Time Trends in Prevalence and Antimicrobial Resistance of Respiratory Pathogens in a Tertiary Hospital in Rome, Italy: A Retrospective Analysis (2018–2023)
Abstract
1. Introduction
2. Results
2.1. Patients and Clinical Specimens
2.2. Trends in Microbial Isolates in the Observation Period
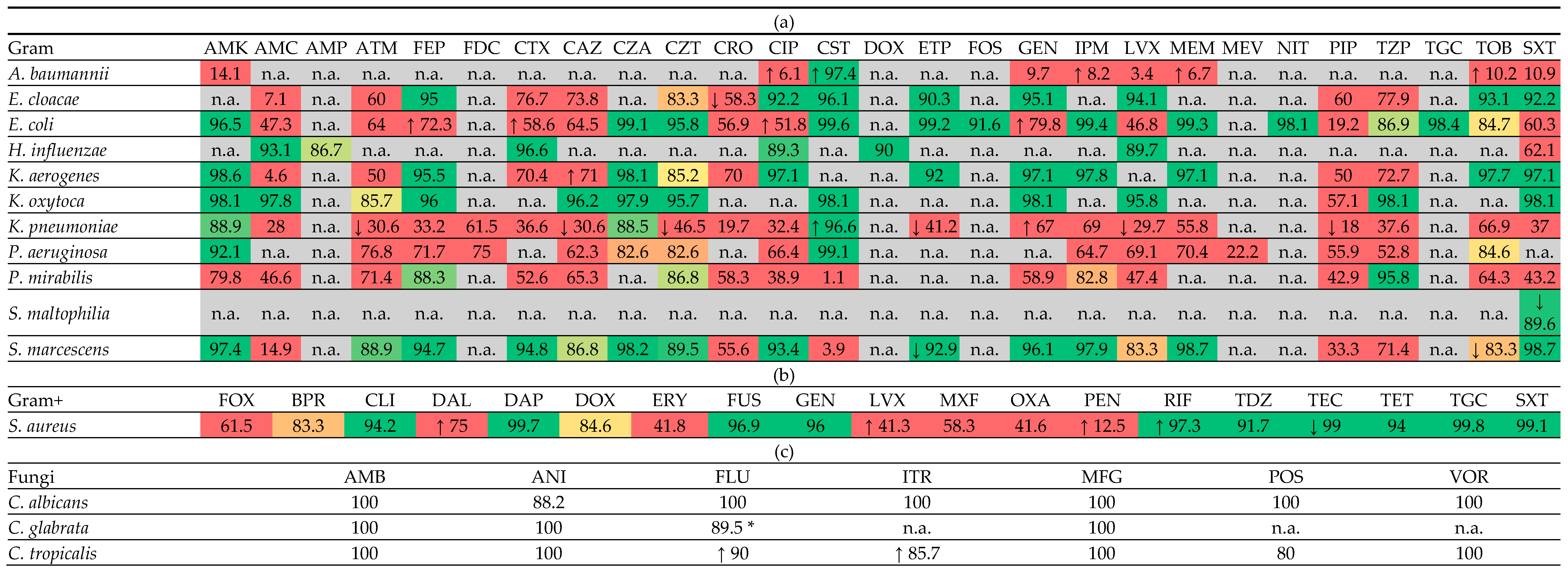
2.3. Resistance Rates
2.4. Resistance Trends
3. Discussion
3.1. Key Results
3.2. Limitations
3.3. Interpretation
3.4. Generalisability
4. Materials and Methods
4.1. Study Design
4.2. Setting
4.3. Sample Selection and Study Size
4.4. Variables
- A unique sample ID assigned by the microbiology laboratory;
- Date of sample collection;
- Patient’s age at the time of sample collection;
- Patient’s gender;
- Hospitalization status and department (if hospitalized), or indication of outpatients’ status;
- Type of respiratory specimen;
- Identified microorganisms (genus and species);
- Antimicrobial (antibiotic or antifungal) tested against each microorganism, with resistance data expressed as Minimum Inhibitory Concentration (MIC).
4.5. Data Source/Measurements
4.6. Bias and Bias Reduction
4.7. Statistical Methods
- Breakpoint—For each antimicrobial agent, the clinical breakpoint defined by the EUCAST 2025 guidelines was reported. This threshold determines whether a microorganism is classified as susceptible, susceptible with increased exposure, or resistant.
- Observed resistance rate “R (%)”—Expressed as a percentage, this value represents the proportion of resistant isolates among all those tested for a specific microorganism–antibiotic (or antifungal) combination across the entire study period.
- Ninety-five percent confidence interval (CI) for the resistance rate “IC95 (R%)”—This interval estimates the range within which the true population resistance rate is expected to lie, with 95% confidence, providing a measure of statistical precision.
- Pearson correlation coefficient “Pearson Coef.”—This coefficient quantifies the strength and direction of the linear relationship between time (in quarters) and the resistance rate. Positive values indicate increasing trends, while negative values suggest decreasing resistance over time.
- Regression slope coefficient “β coef.”—Derived from a univariate linear regression model, this coefficient represents the estimated average change in resistance rate per quarter. It provides a quantitative measure of the rate of increase or decrease in resistance over time.
- Ninety-five percent confidence interval for the regression slope coefficient “IC95 β coef.”—This indicates the statistical uncertainty around the estimated slope. If the interval excludes zero, the trend is considered statistically significant.
- p-value—This assesses the statistical significance of the observed trend. A p-value < 0.05 indicates that the resistance trend over time is unlikely to be due to chance alone and was considered significant.
- R-squared “R2”—The coefficient of determination quantifies how much of the variation in resistance rates is explained by time in the linear regression model. Values closer to 1 reflect a stronger explanatory power of the model.
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| (%) | Percentage |
| AMR | Antimicrobial resistance |
| BAL | Bronchoalveolar lavage |
| BAS | Bronchial aspirate |
| EUCAST | European Committee on Antimicrobial Susceptibility Testing |
| IC95 β coef. | Ninety-five percent confidence interval of the Beta Coefficient |
| inf | Lower bound of the confidence interval (inferior limit) |
| MDR | Multidrug Resistant |
| MIC | Minimum Inhibitory Concentration |
| NAC | Non-albicans Candida |
| Pearson Coef. | Pearson correlation coefficient |
| R2 | Coefficient of determination |
| sup | Upper bound of the confidence interval (superior limit) |
| WHO | World Health Organization |
| β coef. | Beta Coefficient (slope) of the linear regression |
| The following abbreviations are used in Table 3a–c for antibiotic/antifungal agents: | |
| Code | Drug name |
| AMK | Amikacin |
| AMC | Amoxicillin/Clavulanic acid |
| AMB | Amphotericin B |
| AMP | Ampicillin |
| ANI | Anidulafungin |
| ATM | Aztreonam |
| FEP | Cefepime |
| FDC | Cefiderocol |
| CTX | Cefotaxime |
| FOX | Cefoxitin |
| CAZ | Ceftazidime |
| CZA | Ceftazidime/Avibactam |
| BPR | Ceftobiprole |
| CZT | Ceftolozane/Tazobactam |
| CRO | Ceftriaxone |
| CIP | Ciprofloxacin |
| CLI | Clindamycin |
| CST | Colistin |
| DAL | Dalbavancin |
| DAP | Daptomycin |
| DOX | Doxycycline |
| ETP | Ertapenem |
| ERY | Erythromycin |
| FLU | Fluconazole |
| FOS | Fosfomycin |
| FUS | Fusidic acid |
| GEN | Gentamicin |
| IPM | Imipenem |
| ITR | Itraconazole |
| LVX | Levofloxacin |
| MEM | Meropenem |
| MEV | Meropenem/Vaborbactam |
| MFG | Micafungin |
| MXF | Moxifloxacin |
| NIT | Nitrofurantoin |
| OXA | Oxacillin |
| PEN | Penicillin G |
| PIP | Piperacillin |
| TZP | Piperacillin/Tazobactam |
| POS | Posaconazole |
| RIF | Rifampin |
| TDZ | Tedizolid |
| TEC | Teicoplanin |
| TET | Tetracycline |
| TGC | Tigecycline |
| TOB | Tobramycin |
| SXT | Trimethoprim/Sulfamethoxazole |
| VOR | Voriconazole |
Appendix A
| Specimen by Wards | Coronar Care Unit | Emergency | Hematology | Intensive Care Unit | Infectious Diseases | Medicine | Obstetrics and Gynecology | Psychiatry | Surgery | Traumatology | Orthopedics | Total | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| inpatients | Bronchial aspirate | 53 (0.76%) | 288 (4.14%) | 6 (0.09%) | 2449 (35.22%) | 101 (1.45%) | 581 (8.36%) | n.a. | 3 (0.04%) | 30 (0.43%) | 6 (0.09%) | n.a. | 3517 (50.58%) |
| Broncholavage | 16 (0.23%) | 204 (2.93%) | 5 (0.07%) | 718 (10.33%) | 149 (2.14%) | 313 (4.5%) | n.a. | 1 (0.01%) | 37 (0.53%) | 7 (0.1%) | n.a. | 1450 (20.85%) | |
| Oropharigeal swab | 1 (0.01%) | 276 (3.97%) | 3 (0.04%) | 17 (0.24%) | 19 (0.27%) | 65 (0.93%) | n.a. | 2 (0.03%) | 114 (1.64%) | 4 (0.06%) | n.a. | 501 (7.21%) | |
| Sputum | 10 (0.14%) | 275 (3.96%) | 24 (0.35%) | 54 (0.78%) | 172 (2.47%) | 281 (4.04%) | 1 (0.01%) | 1 (0.01%) | 70 (1.01%) | 4 (0.06%) | n.a. | 892 (12.83%) | |
| outpatients | Bronchial aspirate | 2 (0.03%) | n.a. | n.a. | n.a. | n.a. | 255 (3.67%) | n.a. | n.a. | n.a. | n.a. | n.a. | 257 (3.7%) |
| Broncholavage | n.a. | n.a. | n.a. | n.a. | n.a. | 171 (2.46%) | n.a. | n.a. | n.a. | n.a. | n.a. | 171 (2.46%) | |
| Oropharigeal swab | n.a. | n.a. | n.a. | n.a. | n.a. | 25 (0.36%) | n.a. | n.a. | n.a. | n.a. | 5 (0.07%) | 30 (0.43%) | |
| Sputum | n.a. | n.a. | 2 (0.03%) | n.a. | n.a. | 130 (1.87%) | n.a. | n.a. | n.a. | n.a. | 3 (0.04%) | 135 (1.94%) | |
| Total | 82 (1.18%) | 1043 (15%) | 40 (0.58%) | 3238 (46.57%) | 441 (6.34%) | 1821 (26.19%) | 1 (0.01%) | 7 (0.1%) | 251 (3.61%) | 21 (0.3%) | 8 (0.12%) | 6953 (100%) | |
References
- Davies, J.; Davies, D. Origins and Evolution of Antibiotic Resistance. Microbiol. Mol. Biol. Rev. 2010, 74, 417–433. [Google Scholar] [CrossRef]
- Prestinaci, F.; Pezzotti, P.; Pantosti, A. Antimicrobial Resistance: A Global Multifaceted Phenomenon. Pathog. Glob. Health 2015, 109, 309–318. [Google Scholar] [CrossRef]
- Abban, M.K.; Ayerakwa, E.A.; Mosi, L.; Isawumi, A. The Burden of Hospital Acquired Infections and Antimicrobial Resistance. Heliyon 2023, 9, e20561. [Google Scholar] [CrossRef] [PubMed]
- Raghavan, S.; Kim, K.-S. Host Immunomodulation Strategies to Combat Pandemic-Associated Antimicrobial-Resistant Secondary Bacterial Infections. Int. J. Antimicrob. Agents 2024, 64, 107308. [Google Scholar] [CrossRef] [PubMed]
- Salam, M.A.; Al-Amin, M.Y.; Salam, M.T.; Pawar, J.S.; Akhter, N.; Rabaan, A.A.; Alqumber, M.A.A. Antimicrobial Resistance: A Growing Serious Threat for Global Public Health. Healthcare 2023, 11, 1946. [Google Scholar] [CrossRef]
- Bianco, A.; Licata, F.; Nobile, C.G.; Napolitano, F.; Pavia, M. Pattern and Appropriateness of Antibiotic Prescriptions for Upper Respiratory Tract Infections in Primary Care Paediatric Patients. Int. J. Antimicrob. Agents 2022, 59, 106469. [Google Scholar] [CrossRef]
- The Lancet Respiratory Medicine. Antimicrobial Resistance: A Global Health Emergency. In The Lancet Respiratory Medicine; Elsevier: Amsterdam, The Netherlands, 2024; Volume 12, p. 837. [Google Scholar] [CrossRef]
- Guitor, A.K.; Wright, G.D. Antimicrobial Resistance and Respiratory Infections. Chest 2018, 154, 1202–1212. [Google Scholar] [CrossRef]
- Chu, V.T.; Tsitsiklis, A.; Mick, E.; Ambroggio, L.; Kalantar, K.L.; Glascock, A.; Osborne, C.M.; Wagner, B.D.; Matthay, M.A.; DeRisi, J.L.; et al. The Antibiotic Resistance Reservoir of the Lung Microbiome Expands with Age in a Population of Critically Ill Patients. Nat. Commun. 2024, 15, 92. [Google Scholar] [CrossRef]
- Lim, C.; Ashley, E.A.; Hamers, R.L.; Turner, P.; Kesteman, T.; Akech, S.; Corso, A.; Mayxay, M.; Okeke, I.N.; Limmathurotsakul, D.; et al. Surveillance Strategies Using Routine Microbiology for Antimicrobial Resistance in Low- and Middle-Income Countries. Clin. Microbiol. Infect. 2021, 27, 1391–1399. [Google Scholar] [CrossRef]
- EpiCentro Antimicrobial Resistance. Available online: https://www.epicentro.iss.it/en/antimicrobial-resistance/about (accessed on 26 August 2025).
- Maurici, M.; D’Alò, G.L.; Fontana, C.; Santoro, V.; Gaziano, R.; Ciotti, M.; Cicciarella Modica, D.; De Filippis, P.; Sarmati, L.; De Carolis, G.; et al. Microbiology and Clinical Outcome of Hospital-Acquired Respiratory Infections in an Italian Teaching Hospital: A Retrospective Study. Healthcare 2022, 10, 2271. [Google Scholar] [CrossRef]
- Garcia-Clemente, M.; de la Rosa, D.; Máiz, L.; Girón, R.; Blanco, M.; Olveira, C.; Canton, R.; Martinez-García, M.A. Impact of Pseudomonas Aeruginosa Infection on Patients with Chronic Inflammatory Airway Diseases. J. Clin. Med. 2020, 9, 3800. [Google Scholar] [CrossRef]
- Migiyama, Y.; Sakata, S.; Iyama, S.; Tokunaga, K.; Saruwatari, K.; Tomita, Y.; Saeki, S.; Okamoto, S.; Ichiyasu, H.; Sakagami, T. Airway Pseudomonas Aeruginosa Density in Mechanically Ventilated Patients: Clinical Impact and Relation to Therapeutic Efficacy of Antibiotics. Crit. Care 2021, 25, 59. [Google Scholar] [CrossRef]
- Elfadadny, A.; Ragab, R.F.; AlHarbi, M.; Badshah, F.; Ibáñez-Arancibia, E.; Farag, A.; Hendawy, A.O.; De Los Ríos-Escalante, P.R.; Aboubakr, M.; Zakai, S.A.; et al. Antimicrobial Resistance of Pseudomonas Aeruginosa: Navigating Clinical Impacts, Current Resistance Trends, and Innovations in Breaking Therapies. Front. Microbiol. 2024, 15, 1374466. [Google Scholar] [CrossRef]
- Pang, Z.; Raudonis, R.; Glick, B.R.; Lin, T.-J.; Cheng, Z. Antibiotic Resistance in Pseudomonas Aeruginosa: Mechanisms and Alternative Therapeutic Strategies. Biotechnol. Adv. 2019, 37, 177–192. [Google Scholar] [CrossRef] [PubMed]
- EpiCentro Pseudomonas aeruginosa—Sorveglianza dell’Antibiotico-Resistenza—AR-ISS. Available online: https://www.epicentro.iss.it/antibiotico-resistenza/ar-iss-rapporto-pseudomonas-aeruginosa (accessed on 26 August 2025).
- Nduagu, C.E.; Dinçman, G.E.; Çakır, N.; Nduagu, C.E.; Dinçman, G.E.; Çakır, N. Piperacillin/Tazobactam Resistance in Clinical Isolates of Pseudomonas and Klebsiella Species at a University Hospital in North Cyprus: A Retrospective Study. Cyprus J. Med. Sci. 2024, 9, 185–191. [Google Scholar] [CrossRef]
- Harris, S.; Singer, M.; Rowan, K.; Sanderson, C. Delay to Admission to Critical Care and Mortality among Deteriorating Ward Patients in UK Hospitals: A Multicentre, Prospective, Observational Cohort Study. Lancet 2015, 385 (Suppl. 1), S40. [Google Scholar] [CrossRef]
- Lodise, T.P., Jr.; Lomaestro, B.; Drusano, G.L. Piperacillin-Tazobactam for Pseudomonas Aeruginosa Infection: Clinical Implications of an Extended-Infusion Dosing Strategy. Clin. Infect. Dis. 2007, 44, 357–363. [Google Scholar] [CrossRef]
- Khadka, S.; Khan, S.; King, A.; Goldberg, L.R.; Crocombe, L.; Bettiol, S. Poor Oral Hygiene, Oral Microorganisms and Aspiration Pneumonia Risk in Older People in Residential Aged Care: A Systematic Review. Age Ageing 2021, 50, 81–87. [Google Scholar] [CrossRef]
- Parente, D.M.; Cunha, C.B.; Mylonakis, E.; Timbrook, T.T. The Clinical Utility of Methicillin-Resistant Staphylococcus Aureus (MRSA) Nasal Screening to Rule Out MRSA Pneumonia: A Diagnostic Meta-Analysis With Antimicrobial Stewardship Implications. Clin. Infect. Dis. 2018, 67, 1–7. [Google Scholar] [CrossRef]
- Pezzani, M.D.; Arieti, F.; Rajendran, N.B.; Barana, B.; Cappelli, E.; De Rui, M.E.; Galia, L.; Hassoun-Kheir, N.; Argante, L.; Schmidt, J.; et al. Frequency of Bloodstream Infections Caused by Six Key Antibiotic-Resistant Pathogens for Prioritization of Research and Discovery of New Therapies in Europe: A Systematic Review. Clin. Microbiol. Infect. 2024, 30 (Suppl. 1), S4–S13. [Google Scholar] [CrossRef] [PubMed]
- EpiCentro Staphylococcus aureus—Sorveglianza dell’Antibiotico-Resistenza—AR-ISS. Available online: https://www.epicentro.iss.it/antibiotico-resistenza/ar-iss-rapporto-staphylococcus-aureus (accessed on 26 August 2025).
- Touaitia, R.; Mairi, A.; Ibrahim, N.A.; Basher, N.S.; Idres, T.; Touati, A. Staphylococcus Aureus: A Review of the Pathogenesis and Virulence Mechanisms. Antibiotics 2025, 14, 470. [Google Scholar] [CrossRef] [PubMed]
- Palavecino, E.L. Clinical, Epidemiologic, and Laboratory Aspects of Methicillin-Resistant Staphylococcus Aureus Infections. Methods Mol. Biol. 2014, 1085, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Huang, L.; Cai, J.; Zhu, H.; Li, J.; Yu, Y.; Xu, Y.; Shi, G.; Feng, Y. Clinical Characteristics of Respiratory Tract Infection Caused by Klebsiella Pneumoniae in Immunocompromised Patients: A Retrospective Cohort Study. Front. Cell. Infect. Microbiol. 2023, 13, 1137664. [Google Scholar] [CrossRef] [PubMed]
- Asokan, S.; Jacob, T.; Jacob, J.; AlSosowaa, A.A.; Cherian, T.; Peijnenburg, W.J.G.M.; Vijayan, S. Klebsiella Pneumoniae: A Growing Threat in the Era of Antimicrobial Resistance. Microbe 2025, 7, 100333. [Google Scholar] [CrossRef]
- EpiCentro Klebsiella pneumoniae—Sorveglianza dell’Antibiotico-Resistenza—AR-ISS. Available online: https://www.epicentro.iss.it/antibiotico-resistenza/ar-iss-rapporto-klebsiella-pneumoniae (accessed on 26 August 2025).
- Antimicrobial Resistance, Hypervirulent Klebsiella Pneumoniae—Global Situation. Available online: https://www.who.int/emergencies/disease-outbreak-news/item/2024-DON527 (accessed on 26 August 2025).
- Alvisi, G.; Curtoni, A.; Fonnesu, R.; Piazza, A.; Signoretto, C.; Piccinini, G.; Sassera, D.; Gaibani, P. Epidemiology and Genetic Traits of Carbapenemase-Producing Enterobacterales: A Global Threat to Human Health. Antibiotics 2025, 14, 141. [Google Scholar] [CrossRef]
- Pendleton, K.M.; Huffnagle, G.B.; Dickson, R.P. The Significance of Candida in the Human Respiratory Tract: Our Evolving Understanding. Pathog. Dis. 2017, 75, ftx029. [Google Scholar] [CrossRef]
- Delisle, M.-S.; Williamson, D.R.; Perreault, M.M.; Albert, M.; Jiang, X.; Heyland, D.K. The Clinical Significance of Candida Colonization of Respiratory Tract Secretions in Critically Ill Patients. J. Crit. Care 2008, 23, 11–17. [Google Scholar] [CrossRef]
- Hamet, M.; Pavon, A.; Dalle, F.; Pechinot, A.; Prin, S.; Quenot, J.-P.; Charles, P.-E. Candida Spp. Airway Colonization Could Promote Antibiotic-Resistant Bacteria Selection in Patients with Suspected Ventilator-Associated Pneumonia. Intensive Care Med. 2012, 38, 1272–1279. [Google Scholar] [CrossRef]
- De Pascale, G.; Antonelli, M. Candida Colonization of Respiratory Tract: To Treat or Not to Treat, Will We Ever Get an Answer? Intensive Care Med. 2014, 40, 1381–1384. [Google Scholar] [CrossRef]
- Lockhart, S.R.; Chowdhary, A.; Gold, J.A.W. The Rapid Emergence of Antifungal-Resistant Human-Pathogenic Fungi. Nat. Rev. Microbiol. 2023, 21, 818–832. [Google Scholar] [CrossRef]
- Vitiello, A.; Sabbatucci, M.; Boccellino, M.; Ponzo, A.; Langella, R.; Zovi, A. Therapeutic and Unconventional Strategies to Contrast Antimicrobial Resistance: A Literature Review. Discov. Med. 2023, 35, 750–756. [Google Scholar] [CrossRef]
- Fisher, M.C.; Alastruey-Izquierdo, A.; Berman, J.; Bicanic, T.; Bignell, E.M.; Bowyer, P.; Bromley, M.; Brüggemann, R.; Garber, G.; Cornely, O.A.; et al. Tackling the Emerging Threat of Antifungal Resistance to Human Health. Nat. Rev. Microbiol. 2022, 20, 557–571. [Google Scholar] [CrossRef]
- Howard, A.; O’Donoghue, M.; Feeney, A.; Sleator, R.D. Acinetobacter Baumannii: An Emerging Opportunistic Pathogen. Virulence 2012, 3, 243–250. [Google Scholar] [CrossRef] [PubMed]
- Kyriakidis, I.; Vasileiou, E.; Pana, Z.D.; Tragiannidis, A. Acinetobacter Baumannii Antibiotic Resistance Mechanisms. Pathogens 2021, 10, 373. [Google Scholar] [CrossRef]
- EpiCentro Acinetobacter Species—Sorveglianza dell’Antibiotico-Resistenza—AR-ISS. Available online: https://www.epicentro.iss.it/antibiotico-resistenza/ar-iss-rapporto-acinetobacter-species (accessed on 26 August 2025).
- Torres, D.A.; Seth-Smith, H.M.B.; Joosse, N.; Lang, C.; Dubuis, O.; Nüesch-Inderbinen, M.; Hinic, V.; Egli, A. Colistin Resistance in Gram-Negative Bacteria Analysed by Five Phenotypic Assays and Inference of the Underlying Genomic Mechanisms. BMC Microbiol. 2021, 21, 321. [Google Scholar] [CrossRef] [PubMed]
- Yamin, D.; Uskoković, V.; Wakil, A.M.; Goni, M.D.; Shamsuddin, S.H.; Mustafa, F.H.; Alfouzan, W.A.; Alissa, M.; Alshengeti, A.; Almaghrabi, R.H.; et al. Current and Future Technologies for the Detection of Antibiotic-Resistant Bacteria. Diagnostics 2023, 13, 3246. [Google Scholar] [CrossRef]
- Mullins, L.P.; Mason, E.; Winter, K.; Sadarangani, M. Vaccination Is an Integral Strategy to Combat Antimicrobial Resistance. PLoS Pathog. 2023, 19, e1011379. [Google Scholar] [CrossRef]
- Ginsburg, A.S.; Klugman, K.P. Vaccination to Reduce Antimicrobial Resistance. Lancet Glob. Health 2017, 5, e1176–e1177. [Google Scholar] [CrossRef] [PubMed]
- Whonet Microbiology Laboratory Database Software. Available online: https://whonet.org/ (accessed on 26 August 2025).
- Simner, P.J.; Hindler, J.A.; Bhowmick, T.; Das, S.; Johnson, J.K.; Lubers, B.V.; Redell, M.A.; Stelling, J.; Erdman, S.M. What’s New in Antibiograms? Updating CLSI M39 Guidance with Current Trends. J. Clin. Microbiol. 2022, 60, e0221021. [Google Scholar] [CrossRef]
| Variables | 2018 α | 2019 | 2020 | 2021 | 2022 | 2023 α | Total | |
|---|---|---|---|---|---|---|---|---|
| Age | 64.29 ± 16.89 | 64.26 ± 16.96 | 67.4 ± 14.15 | 64.74 ± 15.26 | 64.41 ± 15.12 | 63.44 ± 15.56 | 64.87 ± 15.59 | |
| Gender | Male | 71.64% (n = 384) | 71.1% (n = 967) | 66.84% (n = 752) | 67.59% (n = 1103) | 71.76% (n = 1324) | 71.65% (n = 326) | 69.84% (n = 4856) |
| Female | 28.36% (n = 152) | 28.9% (n = 393) | 33.16% (n = 373) | 32.41% (n = 529) | 28.24% (n = 521) | 28.35% (n = 129) | 30.16% (n = 2097) | |
| Patient | Inpatient | 91.79% (n = 492) | 90.37% (n = 1229) | 91.11% (n = 1025) | 93.26% (n = 1522) | 90.46% (n = 1669) | 92.97% (n = 423) | 91.47% (n = 6360) |
| Outpatient | 8.21% (n = 44) | 9.63% (n = 131) | 8.89% (n = 100) | 6.74% (n = 110) | 9.54% (n = 176) | 7.03% (n = 32) | 8.53% (n = 593) | |
| Specimen Type | BAS | 58.4% (n = 313) | 54.34% (n = 739) | 47.82% (n = 538) | 54.04% (n = 882) | 56.37% (n = 1040) | 57.58% (n = 262) | 54.28% (n = 3774) |
| BAL | 21.64% (n = 116) | 21.69% (n = 295) | 34.22% (n = 385) | 22.12% (n = 361) | 20.33% (n = 375) | 19.56% (n = 89) | 23.31% (n = 1621) | |
| Oropharyngeal swab | 9.33% (n = 50) | 11.03% (n = 150) | 7.29% (n = 82) | 7.23% (n = 118) | 5.31% (n = 98) | 7.25% (n = 33) | 7.64% (n = 531) | |
| Sputum | 10.63% (n = 57) | 12.94% (n = 176) | 10.67% (n = 120) | 16.61% (n = 271) | 17.99% (n = 332) | 15.6% (n = 71) | 14.77% (n = 1027) | |
| Microorganism | Gram- | 59.89% (n = 321) | 64.71% (n = 880) | 63.73% (n = 717) | 61.64% (n = 1006) | 59.02% (n = 1089) | 57.8% (n = 263) | 61.5% (n = 4276) |
| Gram+ | 20.71% (n = 111) | 18.68% (n = 254) | 19.64% (n = 221) | 15.99% (n = 261) | 18.43% (n = 340) | 21.98% (n = 100) | 18.51% (n = 1287) | |
| Fungi | 19.4% (n = 104) | 16.62% (n = 226) | 16.62% (n = 187) | 22.18% (n = 362) | 22.22% (n = 410) | 20.22% (n = 92) | 19.86% (n = 1381) | |
| Others β | 0% (n = 0) | 0% (n = 0) | 0% (n = 0) | 0.18% (n = 3) | 0.33% (n = 6) | 0% (n = 0) | 0.13% (n = 9) | |
| Microorganism | Group | Number of Isolates | (%) | Pearson Coef. γ | β Coef. δ | IC95 β Coef. ε | p-Value | R2 |
|---|---|---|---|---|---|---|---|---|
| P. aeruginosa | Gram − | 1102 | 15.85 | −0.17 | −0.001 | −0.004, 0.002 | 0.49 | 0.03 |
| S. aureus | Gram + | 1043 | 15.00 | −0.04 | −0.000 | −0.004, 0.003 | 0.87 | 0.00 |
| K. pneumoniae | Gram − | 883 | 12.70 | 0.17 | 0.000 | −0.001, 0.004 | 0.49 | 0.03 |
| C. albicans | Fungi | 840 | 12.08 | 0.51 | 0.003 | 0.000, 0.005 | 0.03 | 0.26 |
| A. baumannii | Gram − | 636 | 9.15 | −0.22 | 0.001 | −0.000, 0.001 | 0.36 | 0.05 |
| E. coli | Gram − | 423 | 6.08 | −0.14 | −0.000 | −0.002, 0.001 | 0.57 | 0.02 |
| C. glabrata | Fungi | 202 | 2.91 | 0.35 | 0.001 | −0.000, 0.002 | 0.14 | 0.12 |
| S. maltophilia | Gram − | 169 | 2.43 | 0.03 | 0.000 | −0.001, 0.001 | 0.90 | 0.00 |
| E. cloacae | Gram - | 150 | 2.16 | 0.09 | 0.000 | −0.001, 0.001 | 0.70 | 0.01 |
| C. tropicalis | Fungi | 130 | 1.87 | 0.33 | 0.001 | −0.000, 0.001 | 0.16 | 0.11 |
| P. mirabilis | Gram − | 122 | 1.75 | 0.02 | 0.000 | −0.001, 0.001 | 0.95 | 0.00 |
| K. aerogenes | Gram − | 119 | 1.71 | −0.36 | −0.000 | −0.002, 0.000 | 0.13 | 0.13 |
| S. marcescens | Gram − | 105 | 1.51 | −0.23 | −0.000 | −0.001, 0.000 | 0.34 | 0.05 |
| K. oxytoca | Gram − | 86 | 1.24 | 0.00 | 0.000 | −0.001, 0.001 | 0.99 | 0.00 |
| H. influenzae | Gram − | 62 | 0.89 | 0.09 | 0.000 | −0.000, 0.001 | 0.71 | 0.01 |
| Others Gram- | Gram − | 419 | 6.03 | n.a. | n.a. | n.a. | n.a. | n.a. |
| Others Gram+ | Gram + | 244 | 3.51 | n.a. | n.a. | n.a. | n.a. | n.a. |
| Others NAC | Fungi | 47 | 0.68 | n.a. | n.a. | n.a. | n.a. | n.a. |
| Other fungi (not Candida genus) | Fungi | 162 | 2.33 | n.a. | n.a. | n.a. | n.a. | n.a. |
| Others—no classified | n.a. | 9 | 0.13 | n.a. | n.a. | n.a. | n.a. | n.a. |
| Total | n.a. | 6953 | 100 | n.a. | n.a. | n.a. | n.a. | n.a. |
 |
| Organism | Molecules | Break Points | R(%) | IC95 (R%) | Pearson Coef. γ | β Coef. δ | IC95 β Coef. ε | p-Value | R2 | |
|---|---|---|---|---|---|---|---|---|---|---|
| E. cloacae | ceftriaxone | S ≤ 1 | R ≥ 4 | 41.7 | 16.5–71.4 | 0.827 | 10.57 | 0.611; 20.529 | 0.042 | 0.685 |
| K. pneumoniae | aztreonam | S ≤ 1 | R ≥ 8 | 69.4 | 56.2–80.1 | 0.732 | 3.613 | 1.598; 5.628 | 0.002 | 0.536 |
| K. pneumoniae | ceftazidime/ avibactam | S ≤ 8 | R ≥ 16 | 11.5 | 9.0–14.6 | 0.601 | 0.963 | 0.308; 1.618 | 0.006 | 0.361 |
| K. pneumoniae | ceftolozane/ tazobactam | S ≤ 2 | R ≥ 4 | 53.5 | 49.1–57.8 | 0.594 | 1.895 | 0.583; 3.207 | 0.007 | 0.353 |
| K. pneumoniae | ertapenem | S ≤ 0.5 | R ≥ 1 | 58.8 | 53.5–64.0 | 0.61 | 2.072 | 0.695; 3.449 | 0.006 | 0.372 |
| K. pneumoniae | levofloxacin | S ≤ 0.5 | R ≥ 2 | 70.3 | 65.0–75.1 | 0.568 | 1.641 | 0.424; 2.858 | 0.011 | 0.323 |
| K. pneumoniae | piperacillin | S ≤ 8 | R ≥ 16 | 82 | 69.6–90.2 | 0.742 | 1.988 | 0.913; 3.064 | 0.002 | 0.551 |
| S. aureus | teicoplanin | S ≤ 2 | R ≥ 4 | 1 | 0.5–2.2 | 0.548 | 0.149 | 0.032; 0.266 | 0.015 | 0.3 |
| S. maltophilia | trimethoprim/ sulfamethoxazole | I ≤ 2 | R ≥ 4 | 10.4 | 5.4–18.7 | 0.533 | 1.083 | 0.203; 1.963 | 0.019 | 0.284 |
| S. marcescens | ertapenem | S ≤ 0.5 | R ≥ 1 | 7.1 | 1.2–25.0 | 0.621 | 3.885 | 0.631; 7.138 | 0.023 | 0.386 |
| S. marcescens | tobramycin | S ≤ 2 | R ≥ 4 | 16.7 | 8.0–30.8 | 0.57 | 1.567 | 0.145; 2.989 | 0.033 | 0.325 |
| Organism | Molecules | Break Points | R (%) | IC95 (R%) | Pearson Coef. γ | β Coef. δ | IC95 β Coef. ε | p-Value | R2 | |
|---|---|---|---|---|---|---|---|---|---|---|
| A. baumannii | ciprofloxacin | I ≤ 1 | R ≥ 2 | 93.9 | 91.3–95.9 | −0.532 | −0.863 | −1.565; −0.16 | 0.019 | 0.283 |
| A. baumannii | colistin | S ≤ 2 | R ≥ 4 | 2.6 | 1.4–4.6 | −0.52 | −0.59 | −1.086; −0.094 | 0.023 | 0.27 |
| A. baumannii | imipenem | S ≤ 2 | R ≥ 8 | 91.8 | 88.0–94.5 | −0.579 | −0.941 | −1.619; −0.263 | 0.009 | 0.335 |
| A. baumannii | meropenem | S ≤ 2 | R ≥ 16 | 93.3 | 90.5–95.3 | −0.496 | −0.794 | −1.504; −0.083 | 0.031 | 0.246 |
| A. baumannii | tobramycin | S ≤ 4 | R ≥ 8 | 89.8 | 85.7–92.9 | −0.501 | −1.061 | −2.07; −0.053 | 0.04 | 0.251 |
| C. tropicalis | fluconazole | S ≤ 2 | R ≥ 8 | 10 | 0.5–45.9 | −0.693 | −4.575 | −8.828; −0.323 | 0.038 | 0.48 |
| C. tropicalis | itraconazole | S ≤ 0.125 | R ≥ 0.25 | 14.3 | 0.8–58.0 | −0.758 | −6.154 | −12.244; −0.064 | 0.048 | 0.574 |
| E. coli | cefepime | S ≤ 1 | R ≥ 8 | 27.7 | 22.6–33.5 | −0.462 | −1.193 | −2.363; −0.022 | 0.046 | 0.214 |
| E. coli | cefotaxime | S ≤ 1 | R ≥ 4 | 41.4 | 34.8–48.4 | −0.457 | −2.002 | −3.998; −0.006 | 0.049 | 0.209 |
| E. coli | ciprofloxacin | S ≤ 0.25 | R ≥ 1 | 48.2 | 42.2–54.2 | −0.531 | −1.888 | −3.431; −0.346 | 0.019 | 0.282 |
| E. coli | gentamicin | S ≤ 2 | R ≥ 4 | 20.2 | 15.8–25.4 | −0.627 | −1.706 | −2.792; −0.621 | 0.004 | 0.393 |
| K. aerogenes | ceftazidime/ avibactam | S ≤ 8 | R ≥ 16 | 1.9 | 0.1–11.2 | −0.539 | −1.296 | −2.509; −0.083 | 0.038 | 0.291 |
| K. pneumoniae | colistin | S ≤ 2 | R ≥ 4 | 3.4 | 2.2–5.3 | −0.575 | −0.381 | −0.659; −0.103 | 0.01 | 0.33 |
| K. pneumoniae | gentamicin | S ≤ 2 | R ≥ 4 | 33 | 29.3–36.9 | −0.478 | −1.352 | −2.623; −0.081 | 0.038 | 0.229 |
| S. aureus | dalbavancin | S ≤ 0.25 | R ≥ 0.5 | 25 | 6.7–57.2 | −0.879 | −13.35 | −21.669; −5.03 | 0.009 | 0.773 |
| S. aureus | levofloxacin | I ≤ 1 | R ≥ 2 | 58.7 | 54.9–62.5 | −0.707 | −1.564 | −2.365; −0.763 | 0.001 | 0.499 |
| S. aureus | penicillin G | S ≤ 0.125 | R ≥ 0.25 | 87.5 | 84.6–89.9 | −0.641 | −0.872 | −1.406; −0.338 | 0.003 | 0.411 |
| S. aureus | rifampin | S ≤ 0.064 | R ≥ 0.12 | 2.7 | 1.7–4.3 | −0.638 | −0.392 | −0.634; −0.15 | 0.003 | 0.407 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ingravalle, F.; Maurici, M.; Vinci, A.; Di Carlo, S.; D’Agostini, C.; Pica, F.; Ciotti, M. Time Trends in Prevalence and Antimicrobial Resistance of Respiratory Pathogens in a Tertiary Hospital in Rome, Italy: A Retrospective Analysis (2018–2023). Antibiotics 2025, 14, 932. https://doi.org/10.3390/antibiotics14090932
Ingravalle F, Maurici M, Vinci A, Di Carlo S, D’Agostini C, Pica F, Ciotti M. Time Trends in Prevalence and Antimicrobial Resistance of Respiratory Pathogens in a Tertiary Hospital in Rome, Italy: A Retrospective Analysis (2018–2023). Antibiotics. 2025; 14(9):932. https://doi.org/10.3390/antibiotics14090932
Chicago/Turabian StyleIngravalle, Fabio, Massimo Maurici, Antonio Vinci, Stefano Di Carlo, Cartesio D’Agostini, Francesca Pica, and Marco Ciotti. 2025. "Time Trends in Prevalence and Antimicrobial Resistance of Respiratory Pathogens in a Tertiary Hospital in Rome, Italy: A Retrospective Analysis (2018–2023)" Antibiotics 14, no. 9: 932. https://doi.org/10.3390/antibiotics14090932
APA StyleIngravalle, F., Maurici, M., Vinci, A., Di Carlo, S., D’Agostini, C., Pica, F., & Ciotti, M. (2025). Time Trends in Prevalence and Antimicrobial Resistance of Respiratory Pathogens in a Tertiary Hospital in Rome, Italy: A Retrospective Analysis (2018–2023). Antibiotics, 14(9), 932. https://doi.org/10.3390/antibiotics14090932








