5 (13.5) | 14 (37.8) | 36 (97.3) | 29 (78.3) | 0 (0) | 25 (67.5) | 0 (0) | 0 (0) | |
| p-value | 0.438 | 1 | 0.001 | 0.667 | 0.024 | 0.191 | 0.096 | 0.317 | 0 (0) | 0.047 | 0.216 | 0.141 | |
| Total No. (%) | 123 (99.1) | 124 (100) | 67 (54) | 22 (17.7) | 9 (7.2) | 33 (26.6) | 119 (95.9) | 97 (78.3) | 0 (0) | 87 (70.1) | 2 (1.6) | 3 (2.4) | |
| Healthcare-Associated Infections (n = 67) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Protease | Phospholipases | Elastases | Rhamnolipids | Quorum-Sensing | |||||
| aprA No. (%) | pIcH No. (%) | plcN No. (%) | lasA No. (%) | lasB No. (%) | rhLA No. (%) | lasI No. (%) | rhII No. (%) | ||
| Phenotype of β-lactamase inhibitor antibiotics | Amoxicillin/ clavulanic acid | 64 (95.5) | 66 (98.5) | 61 (91) | 65 (97) | 46 (68.6) | 66 (98.5) | 66 (98.5) | 66 (98.5) |
| Ampicillin/ sulbactam | 65 (97) | 67 (100) | 61 (91) | 66 (98.5) | 47 (70.1) | 67 (100) | 67 (100) | 67 (100) | |
| Piperacillin/ tazobactam | 34 (50.7) | 34 (50.7) | 31 (46.2) | 33 (49.2) | 26 (38.8) | 34 (50.7) | 34 (50.7) | 34 (50.7) | |
| β-lactamase genes | blaSHV | 12 (17.9) | 12 (17.9) | 11 (16.4) | 12 (17.9) | 8 (11.9) | 12 (17.9) | 12 (17.9) | 12 (17.9) |
| blaCITM | 1 (1.4) | 1 (1.4) | 1 (1.4) | 1 (1.4) | 1 (1.4) | 1 (1.4) | 1 (1.4) | 1 (1.4) | |
| blaOXA-1 | 14 (20.8) | 15 (22.3) | 13 (19.4) | 15 (22.3) | 10 (14.9) | 15 (22.3) | 15 (22.3) | 15 (22.3) | |
| blaOXA-2 | 63 (94) | 65 (97) | 59 (88) | 64 (95.5) | 47 (70.1) | 65 (97) | 65 (97) | 65 (97) | |
| blaOXA-4 | 52 (77.6) | 54 (80.5) | 49 (73.1) | 53 (79.1) | 37 (55.2) | 54 (80.5) | 54 (80.5) | 54 (80.5) | |
| blaOXA-10 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |
| blaGES | 49 (73.1) | 50 (74.6) | 45 (67.1) | 50 (74.6) | 41 (61.1) | 50 (74.6) | 50 (74.6) | 50 (74.6) | |
| blaPER | 0 (0) | 1 (1.4) | 0 (0) | 1 (1.4) | 1 (1.4) | 1 (1.4) | 1 (1.4) | 1 (1.4) | |
| blaVEB | 0 (0) | 2 (2.9) | 2 (2.9) | 2 (2.9) | 2 (2.9) | 2 (2.9) | 2 (2.9) | 2 (2.9) | |
| p-value | 0.0004 | 0.0004 | 0.0004 | 0.0004 | 0.0004 | 0.0004 | 0.0004 | 0.0004 | |
| Community-Acquired Infections (n = 57) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Protease | Phospholipases | Elastases | Rhamnolipids | Quorum-Sensing | |||||
| aprA No. (%) | pIcH No. (%) | plcN No. (%) | lasA No. (%) | lasB No. (%) | rhLA No. (%) | lasI No. (%) | rhII No. (%) | ||
| Phenotype of β-lactamase inhibitor antibiotics | Amoxicillin/ clavulanic acid | 56 (98.2) | 57 (100) | 51 (89.4) | 56 (98.2) | 49 (85.9) | 57 (100) | 57 (100) | 57 (100) |
| Ampicillin/ sulbactam | 56 (98.2) | 57 (100) | 51 (89.4) | 56 (98.2) | 49 (85.9) | 57 (100) | 57 (100) | 57 (100) | |
| Piperacillin/ tazobactam | 33 (57.8) | 24 (42.1) | 20 (35) | 33 (57.8) | 31 (54.3) | 24 (42.1) | 33 (57.8) | 33 (57.8) | |
| β-lactamase genes | blaSHV | 10 (17.5) | 10 (17.5) | 10 (17.5) | 10 (17.5) | 8 (14) | 10 (17.5) | 10 (17.5) | 10 (17.5) |
| blaCITM | 8 (14) | 8 (14) | 7 (12.2) | 8 (14) | 7 (12.2) | 8 (14) | 8 (14) | 8 (14) | |
| blaOXA-1 | 18 (31.5) | 18 (31.5) | 15 (26.3) | 18 (31.5) | 16 (28.0) | 18 (31.5) | 18 (31.5) | 18 (31.5) | |
| blaOXA-2 | 54 (94.7) | 55 (96.4) | 49 (85.9) | 54 (94.7) | 46 (80.7) | 55 (96.4) | 55 (96.4) | 55 (96.4) | |
| blaOXA-4 | 45 (78.9) | 45 (78.9) | 40 (70.1) | 44 (77.1) | 37 (64.9) | 45 (78.9) | 45 (78.9) | 45 (78.9) | |
| blaOXA-10 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |
| blaGES | 36 (63.1) | 37 (64.9) | 31 (54.3) | 37 (64.9) | 30 (52.6) | 37 (64.9) | 37 (64.9) | 37 (64.9) | |
| blaPER | 1 (1.7) | 1 (1.7) | 1 (1.7) | 1 (1.7) | 1 (1.7) | 1 (1.7) | 1 (1.7) | 1 (1.7) | |
| blaVEB | 1 (1.7) | 1 (1.7) | 1 (1.7) | 1 (1.7) | 1 (1.7) | 1 (1.7) | 1 (1.7) | 1 (1.7) | |
| p-value | 0.0004 | 0.0004 | 0.0004 | 0.0004 | 0.0004 | 0.0004 | 0.0004 | 0.0004 | |
| Strain Origin (n = 124) | Infection | Phylogroup | MIC for β-Lactamase Inhibitor Antibiotics (0.016–256 μg/mL) | |||||
|---|---|---|---|---|---|---|---|---|
| 1 No. (%) | 2 No. (%) | Not Determined No. (%) | Ticarcillin/ Clavulanic Acid | Piperacillin/ Tazobactam | ||||
| MIC | No. (%) | MIC | No. (%) | |||||
| Healthcare associated infections (n = 67) | Pneumonia (n = 16) | 3 (18.7) | 1 (6.2) | 12 (75) | >256 | 16 (100) | >256 48 32 24 6 4 3 | 6 (37.5) 1 (6.2) 2 (12.5) 2 (12.5) 3 (18.7) 1 (6.2) 1 (6.2) |
| Bacteremia (n = 24) | 4 (16.6) | 2 (8.3) | 18 (75) | >256 64 | 23 (95.8) 1 (4.1) | >256 48 32 24 16 12 8 | 9 (37.5) 4 (16.6) 4 (16.6) 2 (8.3) 2 (8.3) 2 (8.3) 1 (4.1) | |
| Wound infection (n = 27) | 4 (14.8) | 2 (7.4) | 21 (77.7) | >256 | 27 (100) | >256 64 48 32 24 12 3 | 18 (66.6) 1 (3.7) 1 (3.7) 1 (3.7) 1 (3.7) 2 (7.4) 3 (11.1) | |
| Community-acquired infections (n = 57) | Respiratory infection (n = 10) | 1 (10) | 1 (10) | 8 (80) | >256 | 10 (100) | >256 16 12 8 | 7 (70) 1 (10) 1 (10) 1 (10) |
| Catheter associated infection (n = 10) | 2 (20) | 2 (20) | 6 (60) | >256 | 10 (100) | >256 96 48 | 8 (80) 1 (10) 1 (10) | |
| UTI (n = 37) | 8 (21.6) | 0 (0) | 29 (78.3) | >256 | 37 (100) | >256 64 48 32 28 16 12 8 4 | 19 (51.3) 1 (2.7) 2 (5.4) 4 (10.8) 1 (2.7) 2 (5.4) 1 (2.7) 6 (16.2) 1 (2.7) | |
| Total, No. (%) | 22 (17.7) | 8 (6.4) | 94 (75.8) | >256 (R) 64 (S) | 123 (99.1) 1 (0.9) | >256 (R) 3–96 (S) | 67 (54) 57 (46) | |
| Pattern Number | Genotype Combination Related to Inhibitor Resistance Phenotype | Healthcare Associated Infections (n = 67) | Community-Acquired Infections (n = 57) | Phylogroup | Total (n = 124) No. (%) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Pneumonia (n = 16) No. (%) | Bacteremia (n = 24) No. (%) | Wound Infection (n = 27) No. (%) | Respiratory Infection (n = 10) No. (%) | Catheter Associated Infection (n = 10) No. (%) | UTI (n = 37) No. (%) | 1 | 2 | No Deter Mined | |||
| 1 | aprA/pIcH/pIcN/lasA/lasB/rhLA/lasI/rhII/blaOXA-2/blaOXA-4/blaGES/Am-Sul/Amox-Ac.Clavul/Piper-Tazo | 2 (12.5) | 3 (12.5) | 5 (18.5) | 2 (20) | 0 (0) | 2 (5.4) | 2 | 1 | 11 | 14 (11.3) |
| 2 | aprA/pIcH/pIcN/lasA/lasB/rhLA/lasI/rhII/blaOXA-2/blaOXA-4/Am-Sul/Amox-Ac.Clavul/Piper-Tazo | 0 (0) | 1 (4.1) | 0 (0) | 2 (20) | 2 (20) | 5 (13.5) | 2 | 1 | 7 | 10 (8.0) |
| 3 | aprA/pIcH/pIcN/lasA/lasB/rhLA/lasI/rhII/blaOXA-2/blaOXA-4/blaGES/blaOXA-1/Am-Sul/Amox-Ac.Clavul/Piper-Tazo | 0 (0) | 0 (0) | 3 (11.1) | 1 (10) | 1 (10) | 4 (10.8) | 3 | 0 | 6 | 9 (7.2) |
| 4 | aprA/pIcH/pIcN/lasA/lasB/rhLA/lasI/rhII/blaOXA-2/blaOXA-4/blaGES/Am-Sul/Amox-Ac.Clavul | 2 (12.5) | 2 (8.3) | 3 (11.1) | 0 (0) | 0 (0) | 1 (2.7) | 1 | 0 | 7 | 8 (6.4) |
| 5 | aprA/pIcH/pIcN/lasA/lasB/rhLA/lasI/rhII/blaOXA-2/blaGES/Am-Sul/Amox-Ac.Clavul/Piper-Tazo | 0 (0) | 1 (4.1) | 1 (3.7) | 0 (0) | 0 (0) | 3 (8.1) | 0 | 0 | 5 | 5 (4.0) |
| 6 | aprA/pIcH/pIcN/lasA/rhLA/lasI/rhII/blaOXA-2/blaOXA-4/blaGES/Am-Sul/Amox-Ac.Clavul/Piper-Tazo | 0 (0) | 2 (8.3) | 2 (7.4) | 0 (0) | 1 (10) | 0 (0) | 1 | 0 | 4 | 5 (4.0) |
| 7 | aprA/pIcH/pIcN/lasA/rhLA/lasI/rhII/blaOXA-2/blaOXA-4/blaGES/Am-Sul/Amox-Ac.Clavul | 0 (0) | 1 (4.1) | 2 (7.4) | 0 (0) | 0 (0) | 2 (5.4) | 0 | 0 | 5 | 5 (4.0) |
| 8 | aprA/pIcH/pIcN/lasA/lasB/rhLA/lasI/rhII/blaOXA-2/blaGES/Am-Sul/Amox-Ac.Clavul | 2 (12.5) | 1 (4.1) | 0 (0) | 1 (10) | 0 (0) | 0 (0) | 2 | 1 | 1 | 4 (3.2) |
| 9 | aprA/pIcH/lasA/lasB/rhLA/lasI/rhII/blaOXA-2/blaOXA-4/blaGES/Am-Sul/Amox-Ac.Clavul | 1 (6.2) | 1 (4.1) | 0 (0) | 0 (0) | 0 (0) | 1 (2.7) | 0 | 0 | 3 | 3 (2.4) |
| 10 | aprA/pIcH/pIcN/lasA/lasB/rhLA/lasI/rhII/blaOXA-2/Am-Sul/Amox-Ac.Clavul/Piper-Tazo | 2 (12.5) | 0 (0) | 0 (0) | 0 (0) | 1 (10) | 0 (0) | 1 | 0 | 2 | 3 (2.4) |
| 11 | aprA/pIcH/lasA/lasB/rhLA/lasI/rhII/blaOXA-2/blaOXA-4/blaGES/blaOXA-1/Am-Sul/Amox-Ac.Clavul/Piper-Tazo | 1 (6.2) | 0 (0) | 0 (0) | 1 (10) | 0 (0) | 1 (2.7) | 0 | 0 | 3 | 3 (2.4) |
| 12 | aprA/pIcH/pIcN/lasA/rhLA/lasI/rhII/blaOXA-2/blaOXA-4/Am-Sul/Amox-Ac.Clavul/Piper-Tazo | 1 (6.2) | 1 (4.1) | 1 (3.7) | 0 (0) | 0 (0) | 0 (0) | 1 | 0 | 2 | 3 (2.4) |
| 13 | aprA/pIcH/pIcN/lasA/lasB/rhLA/lasI/rhII/blaOXA-2/blaOXA-4/blaGES/blaOXA-1/Am-Sul/Amox-Ac.Clavul | 0 (0) | 0 (0) | 2 (7.4) | 0 (0) | 0 (0) | 1 (2.7) | 2 | 0 | 1 | 3 (2.4) |
| 14 | aprA/pIcH/pIcN/lasA/lasB/rhLA/lasI/rhII/blaOXA-2/blaOXA-4/blaGES/blaOXA-1/blaSHV/Am-Sul/Amox-Ac.Clavul/PiperTazo | 0 (0) | 0 (0) | 1 (3.7) | 0 (0) | 0 (0) | 1 (2.7) | 0 | 0 | 2 | 2 (1.6) |
| 15 | aprA/pIcH/pIcN/lasA/l/rhLA/lasI/rhII/blaOXA-2/Am-Sul/Amox-Ac.Clavul | 0 (0) | 1 (4.1) | 0 (0) | 0 (0) | 0 (0) | 1 (2.7) | 0 | 0 | 2 | 2 (1.6) |
| 16 | aprA/pIcH/pIcN/lasA/lasB/rhLA/lasI/rhII/blaOXA-2/Am-Sul/Amox-Ac.Clavul | 0 (0) | 1 (4.1) | 0 (0) | 1 (10) | 0 (0) | 0 (0) | 0 | 0 | 2 | 2 (1.6) |
| 17 | aprA/pIcH/pIcN/lasA/lasB/rhLA/lasI/rhII/blaOXA-2/blaOXA-4/blaGES/blaOXA-1/blaSHV/Am-Sul/Amox-Ac.Clavul | 1 (6.2) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (2.7) | 0 | 1 | 1 | 2 (1.6) |
| 18 | aprA/pIcH/pIcN/lasA/rhLA/lasI/rhII/blaOXA-2/blaOXA-4/blaGES/blaOXA-1/blaSHV/Am-Sul/Amox-Ac.Clavul | 0 (0) | 1 (4.1) | 0 (0) | 0 (0) | 0 (0) | 1 (2.7) | 0 | 0 | 2 | 2 (1.6) |
| 19 | aprA/pIcH/pIcN/lasA/rhLA/lasI/rhII/blaOXA-2/blaOXA-4/blaGES/blaOXA-1/Am-Sul/Amox-Ac.Clavul | 0 (0) | 1 (4.1) | 0 (0) | 0 (0) | 0 (0) | 1 (2.7) | 0 | 1 | 1 | 2 (1.6) |
| 19–55 | Other combinations | 4 (25) | 7 (29.1) | 7 (25.9) | 2 (20) | 5 (50) | 12 (32.4) | 7 | 3 | 27 | 37 (29.8) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Paniagua-Contreras, G.L.; Olvera-Navarro, E.; Herrera-Gabriel, J.P.; González-Vega, L.V.; García-Cortés, L.R.; Moreno-Noguez, M.; Martínez-Gregorio, H.; Vaca-Paniagua, F.; Fernández-Presas, A.M.; Monroy-Pérez, E. Genetic Analysis of Virulence and β-Lactamase Determinants Related to β-Lactamase Inhibitors in Pseudomonas aeruginosa Strains from Nosocomial Infections. Antibiotics 2026, 15, 16. https://doi.org/10.3390/antibiotics15010016
Paniagua-Contreras GL, Olvera-Navarro E, Herrera-Gabriel JP, González-Vega LV, García-Cortés LR, Moreno-Noguez M, Martínez-Gregorio H, Vaca-Paniagua F, Fernández-Presas AM, Monroy-Pérez E. Genetic Analysis of Virulence and β-Lactamase Determinants Related to β-Lactamase Inhibitors in Pseudomonas aeruginosa Strains from Nosocomial Infections. Antibiotics. 2026; 15(1):16. https://doi.org/10.3390/antibiotics15010016
Chicago/Turabian StylePaniagua-Contreras, Gloria Luz, Elizabeth Olvera-Navarro, Jennefer Paloma Herrera-Gabriel, Laura Verónica González-Vega, Luis Rey García-Cortés, Moisés Moreno-Noguez, Héctor Martínez-Gregorio, Felipe Vaca-Paniagua, Ana María Fernández-Presas, and Eric Monroy-Pérez. 2026. "Genetic Analysis of Virulence and β-Lactamase Determinants Related to β-Lactamase Inhibitors in Pseudomonas aeruginosa Strains from Nosocomial Infections" Antibiotics 15, no. 1: 16. https://doi.org/10.3390/antibiotics15010016
APA StylePaniagua-Contreras, G. L., Olvera-Navarro, E., Herrera-Gabriel, J. P., González-Vega, L. V., García-Cortés, L. R., Moreno-Noguez, M., Martínez-Gregorio, H., Vaca-Paniagua, F., Fernández-Presas, A. M., & Monroy-Pérez, E. (2026). Genetic Analysis of Virulence and β-Lactamase Determinants Related to β-Lactamase Inhibitors in Pseudomonas aeruginosa Strains from Nosocomial Infections. Antibiotics, 15(1), 16. https://doi.org/10.3390/antibiotics15010016







