Whole-Genome Sequencing and Antibiotic Resistance Profiling of Helicobacter pylori Isolates from a Tertiary Hospital in Southern Thailand
Abstract
1. Introduction
2. Results
2.1. Isolation and Identification of H. pylori
2.2. Antimicrobial Susceptibility Profiles
2.3. Genomic Features of the H. pylori Isolates
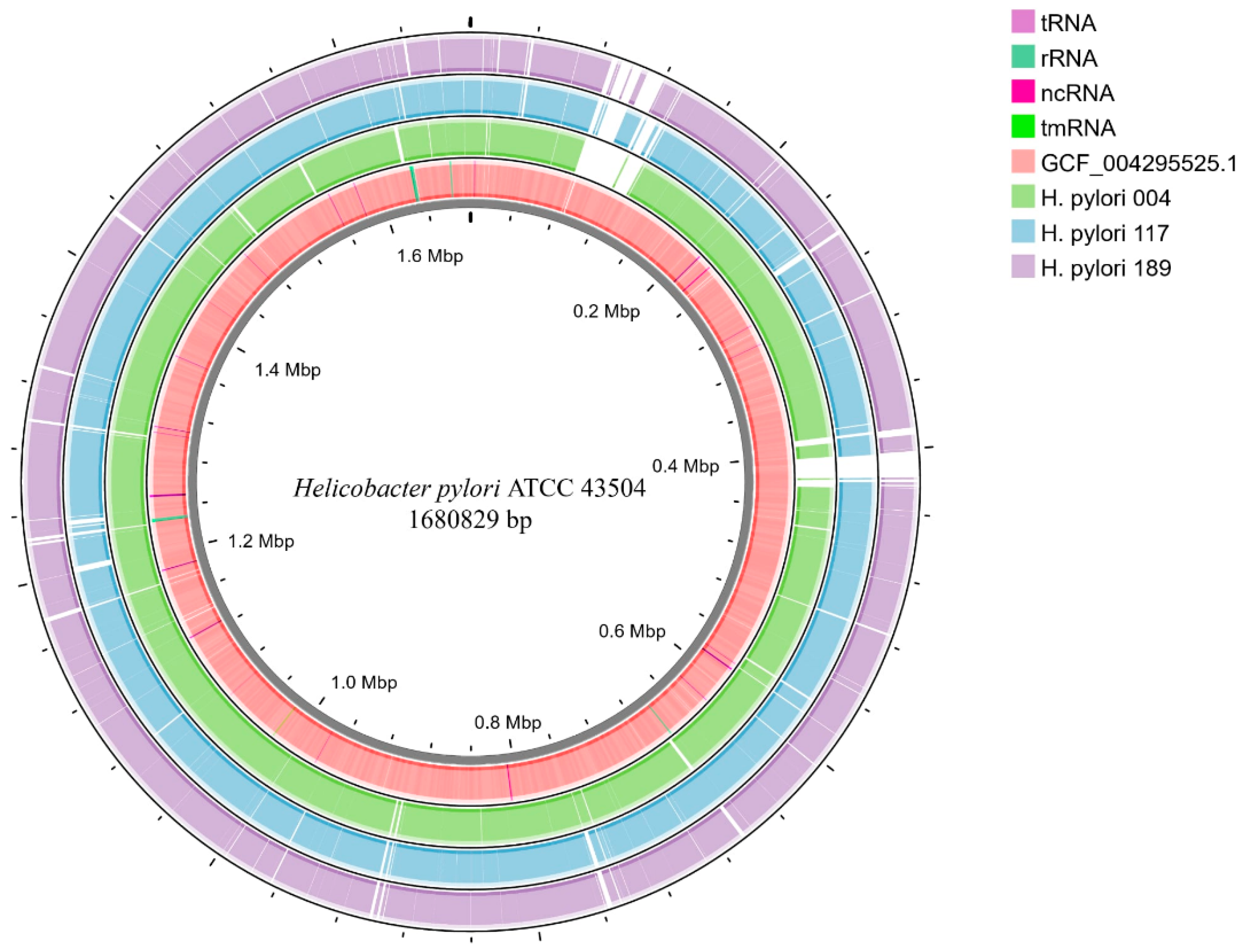
2.4. Comparative Genomics
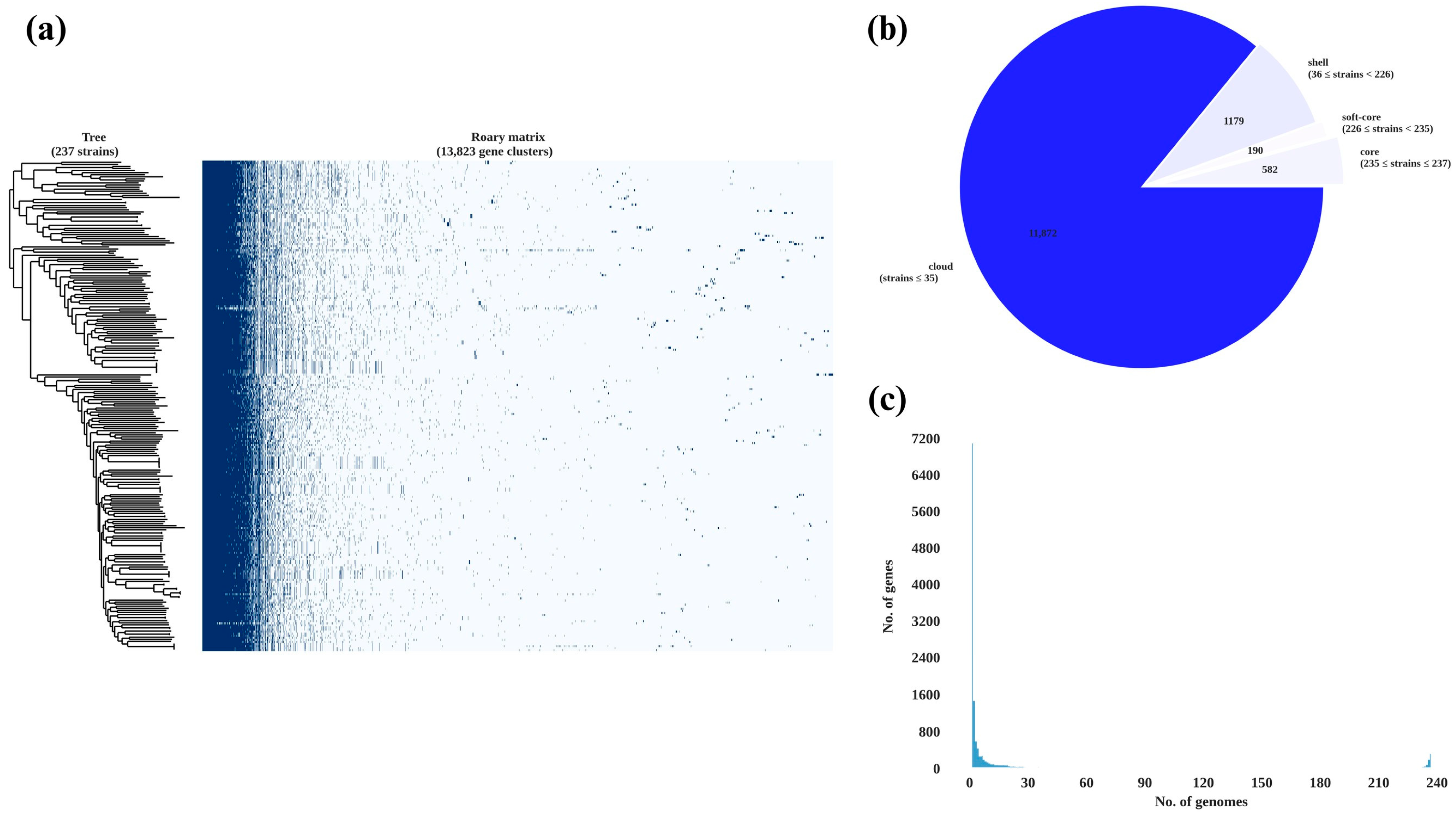
2.5. Pan-Genome Analysis and Phylogenetic Relationships
2.6. AMR Gene Profiling
2.7. Virulence Gene Profiling
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. Tissue Biopsy
4.3. H. pylori Isolation and Culture
4.4. MALDI-TOF MS Identification
4.5. Antibiotic Susceptibility Testing
4.6. Genomic Analysis
4.6.1. DNA Extraction and Whole-Genome Sequencing
4.6.2. Pan-Genome Analysis and Comparative Genomics
4.6.3. Analysis of Gene Correlations with Gastric Cancer
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AMR | antimicrobial resistance |
| MIC | minimal inhibitory concentration |
| MALDI-TOF MS | matrix-assisted laser desorption ionisation-time-of-flight mass spectrometry |
References
- McColl, K.E.L. Clinical practice. Helicobacter pylori infection. N. Engl. J. Med. 2010, 362, 1597–1604. [Google Scholar] [CrossRef]
- FitzGerald, R.; Smith, S.M. An overview of Helicobacter pylori infection. Methods Mol. Biol. 2021, 2283, 1–14. [Google Scholar] [CrossRef]
- Suerbaum, S.; Michetti, P. Helicobacter pylori infection. N. Engl. J. Med. 2002, 347, 1175–1186. [Google Scholar] [CrossRef]
- Ali, A.; AlHussaini, K.I. Helicobacter pylori: A contemporary perspective on pathogenesis, diagnosis and treatment strategies. Microorganisms 2024, 12, 222. [Google Scholar] [CrossRef]
- Møller, H.; Heseltine, E.; Vainio, H. Working group report on schistosomes, liver flukes and Helicobacter pylori. Int. J. Cancer 1995, 60, 587–589. [Google Scholar] [CrossRef]
- Malfertheiner, P.; Camargo, M.C.; El-Omar, E.; Liou, J.M.; Peek, R.; Schulz, C.; Smith, S.I.; Suerbaum, S. Helicobacter pylori infection. Nat. Rev. Dis. Primers 2023, 9, 19. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.C.; Malfertheiner, P.; Yu, H.T.; Kuo, C.L.; Chang, Y.Y.; Meng, F.T.; Wu, Y.X.; Hsiao, J.L.; Chen, M.J.; Lin, K.P.; et al. Global prevalence of Helicobacter pylori infection and incidence of gastric cancer between 1980 and 2022. Gastroenterology 2024, 166, 605–619. [Google Scholar] [CrossRef] [PubMed]
- Vilaichone, R.K.; Ratanachu-Ek, T.; Gamnarai, P.; Chaithongrat, S.; Uchida, T.; Yamaoka, Y.; Mahachai, V. Extremely high prevalence of metronidazole-resistant Helicobacter pylori strains in mountain people (Karen and Hmong) in Thailand. Am. J. Trop. Med. Hyg. 2016, 94, 717–720. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Vilaichone, R.K.; Gumnarai, P.; Ratanachu-Ek, T.; Mahachai, V. Nationwide survey of Helicobacter pylori antibiotic resistance in Thailand. Diagn. Microbiol. Infect. Dis. 2013, 77, 346–349. [Google Scholar] [CrossRef]
- Yi, M.; Chen, S.; Yi, X.; Zhang, F.; Zhou, X.; Zeng, M.; Song, H. Helicobacter pylori infection process: From the molecular world to clinical treatment. Front. Microbiol. 2025, 16, 1541140. [Google Scholar] [CrossRef]
- de Brito, B.B.; da Silva, F.A.F.; Soares, A.S.; Pereira, V.A.; Santos, M.L.C.; Sampaio, M.M.; Neves, P.H.M.; de Melo, F.F. Pathogenesis and clinical management of Helicobacter pylori gastric infection. World J. Gastroenterol. 2019, 25, 5578–5589. [Google Scholar] [CrossRef]
- Machlowska, J.; Baj, J.; Sitarz, M.; Maciejewski, R.; Sitarz, R. Gastric cancer: Epidemiology, risk factors, classification, genomic characteristics and treatment strategies. Int. J. Mol. Sci. 2020, 21, 4012. [Google Scholar] [CrossRef] [PubMed]
- Savoldi, A.; Carrara, E.; Graham, D.Y.; Conti, M.; Tacconelli, E. Prevalence of antibiotic resistance in Helicobacter pylori: A systematic review and meta-analysis in World Health Organization regions. Gastroenterology 2018, 155, 1372–1382.e17. [Google Scholar] [CrossRef] [PubMed]
- Megraud, F.; Coenen, S.; Versporten, A.; Kist, M.; Lopez-Brea, M.; Hirschl, A.M.; Andersen, L.P.; Goossens, H.; Glupczynski, Y.; Study Group participants. Helicobacter pylori resistance to antibiotics in Europe and its relationship to antibiotic consumption. Gut 2013, 62, 34–42. [Google Scholar] [CrossRef]
- Tshibangu-Kabamba, E.; Yamaoka, Y. Helicobacter pylori infection and antibiotic resistance—From biology to clinical implications. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 613–629. [Google Scholar] [CrossRef]
- Jeong, J.Y.; Mukhopadhyay, A.K.; Akada, J.K.; Dailidiene, D.; Hoffman, P.S.; Berg, D.E. Roles of FrxA and RdxA nitroreductases of Helicobacter pylori in susceptibility and resistance to metronidazole. J. Bacteriol. 2001, 183, 5155–5162. [Google Scholar] [CrossRef]
- Gong, X.; Wang, Y.; An, Y.; Li, Z.; Liu, D.; Yong, X. The crosstalk between efflux pump and resistance gene mutation in Helicobacter pylori. Gut Microbes 2024, 16, 2379439. [Google Scholar] [CrossRef]
- Rimbara, E.; Noguchi, N.; Kawai, T.; Sasatsu, M. Mutations in penicillin-binding proteins 1, 2 and 3 are responsible for amoxicillin resistance in Helicobacter pylori. J. Antimicrob. Chemother. 2008, 61, 995–998. [Google Scholar] [CrossRef] [PubMed]
- Attaran, B.; Salehi, N.; Ghadiri, B.; Esmaeili, M.; Kalateh, S.; Tashakoripour, M.; Eshagh Hosseini, M.; Mohammadi, M. The penicillin binding protein 1A of Helicobacter pylori, its amoxicillin binding site and access routes. Gut Pathog. 2021, 13, 43. [Google Scholar] [CrossRef]
- Hatakeyama, M. Helicobacter pylori CagA and gastric cancer: A paradigm for hit-and-run carcinogenesis. Cell Host Microbe 2014, 15, 306–316. [Google Scholar] [CrossRef]
- Gareayaghi, N.; Kocazeybek, B. Detection of A2143G, A2142C, and A2142G point mutations with real-time PCR in stool specimens from children infected with Helicobacter pylori. Diagnostics 2022, 12, 2119. [Google Scholar] [CrossRef]
- Breurec, S.; Guillard, B.; Hem, S.; Brisse, S.; Dieye, F.B.; Huerre, M.; Oung, C.; Raymond, J.; Tan, T.S.; Thiberge, J.M.; et al. Evolutionary history of Helicobacter pylori sequences reflect past human migrations in Southeast Asia. PLoS ONE 2011, 6, e22058. [Google Scholar] [CrossRef] [PubMed]
- Ailloud, F.; Didelot, X.; Woltemate, S.; Pfaffinger, G.; Overmann, J.; Bader, R.C.; Schulz, C.; Malfertheiner, P.; Suerbaum, S. Within-host evolution of Helicobacter pylori shaped by niche-specific adaptation, intragastric migrations and selective sweeps. Nat. Commun. 2019, 10, 2273. [Google Scholar] [CrossRef]
- Chen, C.; Zhang, Y.; Wu, H.; Qiao, J.; Caiyin, Q. Advances in diversity, evolutionary dynamics and biotechnological potential of restriction-modification systems. Microorganisms 2025, 13, 1126. [Google Scholar] [CrossRef] [PubMed]
- Baquero, F.; Martínez, J.L.; F Lanza, V.F.; Rodríguez-Beltrán, J.; Galán, J.C.; San Millán, A.; Cantón, R.; Coque, T.M. Evolutionary pathways and trajectories in antibiotic resistance. Clin. Microbiol. Rev. 2021, 34, e0005019. [Google Scholar] [CrossRef]
- Barber, M.F.; Fitzgerald, J.R. Mechanisms of host adaptation by bacterial pathogens. FEMS Microbiol. Rev. 2024, 48, fuae019. [Google Scholar] [CrossRef]
- Emamalipour, M.; Seidi, K.; Zununi Vahed, S.; Jahanban-Esfahlan, A.; Jaymand, M.; Majdi, H.; Amoozgar, Z.; Chitkushev, L.T.; Javaheri, T.; Jahanban-Esfahlan, R.; et al. Horizontal gene transfer: From evolutionary flexibility to disease progression. Front. Cell Dev. Biol. 2020, 8, 229. [Google Scholar] [CrossRef]
- Amieva, M.; Peek, R.M., Jr. Pathobiology of Helicobacter pylori-induced gastric cancer. Gastroenterology 2016, 150, 64–78. [Google Scholar] [CrossRef]
- Saeidi, Y.; Pournajaf, A.; Gholami, M.; Hasannejad-Bibalan, M.; Yaghoubi, S.; Khodabandeh, M.; Emadi, B.; Ferdosi-Shahandashti, E.; Rajabnia, R. Determination of Helicobacter pylori virulence-associated genes in duodenal ulcer and gastric biopsies. Med. J. Islam. Repub. Iran 2017, 31, 95. [Google Scholar] [CrossRef]
- Shiota, S.; Suzuki, R.; Yamaoka, Y. The significance of virulence factors in Helicobacter pylori. J. Dig. Dis. 2013, 14, 341–349. [Google Scholar] [CrossRef] [PubMed]
- Kharel, S.; Bist, A.; Shrestha, S.; Homagain, S. Helicobacter pylori healthy South Asians. JGH Open 2020, 4, 1037–1046. [Google Scholar] [CrossRef]
- Tongtawee, A.P.T.; Simawaranon, T.; Wattanawongdon, W. Helicobacter pylori infection and gastric cancer worldwide: Thailand distribution and burden disease. Suranaree J. Sci. Technol. 2017, 24, 311–329. [Google Scholar]
- McFarland, J. The nephelometer: An instrument for estimating the number of bacteria in suspensions used for calculating the opsonic index and for vaccines. JAMA 1907, 49, 1176–1178. [Google Scholar] [CrossRef]
- The European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters, Version 14.0, 2024. Available online: https://www.eucast.org (accessed on 27 May 2025).
- Chukamnerd, A.; Jeenkeawpiam, K.; Chusri, S.; Pomwised, R.; Singkhamanan, K.; Surachat, K. BacSeq: A user-friendly automated pipeline for whole-genome sequence analysis of bacterial genomes. Microorganisms 2023, 11, 1769. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Chaumeil, P.A.; Mussig, A.J.; Hugenholtz, P.; Parks, D.H. GTDB-Tk: A toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics 2019, 36, 1925–1927. [Google Scholar] [CrossRef] [PubMed]
- Grant, J.R.; Enns, E.; Marinier, E.; Mandal, A.; Herman, E.K.; Chen, C.Y.; Graham, M.; Van Domselaar, G.; Stothard, P. Proksee: In-depth characterization and visualization of bacterial genomes. Nucleic Acids Res. 2023, 51, W484–W492. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Yang, J.; Yu, J.; Yao, Z.; Sun, L.; Shen, Y.; Jin, Q. VFDB: A reference database for bacterial virulence factors. Nucleic Acids Res. 2005, 33, D325–D328. [Google Scholar] [CrossRef] [PubMed]
- McArthur, A.G.; Waglechner, N.; Nizam, F.; Yan, A.; Azad, M.A.; Baylay, A.J.; Bhullar, K.; Canova, M.J.; De Pascale, G.; Ejim, L.; et al. The comprehensive antibiotic resistance database. Antimicrob. Agents Chemother. 2013, 57, 3348–3357. [Google Scholar] [CrossRef]
- Brown, C.L.; Mullet, J.; Hindi, F.; Stoll, J.E.; Gupta, S.; Choi, M.; Keenum, I.; Vikesland, P.; Pruden, A.; Zhang, L. mobileOG-db: A manually curated database of protein families mediating the life cycle of bacterial mobile genetic elements. Appl. Environ. Microbiol. 2022, 88, e0099122. [Google Scholar] [CrossRef]
- Starikova, E.V.; Tikhonova, P.O.; Prianichnikov, N.A.; Rands, C.M.; Zdobnov, E.M.; Ilina, E.N.; Govorun, V.M. Phigaro: High-throughput prophage sequence annotation. Bioinformatics 2020, 36, 3882–3884. [Google Scholar] [CrossRef] [PubMed]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.G.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Bardou, P.; Mariette, J.; Escudié, F.; Djemiel, C.; Klopp, C. jvenn: An interactive Venn diagram viewer. BMC Bioinform. 2014, 15, 293. [Google Scholar] [CrossRef] [PubMed]


| Isolate | MIC (μg/mL) | |||
|---|---|---|---|---|
| AMX | LVX | MTZ | CLR | |
| H. pylori 004 | 0.75 (R) | >32 (R) | >256 (R) | 0.094 (S) |
| H. pylori 117 | >256 (R) | 0.19 (S) | >256 (R) | 0.032 (S) |
| H. pylori 189 | 0.023 (S) | 1 (S) | >256 (R) | 0.047 (S) |
| Feature | H. pylori 004 | H. pylori 117 | H. pylori 189 |
|---|---|---|---|
| Bases (bp) | 1,587,334 | 1,559,464 | 1,595,107 |
| Contigs (N) | 29 | 25 | 32 |
| L50 (N) | 6 | 4 | 7 |
| N50 (N) | 82,149 | 136,963 | 108,320 |
| GC content (%) | 38.9 | 38.9 | 39.1 |
| CDSs (N) | 1587 | 1555 | 1625 |
| rRNAs (N) | 2 | 2 | 2 |
| tRNAs (N) | 36 | 36 | 36 |
| tmRNAs (N) | 1 | 1 | 1 |
| Classification | H. pylori | H. pylori | H. pylori |
| FastANI_reference | GCF_009689985.1 | GCF_000277405.1 | GCF_000277405.1 |
| H. pylori Strain | RGI Criteria | ARO Term | SNP | Detection Criteria | % Identity of Matching Region | % Length of Reference Sequence | Drug Class | Resistance Mechanism | AMR Gene Family | Antibiotic | AST Source |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 004 | Strict | vanT in vanG cluster | NA | Protein homolog model | 32.26 | 52.95 | Glycopeptide antibiotic | Antibiotic target alteration | Glycopeptide resistance gene cluster; vanT | Vancomycin | - |
| Strict | Helicobacter pylori frxA mutation conferring resistance to metronidazole | A16T, Y62D | Protein variant model | 96.77 | 100 | Nitroimidazole antibiotic | Antibiotic target alteration | Antibiotic-resistant Helicobacter pylori nitroreductase | Metronidazole | Curated-R | |
| Strict | Helicobacter pylori pbp2 mutants conferring resistance to amoxicillin | S494H, E572G | Protein variant model | 98.13 | 100 | Cephalosporin; penicillin beta-lactam | Antibiotic target alteration | Penicillin-binding protein mutations conferring resistance to beta-lactam antibiotics | Amoxicillin | Curated-R | |
| Strict | Helicobacter pylori rpoB mutation conferring resistance to rifampicin | I837V | Protein variant model | 99.1 | 100 | Fluoroquinolone antibiotic; rifamycin antibiotic | Antibiotic target alteration; antibiotic target replacement | Rifamycin-resistant beta-subunit of RNA polymerase (rpoB) | Levofloxacin; rifampin | Curated-R | |
| Strict | Helicobacter pylori rdxA mutation conferring resistance to metronidazole | T31E, C49T, D59N | Protein variant model | 95.24 | 100 | Nitroimidazole antibiotic | Antibiotic target alteration | Antibiotic-resistant Helicobacter pylori nitroreductase | Metronidazole | Curated-R | |
| Strict | Helicobacter pylori 23S rRNA with mutation conferring resistance to clarithromycin | c1707t, a2144g | rRNA gene variant model | 99.19 | 99.93 | Macrolide antibiotic | Antibiotic target alteration | 23S rRNA with mutation conferring resistance to macrolide antibiotics | Clarithromycin | Curated-R | |
| 117 | Strict | vanTr gene in vanL cluster | NA | Protein homolog model | 36 | 103.01 | Glycopeptide antibiotic | antibiotic target alteration | glycopeptide resistance gene cluster; vanT | vancomycin | - |
| Strict | Helicobacter pylori pbp2 mutants conferring resistance to amoxicillin | S494H, E572G | Protein variant model | 98.13 | 100.51 | Cephalosporin; penicillin beta-lactam | antibiotic target alteration | Penicillin-binding protein mutations conferring resistance to beta-lactam antibiotics | amoxicillin | Curated-R | |
| Strict | Helicobacter pylori pbp3 conferring resistance to amoxicillin | D2N | Protein variant model | 95.93 | 100 | Cephalosporin; penicillin beta-lactam | Antibiotic target alteration | Penicillin-binding protein mutations conferring resistance to beta-lactam antibiotics | Amoxicillin | Curated-R | |
| Strict | Helicobacter pylori rdxA mutation conferring resistance to metronidazole | T31E, C49T, D59N | Protein variant model | 94.29 | 100 | Nitroimidazole antibiotic | Antibiotic target alteration | Antibiotic-resistant Helicobacter pylori nitroreductase | Metronidazole | Curated-R | |
| Strict | Helicobacter pylori frxA mutation conferring resistance to metronidazole | Y62D | Protein variant model | 100 | 100 | Nitroimidazole antibiotic | Antibiotic target alteration | Antibiotic-resistant Helicobacter pylori nitroreductase | Metronidazole | Curated-R | |
| Strict | Helicobacter pylori 23S rRNA with mutation conferring resistance to clarithromycin | c1707t, a2144g | rRNA gene variant model | 99.54 | 94.45 | Macrolide antibiotic | Antibiotic target alteration | 23S rRNA with mutation conferring resistance to macrolide antibiotics | Clarithromycin | Curated-R | |
| 189 | Strict | vanT in vanG cluster | NA | Protein homolog model | 32.88 | 51.69 | Glycopeptide antibiotic | Antibiotic target alteration | Glycopeptide resistance gene cluster; vanT | Vancomycin | - |
| Strict | Helicobacter pylori pbp2 mutants conferring resistance to amoxicillin | S494H, E572G | Protein variant model | 98.13 | 100 | Cephalosporin; penicillin beta-lactam | Antibiotic target alteration | Penicillin-binding protein mutations conferring resistance to beta-lactam antibiotics | Amoxicillin | Curated-R | |
| Strict | Helicobacter pylori rdxA mutation conferring resistance to metronidazole | T31E, H97T, P106S, C49T, D59N, K64N | Protein variant model | 95.71 | 100 | nitroimidazole antibiotic | Antibiotic target alteration | Antibiotic-resistant Helicobacter pylori nitroreductase | Metronidazole | Curated-R | |
| Strict | Helicobacter pylori 23S rRNA with mutation conferring resistance to clarithromycin | c1707t, a2144g | rRNA gene variant model | 99.19 | 99.97 | macrolide antibiotic | antibiotic target alteration | 23S rRNA with mutation conferring resistance to macrolide antibiotics | Clarithromycin | Curated-R |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Romyasamit, C.; Kaewdech, A.; Sripongpun, P.; Chamroonkul, N.; Surachat, K.; Suwannasin, S.; Leepromma, Y.; Saki, M.; Samaeng, M.; Sornsenee, P. Whole-Genome Sequencing and Antibiotic Resistance Profiling of Helicobacter pylori Isolates from a Tertiary Hospital in Southern Thailand. Antibiotics 2025, 14, 944. https://doi.org/10.3390/antibiotics14090944
Romyasamit C, Kaewdech A, Sripongpun P, Chamroonkul N, Surachat K, Suwannasin S, Leepromma Y, Saki M, Samaeng M, Sornsenee P. Whole-Genome Sequencing and Antibiotic Resistance Profiling of Helicobacter pylori Isolates from a Tertiary Hospital in Southern Thailand. Antibiotics. 2025; 14(9):944. https://doi.org/10.3390/antibiotics14090944
Chicago/Turabian StyleRomyasamit, Chonticha, Apichat Kaewdech, Pimsiri Sripongpun, Naichaya Chamroonkul, Komwit Surachat, Sirikan Suwannasin, Yosita Leepromma, Morteza Saki, Maseetoh Samaeng, and Phoomjai Sornsenee. 2025. "Whole-Genome Sequencing and Antibiotic Resistance Profiling of Helicobacter pylori Isolates from a Tertiary Hospital in Southern Thailand" Antibiotics 14, no. 9: 944. https://doi.org/10.3390/antibiotics14090944
APA StyleRomyasamit, C., Kaewdech, A., Sripongpun, P., Chamroonkul, N., Surachat, K., Suwannasin, S., Leepromma, Y., Saki, M., Samaeng, M., & Sornsenee, P. (2025). Whole-Genome Sequencing and Antibiotic Resistance Profiling of Helicobacter pylori Isolates from a Tertiary Hospital in Southern Thailand. Antibiotics, 14(9), 944. https://doi.org/10.3390/antibiotics14090944










