New Insights in blaKPC Gene Mobilization in Pseudomonas aeruginosa: Acquisition of blaKPC-3 and Identification of a New Tn2-like NTE Mobilizing blaKPC-2
Abstract
1. Introduction
2. Results
2.1. The blaKPC-Harboring P. aeruginosa Isolates Increase Their Frequency in the Colombian Hospital Settings
2.2. The Pandemic High-Risk Clones ST111 and ST235 Continue Being the Catcher of the blaKPC Gene
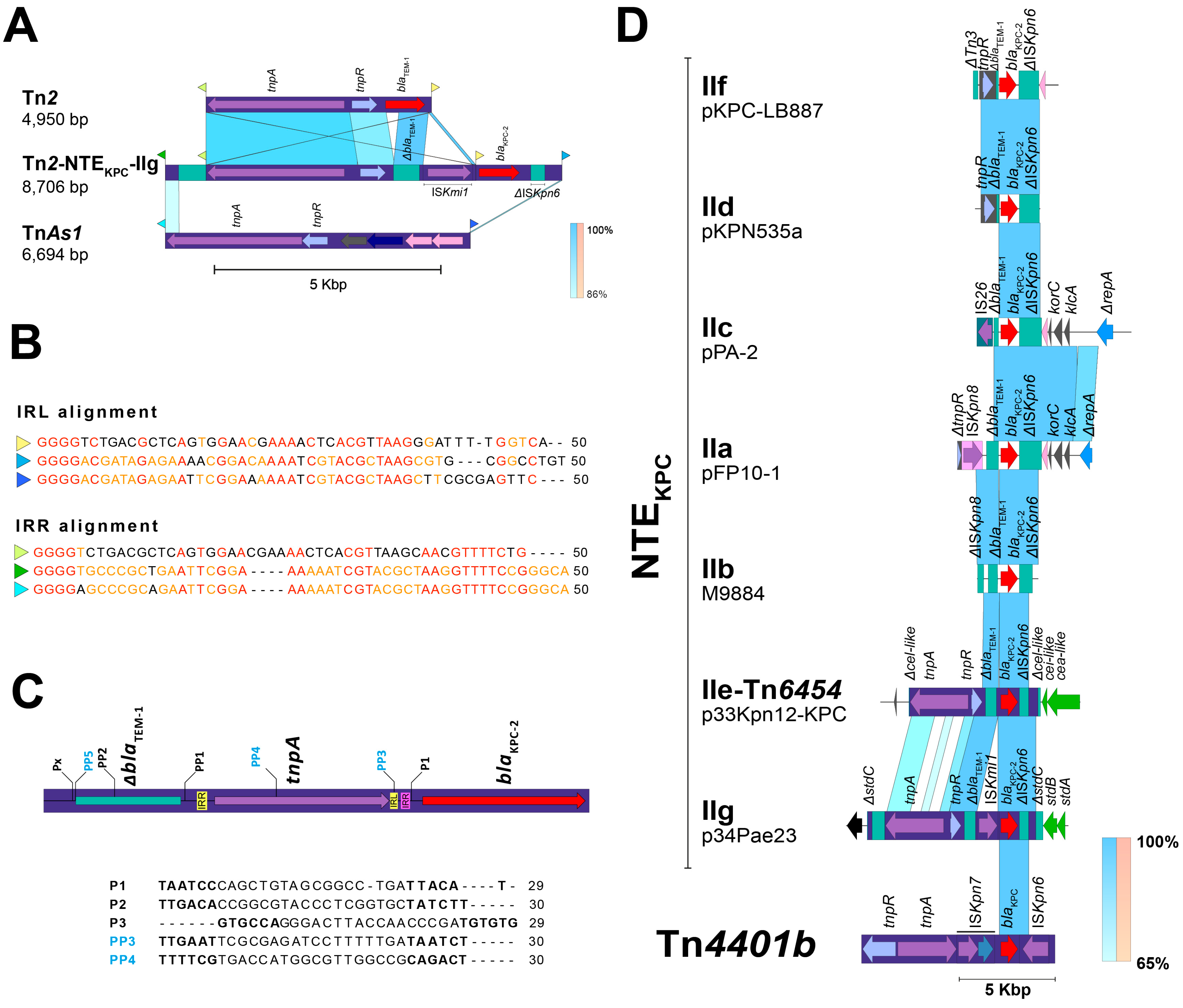
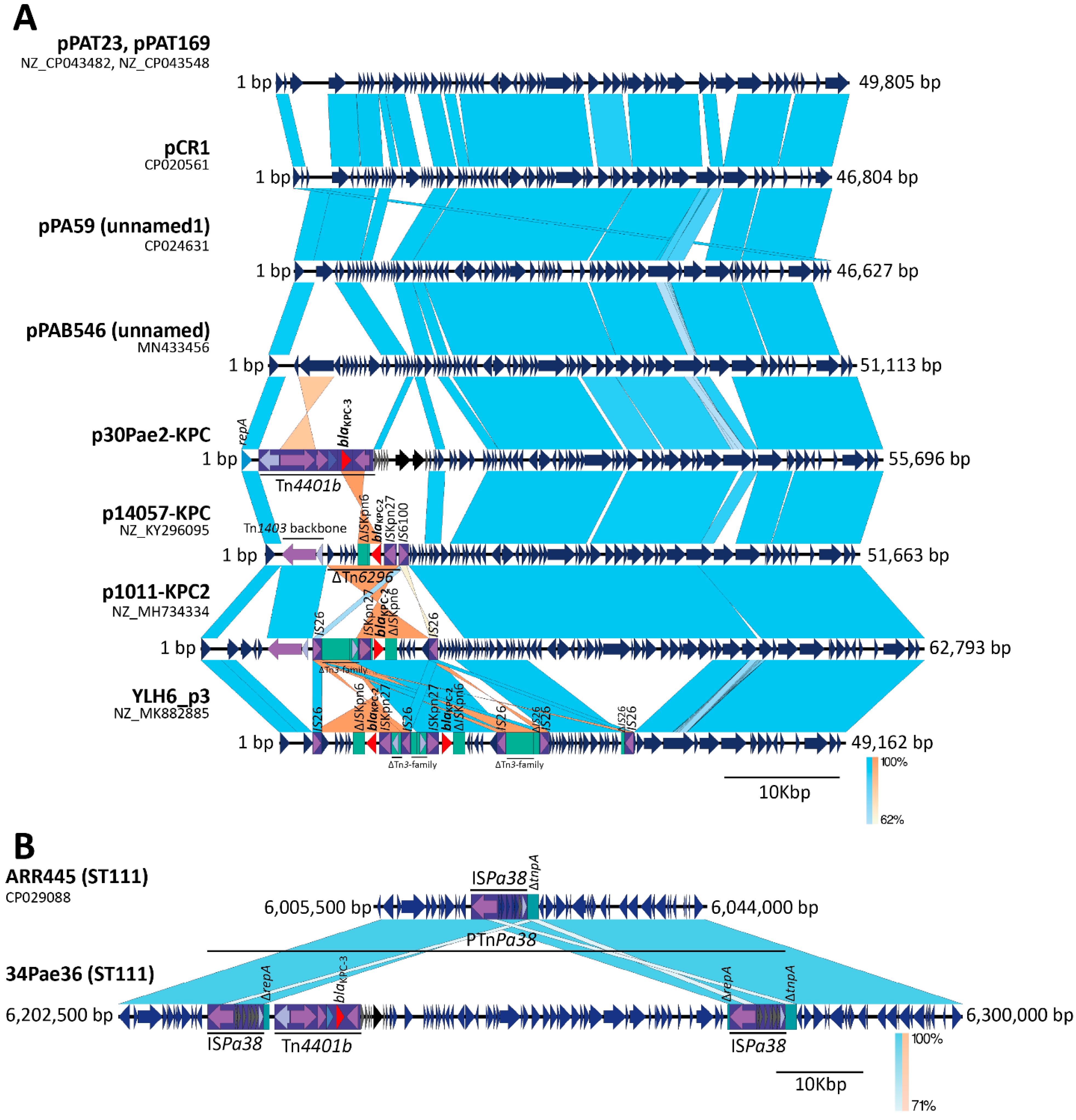
2.3. The blaKPC Gene Is Being Mobilized for New and Different Genetic Platforms
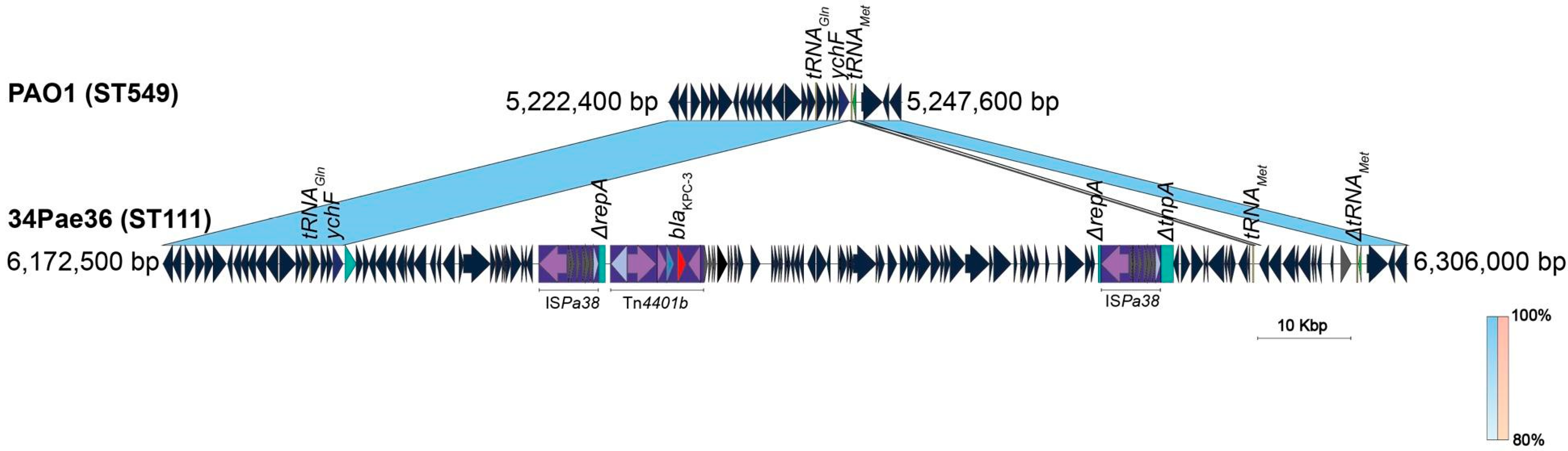
2.4. ISPa38/TnPa38, a New Player Moving Resistance Genes in P. aeruginosa
3. Discussion
4. Materials and Methods
4.1. Source of the Bacterial Isolates, Susceptibility Profiles and Molecular Detection of Carbapenemase Genes
4.2. Genetic Relationship Establishment
4.3. Whole Genome Sequencing (WGS)
4.4. Comparative Genomics Analysis
4.5. Molecular Identification of blaKPC Mobilization Platforms
4.6. Accession Numbers of the Genome Sequences
4.7. Ethics Committee Approbation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yoon, E.J.; Jeong, S.H. Mobile Carbapenemase Genes in Pseudomonas aeruginosa. Front. Microbiol. 2021, 12, 614058. [Google Scholar] [CrossRef] [PubMed]
- Pfizer. Antimicrobial Testing Leadership and Surveillance; Pfizer: New York, NY, USA, 2023. [Google Scholar]
- Yigit, H.; Queenan, A.M.; Anderson, G.J.; Domenech-Sanchez, A.; Biddle, J.W.; Steward, C.D.; Alberti, S.; Bush, K.; Tenover, F.C. Novel carbapenem-hydrolyzing β-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2001, 45, 1151–1161. [Google Scholar] [CrossRef]
- Villegas, M.V.; Lolans, K.; Correa, A.; Kattan, J.N.; Lopez, J.A.; Quinn, J.P. First identification of Pseudomonas aeruginosa isolates producing a KPC-type carbapenem-hydrolyzing β-lactamase. Antimicrob. Agents Chemother. 2007, 51, 1553–1555. [Google Scholar] [CrossRef]
- Akpaka, P.E.; Swanston, W.H.; Ihemere, H.N.; Correa, A.; Torres, J.A.; Tafur, J.D.; Montealegre, M.C.; Quinn, J.P.; Villegas, M.V. Emergence of KPC-producing Pseudomonas aeruginosa in Trinidad and Tobago. J. Clin. Microbiol. 2009, 47, 2670–2671. [Google Scholar] [CrossRef]
- Poirel, L.; Nordmann, P.; Lagrutta, E.; Cleary, T.; Munoz-Price, L.S. Emergence of KPC-Producing Pseudomonas aeruginosa in the United States. Antimicrob. Agents Chemother. 2010, 54, 3072. [Google Scholar] [CrossRef]
- Ge, C.; Wei, Z.; Jiang, Y.; Shen, P.; Yu, Y.; Li, L. Identification of KPC-2-producing Pseudomonas aeruginosa isolates in China. J. Antimicrob. Chemother. 2011, 66, 1184–1186. [Google Scholar] [CrossRef] [PubMed]
- Robledo, I.E.; Aquino, E.E.; Vázquez, G.J. Detection of the KPC gene in Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Acinetobacter baumannii during a PCR-based nosocomial surveillance study in Puerto Rico. Antimicrob. Agents Chemother. 2011, 55, 2968–2970. [Google Scholar] [CrossRef]
- Jácome, P.R.L.D.A.; Alves, Ĺ.R.; Cabral, A.B.; Lopes, A.C.S.; Maciel, M.A.V. First report of KPC-producing Pseudomonas aeruginosa in Brazil. Antimicrob. Agents Chemother. 2012, 56, 4990. [Google Scholar] [CrossRef] [PubMed]
- de Paula-Petroli, S.B.; Campana, E.H.; Bocchi, M.; Bordinhão, T.; Picão, R.C.; Yamada-Ogatta, S.F.; Carrara-Marroni, E.F. Early detection of a hypervirulent KPC-2-producing Pseudomonas aeruginosa ST235 in Brazil. J. Glob. Antimicrob. Resist. 2018, 12, 153–154. [Google Scholar] [CrossRef]
- Hagemann, J.B.; Pfennigwerth, N.; Gatermann, S.G.; von Baum, H.; Essig, A. KPC-2 carbapenemase-producing Pseudomonas aeruginosa reaching Germany. J. Antimicrob. Chemother. 2018, 73, 1812–1814. [Google Scholar] [CrossRef]
- Naas, T.; Bonnin, R.A.; Cuzon, G.; Villegas, M.V.; Nordmann, P. Complete sequence of two KPC-harbouring plasmids from Pseudomonas aeruginosa. J. Antimicrob. Chemother. 2013, 68, 1757–1762. [Google Scholar] [CrossRef]
- Rada, A.M.; De La Cadena, E.; Agudelo, C.; Capataz, C.; Orozco, N.; Pallares, C.; Dinh, A.Q.; Panesso, D.; Ríos, R.; Diaz, L.; et al. Dynamics of bla KPC-2 Dissemination from Non-CG258 Klebsiella pneumoniaea to Other Enterobacterales via IncN Plasmids in an Area of High Endemicity. Antimicrob. Agents Chemother. 2020, 64, e01743-20. [Google Scholar] [CrossRef]
- Abril, D.; Vergara, E.; Palacios, D.; Leal, A.L.; Marquez-Ortiz, R.A.; Madroñero, J.; Rozo, Z.L.C.; De La Rosa, Z.; Nieto, C.A.; Vanegas, N.; et al. Within patient genetic diversity of bla KPC harboring Klebsiella pneumoniaea in a Colombian hospital and identification of a new NTEKPC platform. Sci. Rep. 2021, 11, 21409. [Google Scholar] [CrossRef] [PubMed]
- Ibáñez-Prada, E.D.; Bustos, I.G.; Gamboa-Silva, E.; Josa, D.F.; Mendez, L.; Fuentes, Y.V.; Serrano-Mayorga, C.C.; Baron, O.; Ruiz-Cuartas, A.; Silva, E.; et al. Molecular characterization and descriptive analysis of carbapenemase-producing Gram-negative rod infections in Bogota, Colombia. Microbiol. Spectr. 2024, 12, e01714-23. [Google Scholar] [CrossRef]
- Abril, D.; Marquez-Ortiz, R.A.; Castro-Cardozo, B.; Moncayo-Ortiz, J.I.; Olarte Escobar, N.M.; Corredor Rozo, Z.L.; Reyes, N.; Tovar, C.; Sánchez, H.F.; Castellanos, J.; et al. Genome plasticity favours double chromosomal Tn4401b-bla KPC-2 transposon insertion in the Pseudomonas aeruginosa ST235 clone. BMC Microbiol. 2019, 19, 45. [Google Scholar] [CrossRef] [PubMed]
- Saha, R.; Farrance, C.E.; Verghese, B.; Hong, S.; Donofrio, R.S. Klebsiella michiganensis sp. nov., a new bacterium isolated from a tooth brush holder. Curr. Microbiol. 2013, 66, 72–78. [Google Scholar] [CrossRef]
- Zhu, X.; Li, P.; Qian, C.; Liu, H.; Lin, H.; Zhang, X.; Li, Q.; Lu, J.; Lin, X.; Xu, T.; et al. Prevalence of Aminoglycoside Resistance Genes and Molecular Characterization of a Novel Gene, aac(3)-IIg, among Clinical Isolates of the Enterobacter cloacae Complex from a Chinese Teaching Hospital. Antimicrob. Agents Chemother. 2020, 64, e00852-20. [Google Scholar] [CrossRef] [PubMed]
- Dalrymple’, B.; Arber, W. Promotion of RNA transcription on the insertion element IS30 of E. coli K12. EMBO J. 1985, 4, 2687–2693. [Google Scholar] [CrossRef]
- Cejas, D.; Elena, A.; González-Espinosa, F.E.; Pallecchi, L.; Vay, C.; Rossolini, G.M.; Gutkind, G.; Di Pilato, V.; Radice, M. Characterisation of blaKPC-2–harbouring plasmids recovered from Pseudomonas aeruginosa ST654 and ST235 high-risk clones. J. Glob. Antimicrob. Resist. 2022, 29, 310–312. [Google Scholar] [CrossRef]
- Guérillot, R.; Siguier, P.; Gourbeyre, E.; Chandler, M.; Glaser, P. The diversity of prokaryotic DDE transposases of the mutator superfamily, insertion specificity, and association with conjugation machineries. Genome Biol. Evol. 2014, 6, 260–272. [Google Scholar] [CrossRef]
- Nicolas, E.; Lambin, M.; Dandoy, D.; Galloy, C.; Nguyen, N.; Oger, C.A.; Halletm, B. The Tn3-family of Replicative Transposons. Microbiol. Spectr. 2015, 3, 693–726. [Google Scholar] [CrossRef] [PubMed]
- Molina, L.; Bernal, P.; Udaondo, Z.; Segura, A.; Ramos, J.L. Complete genome sequence of a Pseudomonas putida clinical isolate, strain H8234. Genome Announc. 2013, 1, e00496-13. [Google Scholar] [CrossRef]
- Lawandi, A.; Yek, C.; Kadri, S.S. IDSA guidance and ESCMID guidelines: Complementary approaches toward a care standard for MDR Gram-negative infections. Clin. Microbiol. Infect. 2022, 28, 465–469. [Google Scholar] [CrossRef]
- Paul, M.; Carrara, E.; Retamar, P.; Tängdén, T.; Bitterman, R.; Bonomo, R.A.; de Waele, J.; Daikos, G.L.; Akova, M.; Harbarth, S.; et al. European Society of Clinical Microbiology and Infectious Diseases (ESCMID) guidelines for the treatment of infections caused by multidrug-resistant Gram-negative bacilli (endorsed by European society of intensive care medicine). Clin. Microbiol. Infect. 2022, 28, 521–547. [Google Scholar] [CrossRef]
- Zhu, Y.; Chen, J.; Shen, H.; Chen, Z.; Yang, Q.W.; Zhu, J.; Li, X.; Yang, Q.; Zhao, F.; Ji, J.; et al. Emergence of Ceftazidime-and Avibactam-Resistant Klebsiella pneumoniaea Carbapenemase-Producing Pseudomonas aeruginosa in China. mSystems 2021, 6, e00787-21. [Google Scholar] [CrossRef]
- Rubinstein, E.; Lalani, T.; Corey, G.R.; Kanafani, Z.A.; Nannini, E.C.; Rocha, M.G.; Rahav, G.; Niederman, M.S.; Kollef, M.H.; Shorr, A.F.; et al. Telavancin versus vancomycin for hospital-acquired pneumonia due to gram-positive pathogens. Clin. Infect. Dis. 2011, 52, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Martinez, E.; Pérez, J.E.; Buelvas, F.; Tovar, C.; Vanegas, N.; Stokes, H.W. Establishment and multi drug resistance evolution of ST235 Pseudomonas aeruginosa strains in the intensive care unit of a Colombian hospital. Res. Microbiol. 2014, 165, 852–856. [Google Scholar] [CrossRef] [PubMed]
- del Barrio-Tofiño, E.; López-Causapé, C.; Oliver, A. Pseudomonas aeruginosa epidemic high-risk clones and their association with horizontally-acquired β-lactamases: 2020 update. Int. J. Antimicrob. Agents 2020, 56, 106196. [Google Scholar] [CrossRef]
- Pirnay, J.P.; Bilocq, F.; Pot, B.; Cornelis, P.; Zizi, M.; Van Eldere, J.; Deschaght, P.; Vaneechoutte, M.; Jennes, S.; Pitt, T.; et al. Pseudomonas aeruginosa population structure revisited. PLoS ONE 2009, 4, e7740. [Google Scholar] [CrossRef]
- Rada, A.M.; De La Cadena, E.; Agudelo, C.A.; Pallares, C.; Restrepo, E.; Correa, A.; Villegas, M.V.; Capataz, C. Genetic Diversity of Multidrug-Resistant Pseudomonas aeruginosa Isolates Carrying blaVIM–2 and blaKPC–2 Genes That Spread on Different Genetic Environment in Colombia. Front. Microbiol. 2021, 12, 663020. [Google Scholar] [CrossRef]
- Vanegas, J.M.; Cienfuegos, A.V.; Ocampo, A.M.; López, L.; Del Corral, H.; Roncancio, G.; Sierra, P.; Echeverri-Toro, L.; Ospina, S.; Maldonado, N.; et al. Similar frequencies of Pseudomonas aeruginosa isolates producing KPC and VIM carbapenemases in diverse genetic clones at tertiary-care hospitals in Medellín, Colombia. J. Clin. Microbiol. 2014, 52, 3978–3986. [Google Scholar] [CrossRef] [PubMed]
- Pacheco, T.; Bustos-Cruz, R.H.; Abril, D.; Arias, S.; Uribe, L.; Rincón, J.; García, J.-C.; Escobar-Perez, J. Pseudomonas aeruginosa coharboring blaKPC-2 and blaVIM-2 carbapenemase genes. Antibiotics 2019, 8, 98. [Google Scholar] [CrossRef] [PubMed]
- Abril, D.; Lesmes-Leon, D.N.; Marquez-Ortiz, R.A.; Leal, A.L.; Tovar-Acero, C.; Corredor Rozo, Z.L.; Gómez, N.V.; Escobar-Perez, J.; Hudson, A.O. Draft genome of the Klebsiella pneumoniaea 24Kpn33 and complete sequence of its pCOL-1, a plasmid related to the bla KPC-2 acquisition in Pseudomonas aeruginosa. Microbiol. Resour. Announc. 2024, 13, e00071-24. [Google Scholar] [CrossRef]
- Correa, A.; Del Campo, R.; Perenguez, M.; Blanco, V.M.; Rodríguez-Baños, M.; Perez, F.; Maya, J.J.; Rojas, L.; Cantón, R.; Arias, C.A.; et al. Dissemination of High-Risk Clones of Extensively Drug-Resistant Pseudomonas aeruginosa in Colombia. Antimicrob. Agents Chemother. 2015, 59, 2421–2425. [Google Scholar] [CrossRef]
- Woodford, N.; Turton, J.F.; Livermore, D.M. Multiresistant Gram-negative bacteria: The role of high-risk clones in the dissemination of antibiotic resistance. FEMS Microbiol. Rev. 2011, 35, 736–755. [Google Scholar] [CrossRef]
- Kos, V.N.; Déraspe, M.; McLaughlin, R.E.; Whiteaker, J.D.; Roy, P.H.; Alm, R.A.; Corbeil, J.; Gardner, H. The resistome of Pseudomonas seudomonas aeruginosa in relationship to phenotypic susceptibility. Antimicrob. Agents Chemother. 2015, 59, 427–436. [Google Scholar] [CrossRef]
- Kocsis, B.; Toth, A.; Gulyas, D.; Ligeti, B.; Katona, K.; Rokusz, L.; Szabo, D. Acquired qnrVC1 and bla NDM-1 resistance markers in an international high-risk Pseudomonas aeruginosa ST773 clone. J. Med. Microbiol. 2019, 68, 336–338. [Google Scholar] [CrossRef] [PubMed]
- Crespo, M.P.; Woodford, N.; Sinclair, A.; Kaufmann, M.E.; Turton, J.; Glover, J.; Velez, J.D.; CastañeDa, C.R.; Recalde, M.; Livermore, D.M. Outbreak of carbapenem-resistant Pseudomonas aeruginosa producing VIM-8, a novel metallo-β-lactamase, in a tertiary care center in Cali, Colombia. J. Clin. Microbiol. 2004, 42, 5094–5101. [Google Scholar] [CrossRef]
- Cai, H.; Zhu, Y.; Hu, D.; Li, Y.; Leptihn, S.; Loh, B.; Hua, X.; Yu, Y. Co-harboring of Novel blaKPC–2 Plasmid and Integrative and Conjugative Element Carrying Tn6203 in Multidrug-Resistant Pseudomonas aeruginosa. Front. Microbiol. 2021, 12, 674974. [Google Scholar] [CrossRef]
- Wozniak, A.; Figueroa, C.; Moya-Flores, F.; Guggiana, P.; Castillo, C.; Rivas, L.; Munita, J.M.; García, P.C. A multispecies outbreak of carbapenem-resistant bacteria harboring the bla KPC gene in a non-classical transposon element. BMC Microbiol. 2021, 21, 107. [Google Scholar] [CrossRef]
- Forero-Hurtado, D.; Corredor-Rozo, Z.L.; Ruiz-Castellanos, J.S.; Márquez-Ortiz, R.A.; Abril, D.; Vanegas, N.; Lafaurie, G.I.; Chambrone, L.; Escobar-Pérez, J. Worldwide Dissemination of blaKPC Gene by Novel Mobilization Platforms in Pseudomonas aeruginosa: A Systematic Review. Antibiotics 2023, 12, 658. [Google Scholar] [CrossRef]
- Contreras-Valero, J.F.; Gualtero-Trujillo, S.M.; Cortés-Fraile, G.C.; Hernández-Garzón, S.; Manrique-Marín, N.; Narváez-Chaves, M.Á.; Valderrama-Beltrán, S.L. Epidemiological and clinical characteristics of patients with carbapenem-resistant Enterobacterales in a university hospital of Colombia: Enzyme coproductions in rise. Heliyon 2024, 10, e33698. [Google Scholar] [CrossRef]
- Saavedra, S.Y.; Bernal, J.F.; Montilla-Escudero, E.; Arévalo, S.A.; Prada, D.A.; Valencia, M.F.; Moreno, J.; Hidalgo, A.M.; García-Vega, Á.S.; Abrudan, M.; et al. Complexity of Genomic Epidemiology of Carbapenem-Resistant Klebsiella pneumoniaea Isolates in Colombia Urges the Reinforcement of Whole Genome Sequencing-Based Surveillance Programs. Clin. Infect. Dis. 2021, 73, S290–S299. [Google Scholar] [CrossRef]
- Stillwell, T.; Green, M.; Barbadora, K.; Ferrelli, J.G.; Roberts, T.L.; Weissman, S.J.; Nowalk, A. Outbreak of KPC-3 producing carbapenem- resistant Klebsiella pneumoniaea in a US pediatric hospital. J. Pediatr. Infect. Dis. Soc. 2015, 4, 330–338. [Google Scholar] [CrossRef]
- García, C.J.C.; Amaya, S.; Briceño, C.W.; Rincón, C.; Pinzón, J. Risk factors for carbapenem-resistant bacterial infection or colonization: A case control study. Acta Colomb. Cuid. Intensivo 2017, 17, 29–35. [Google Scholar] [CrossRef]
- Lapp, Z.; Octaria, R.; O’Malley, S.M.; Nguyen, T.N.; Wolford, H.; Crawford, R.; Moore, C.; Vagnone, P.S.; Noel, D.; Duffy, N.; et al. Distinct Origins and Transmission Pathways of blaKPC Enterobacterales across Three U.S. States. J. Clin. Microbiol. 2023, 61, e00259-23. [Google Scholar] [PubMed]
- Centers for Disease Control and Prevention (CDC). The Core Elements of Hospital Antibiotic Stewardship Programs; Centers for Disease Control and Prevention (CDC): Atlanta, GA, USA, 2019.
- World Health Organization (WHO). Antimicrobial Stewardship Programmes in Health-Care Facilities in Low- and Middle-Income Countries. A WHO Practical Toolkit; World Health Organization (WHO): Geneva, Switzerland, 2019. [Google Scholar]
- Timbrook, T.T.; Morton, J.B.; McConeghy, K.W.; Caffrey, A.R.; Mylonakis, E.; LaPlante, K.L. The Effect of Molecular Rapid Diagnostic Testing on Clinical Outcomes in Bloodstream Infections: A Systematic Review and Meta-analysis. Clin. Infect. Dis. 2017, 64, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Schuetz, P.; Wirz, Y.; Sager, R.; Christ-Crain, M.; Stolz, D.; Tamm, M.; Bouadma, L.; Luyt, C.E.; Wolff, M.; Chastre, J.; et al. Effect of procalcitonin-guided antibiotic treatment on mortality in acute respiratory infections: A patient level meta-analysis. Lancet Infect. Dis. 2018, 18, 95–107. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization (WHO). Guidelines on Core Components of Infection Prevention and Control Programmes at the National and Acute Health Care Facility Level; World Health Organization (WHO): Geneva, Switzerland, 2016. [Google Scholar]
- Centers for Disease Control and Prevention (CDC). HAI Prevention and Control for Healthcare; Centers for Disease Control and Prevention (CDC): Atlanta, GA, USA, 2024.
- World Health Organization (WHO). Global Action Plan on Antimicrobial Resistance; World Health Organization (WHO): Geneva, Switzerland, 2016. [Google Scholar]
- Barlam, T.F.; Cosgrove, S.E.; Abbo, L.M.; MacDougall, C.; Schuetz, A.N.; Septimus, E.J.; Srinivasan, A.; Dellit, T.H.; Falck-Ytter, Y.T.; Fishman, N.O.; et al. Implementing an Antibiotic Stewardship Program: Guidelines by the Infectious Diseases Society of America and the Society for Healthcare Epidemiology of America. Clin. Infect. Dis. 2016, 62, e51–e77. [Google Scholar] [CrossRef]
- Tabah, A.; Bassetti, M.; Kollef, M.H.; Zahar, J.R.; Paiva, J.A.; Timsit, J.F.; Roberts, J.A.; Schouten, J.; Giamarellou, H.; Rello, J.; et al. Antimicrobial de-escalation in critically ill patients: A position statement from a task force of the European Society of Intensive Care Medicine (ESICM) and European Society of Clinical Microbiology and Infectious Diseases (ESCMID) Critically Ill Patients Study Group (ESGCIP). Intensive Care Med. 2020, 46, 245–265. [Google Scholar]
- Kalil, A.C.; Metersky, M.L.; Klompas, M.; Muscedere, J.; Sweeney, D.A.; Palmer, L.B.; Napolitano, L.M.; O’Grady, N.P.; Bartlett, J.G.; Carratalà, J.; et al. Management of Adults With Hospital-acquired and Ventilator-associated Pneumonia: 2016 Clinical Practice Guidelines by the Infectious Diseases Society of America and the American Thoracic Society. Clin. Infect. Dis. 2016, 63, e61–e111. [Google Scholar] [CrossRef] [PubMed]
- Spellberg, B. The New Antibiotic Mantra-”Shorter Is Better”. JAMA Intern. Med. 2016, 176, 1254–1255. [Google Scholar] [CrossRef]
- Baur, D.; Gladstone, B.P.; Burkert, F.; Carrara, E.; Foschi, F.; Döbele, S.; Tacconelli, E. Effect of antibiotic stewardship on the incidence of infection and colonisation with antibiotic-resistant bacteria and Clostridium difficile infection: A systematic review and meta-analysis. Lancet Infect. Dis. 2017, 17, 990–1001. [Google Scholar] [CrossRef]
- World Health Organization (WHO). Global Antimicrobial Resistance and Use Surveillance System (GLASS) Report: 2022; World Health Organization (WHO): Geneva, Switzerland, 2022. [Google Scholar]
- Centers for Disease Control and Prevention (CDC). COVID-19: U.S. Impact on Antimicrobial Resistance, Special Report 2022; Centers for Disease Control and Prevention (CDC): Atlanta, GA, USA, 2022.
- McCullough, A.R.; Parekh, S.; Rathbone, J.; Del Mar, C.B.; Hoffmann, T.C. A systematic review of the public’s knowledge and beliefs about antibiotic resistance. J. Antimicrob. Chemother. 2016, 71, 27–33. [Google Scholar] [CrossRef]
- World Health Organization (WHO). Antimicrobial Resistance: Fact Sheet on Sustainable Development Goals (SDGs): Health Targets; World Health Organization (WHO): Geneva, Switzerland, 2021. [Google Scholar]
- Laxminarayan, R.; Matsoso, P.; Pant, S.; Brower, C.; Røttingen, J.A.; Klugman, K.; Davies, S. Access to effective antimicrobials: A worldwide challenge. Lancet 2016, 387, 168–175. [Google Scholar] [CrossRef]
- Forde, B.M.; Bergh, H.; Cuddihy, T.; Hajkowicz, K.; Hurst, T.; Playford, E.G.; Henderson, B.C.; Runnegar, N.; Clark, J.; Jennison, A.V.; et al. Clinical Implementation of Routine Whole-genome Sequencing for Hospital Infection Control of Multi-drug Resistant Pathogens. Clin. Infect. Dis. 2023, 76, E1277–84. [Google Scholar] [CrossRef]
- Santos-Marques, C.; Ferreira, H.; Gonçalves Pereira, S. Infection prevention and control strategies against carbapenem resistant Enterobacteriaceae—A systematic review. J. Infect. Prev. 2022, 23, 167–185. [Google Scholar] [CrossRef]
- Ji, B.; Ye, W. Prevention and control of hospital-acquired infections with multidrug-resistant organism A review. Medicine 2024, 103, E37018. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, J.; Widen, R.H.; Pignatari, A.C.C.; Kubasek, C.; Silbert, S. Rapid detection of carbapenemase genes by multiplex real-time PCR. J. Antimicrob. Chemother. 2012, 67, 906–909. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention (CDC). Standard Operating Procedure for Pulsenet pfge of Escherichia coli o157:h7, Escherichia coli Non-o157 (stec), Salmonella serotypes, Shigella sonneiand Shigella flexneri Code: PNL05; Centers for Disease Control and Prevention (CDC): Atlanta, GA, USA, 2013.
- Tenover, F.C.; Arbeit, R.D.; Goering, R.V.; Mickelsen, P.A.; Murray, B.E.; Persing, D.H.; Swaminathan, B. Guest commentary Interpreting Chromosomal DNA Restriction Patterns Produced by Pulsed-Field Gel Electrophoresis: Criteria for Bacterial Strain Typing. J. Clin. Microbiol. 1995, 33, 2233–2239. [Google Scholar] [CrossRef]
- Curran, B.; Jonas, D.; Grundmann, H.; Pitt, T.; Dowson, C.G. Development of a multilocus sequence typing scheme for the opportunistic pathogen Pseudomonas aeruginosa. J. Clin. Microbiol. 2004, 42, 5644–5649. [Google Scholar] [CrossRef] [PubMed]
- Marquez-Ortiz, R.A.; Haggerty, L.; Olarte, N.; Duarte, C.; Garza-Ramos, U.; Silva-Sanchez, J.; Castro, B.E.; Sim, E.M.; Beltran, M.; Moncada, M.V.; et al. Genomic epidemiology of NDM-1-encoding plasmids in latin American clinical isolates reveals insights into the evolution of multidrug resistance. Genome Biol. Evol. 2017, 9, 1725–1741. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Carver, T.J.; Rutherford, K.M.; Berriman, M.; Rajandream, M.A.; Barrell, B.G.; Parkhill, J. ACT: The Artemis comparison tool. Bioinformatics 2005, 21, 3422–3423. [Google Scholar] [CrossRef]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.-L.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef]
- Sullivan, M.J.; Petty, N.K.; Beatson, S.A. Easyfig: A genome comparison visualizer. Bioinformatics 2011, 27, 1009–1010. [Google Scholar] [CrossRef]
- Bertelli, C.; Laird, M.R.; Williams, K.P.; Lau, B.Y.; Hoad, G.; Winsor, G.L.; Brinkman, F.S.L.; Simon Fraser University Research Computing Group. IslandViewer 4: Expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 2017, 45, W30–W35. [Google Scholar] [CrossRef] [PubMed]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef] [PubMed]


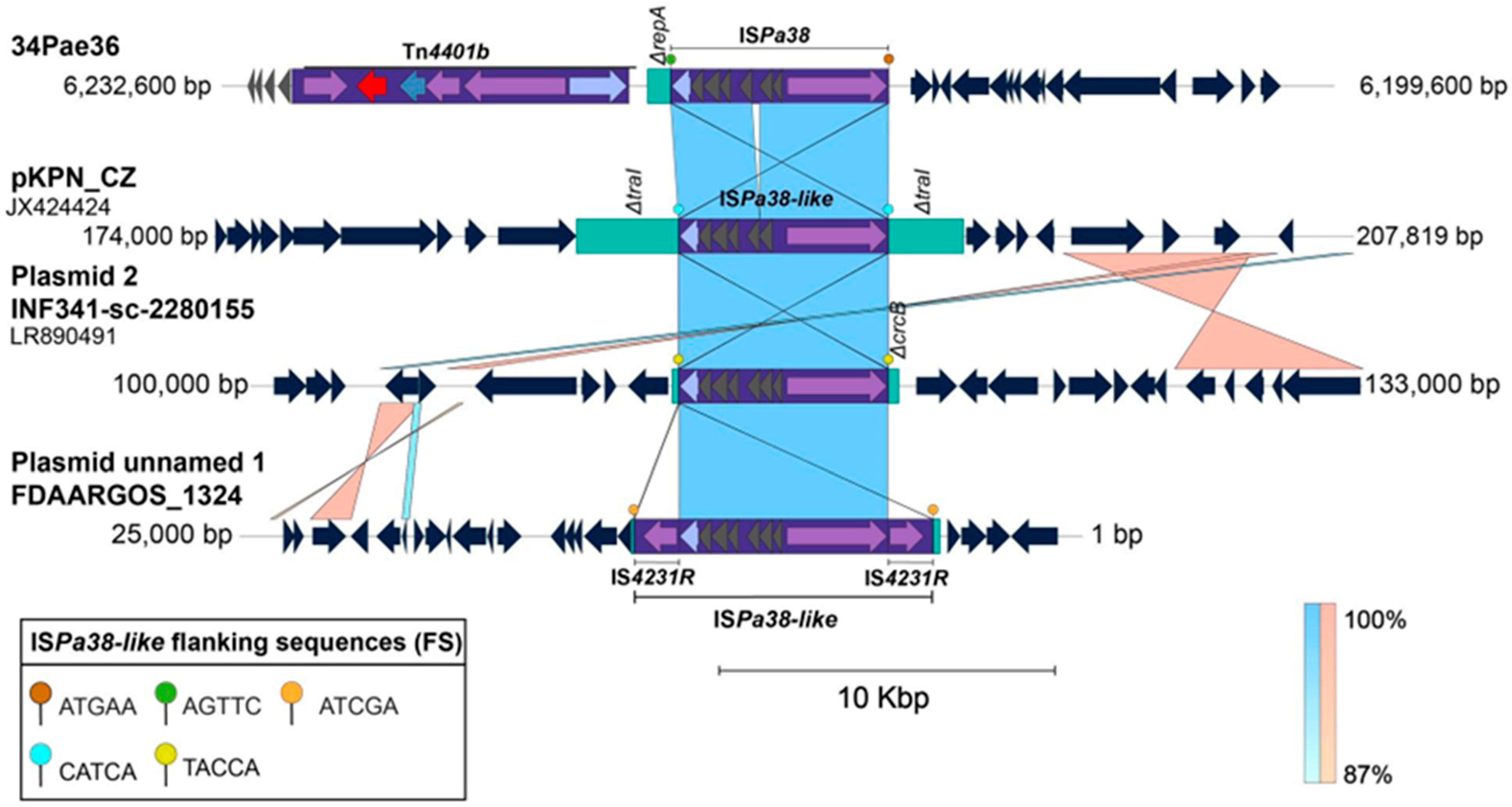
| Insertion | Strain | Flanking Sequences | Chromosomal Position | Insertion Site | Genetic Element | Sequence Type | |
|---|---|---|---|---|---|---|---|
| Left | Right | ||||||
| 1 | 34Pae36 and 30Pae2 | ATGAA | AGTTC | 6,273,055–6,279,529 | Intergenic region of IPC90_06220 and ΔtnpA * | Prophage | ST111 |
| AGTTC | TTCCA | 6,212,818–6,219,281 | |||||
| AG1 | ATGAA | TTCCA | 6,049,321–6,055,784 | Intergenic region of IPC90_06220 and ΔtnpA | Prophage | ST111 | |
| AR445 | ATGAA | TTCCA | 6,016,755–6,022,321 | Intergenic region of IPC90_06220 and ΔtnpA | Prophage | ST111 | |
| Carb01 | ATGAA | TTCCA | 6,373,687–6,381,055 | Intergenic region of IPC90_06220 and ΔtnpA | Prophage | ST111 | |
| PA38182 | ATGAA | TTCCA | 6,623,388–6,616,919 | Intergenic region of IPC90_06220 and ΔtnpA | Prophage | ST111 | |
| Pa5486 | ATGAA | TTCCA | 6,080,366–6,086,830 | Intergenic region of IPC90_06220 and ΔtnpA | Prophage | ST111 | |
| PaAI2 | ATGAA | TTCCA | 6,054,509–6,060,982 | Intergenic region of IPC90_06220 and ΔtnpA | Prophage | ST111 | |
| Y82 * | ATGAA | TTCCA | 5,102,848–5,109,262 | Intergenic region of IPC90_06220 and ΔtnpA | Prophage | ST111 | |
| RIVM-EMC2982 | ATGAA | TTCCA | 1,117,512–1,123,981 | Intergenic region of IPC90_06220 and ΔtnpA | Prophage | ST111 | |
| 2 | AR_0440 ** | - | AGGTA | –4,250,295 | Intergenic region | Genomic Island | ST357 |
| 3 | CF39S | AGGTA | TACCA | 3,334,102–3,340,563 | Intergenic region | Pathogenicity Island | ST175 |
| PcyII-29 | AGGTA | TACC–A | 3,181,009–3,187,470 | Intergenic region | Pathogenicity Island | ST175 | |
| DK2 | AGGTA | TACCA | 3,066,754–3,073,219 | Intergenic region | Pathogenicity Island | ST386 | |
| PABCH0 | AGGTA | TACCA | 3,046,061–3,412,525 | Intergenic region | Pathogenicity Island | ST155 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abril, D.; Bravo-Ojeda, J.; Garcia, J.-C.; Leal-Castro, A.L.; Saavedra-Trujillo, C.H.; Madroñero, J.; Bustos, R.-H.; Marquez-Ortiz, R.A.; Rozo, Z.L.C.; Gómez, N.V.; et al. New Insights in blaKPC Gene Mobilization in Pseudomonas aeruginosa: Acquisition of blaKPC-3 and Identification of a New Tn2-like NTE Mobilizing blaKPC-2. Antibiotics 2025, 14, 947. https://doi.org/10.3390/antibiotics14090947
Abril D, Bravo-Ojeda J, Garcia J-C, Leal-Castro AL, Saavedra-Trujillo CH, Madroñero J, Bustos R-H, Marquez-Ortiz RA, Rozo ZLC, Gómez NV, et al. New Insights in blaKPC Gene Mobilization in Pseudomonas aeruginosa: Acquisition of blaKPC-3 and Identification of a New Tn2-like NTE Mobilizing blaKPC-2. Antibiotics. 2025; 14(9):947. https://doi.org/10.3390/antibiotics14090947
Chicago/Turabian StyleAbril, Deisy, Juan Bravo-Ojeda, Julio-Cesar Garcia, Aura Lucia Leal-Castro, Carlos Humberto Saavedra-Trujillo, Johana Madroñero, Rosa-Helena Bustos, Ricaurte Alejandro Marquez-Ortiz, Zayda Lorena Corredor Rozo, Natasha Vanegas Gómez, and et al. 2025. "New Insights in blaKPC Gene Mobilization in Pseudomonas aeruginosa: Acquisition of blaKPC-3 and Identification of a New Tn2-like NTE Mobilizing blaKPC-2" Antibiotics 14, no. 9: 947. https://doi.org/10.3390/antibiotics14090947
APA StyleAbril, D., Bravo-Ojeda, J., Garcia, J.-C., Leal-Castro, A. L., Saavedra-Trujillo, C. H., Madroñero, J., Bustos, R.-H., Marquez-Ortiz, R. A., Rozo, Z. L. C., Gómez, N. V., & Escobar-Pérez, J. (2025). New Insights in blaKPC Gene Mobilization in Pseudomonas aeruginosa: Acquisition of blaKPC-3 and Identification of a New Tn2-like NTE Mobilizing blaKPC-2. Antibiotics, 14(9), 947. https://doi.org/10.3390/antibiotics14090947








