Surveillance and Characterization of Vancomycin-Resistant and Vancomycin-Variable Enterococci in a Hospital Setting
Abstract
1. Introduction
2. Results
2.1. Collection and Phenotypic Characterization of Isolates
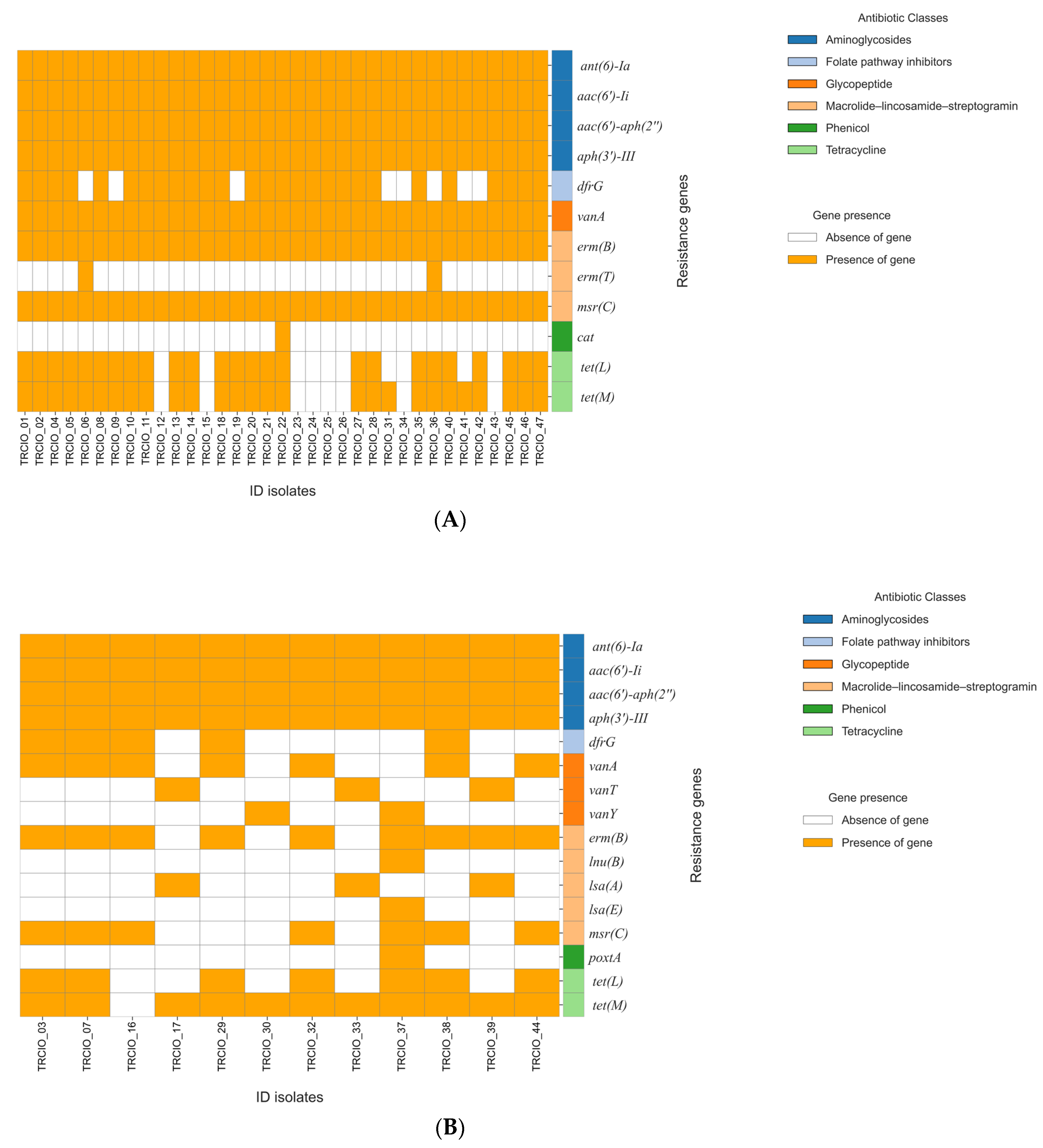
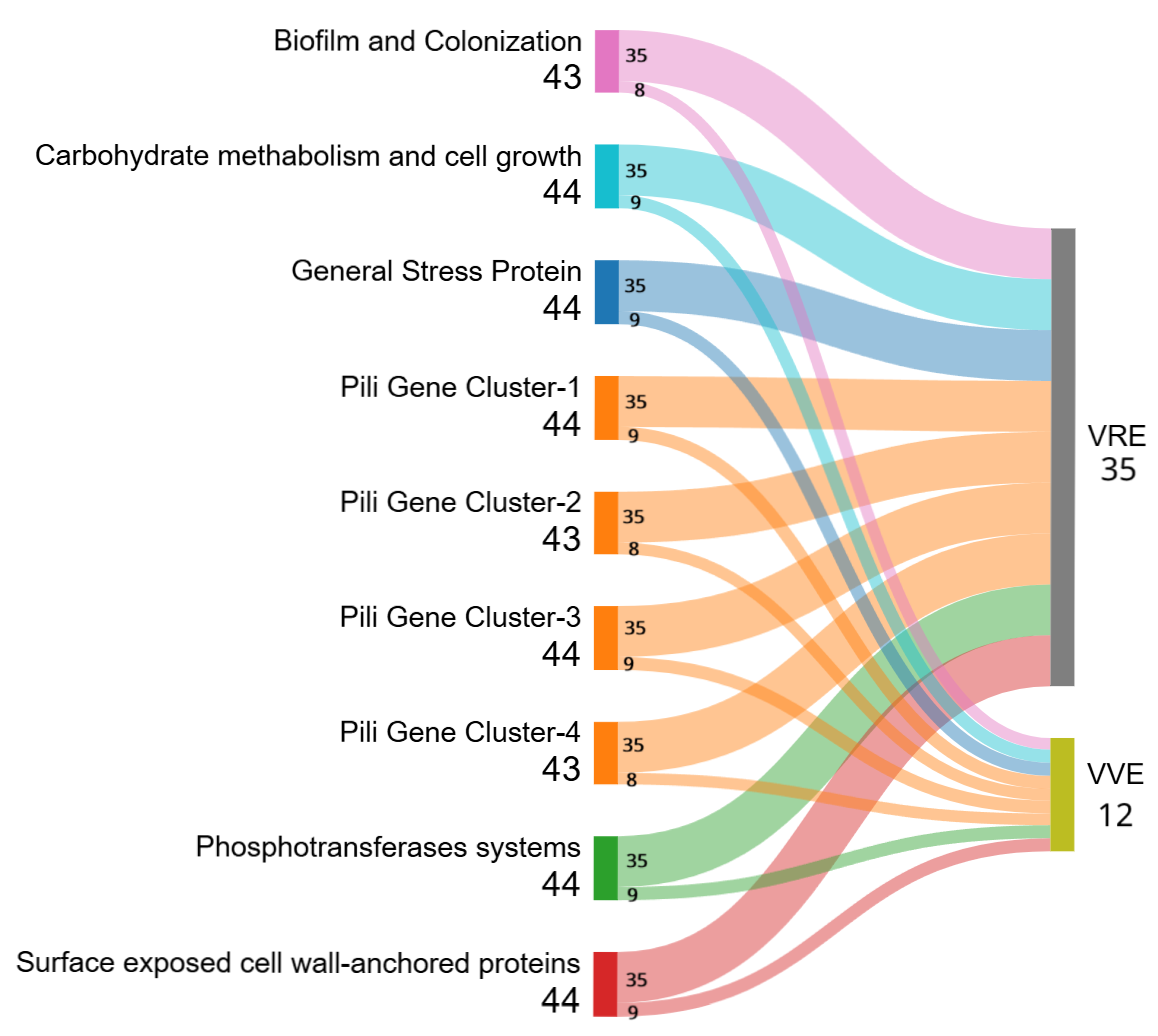
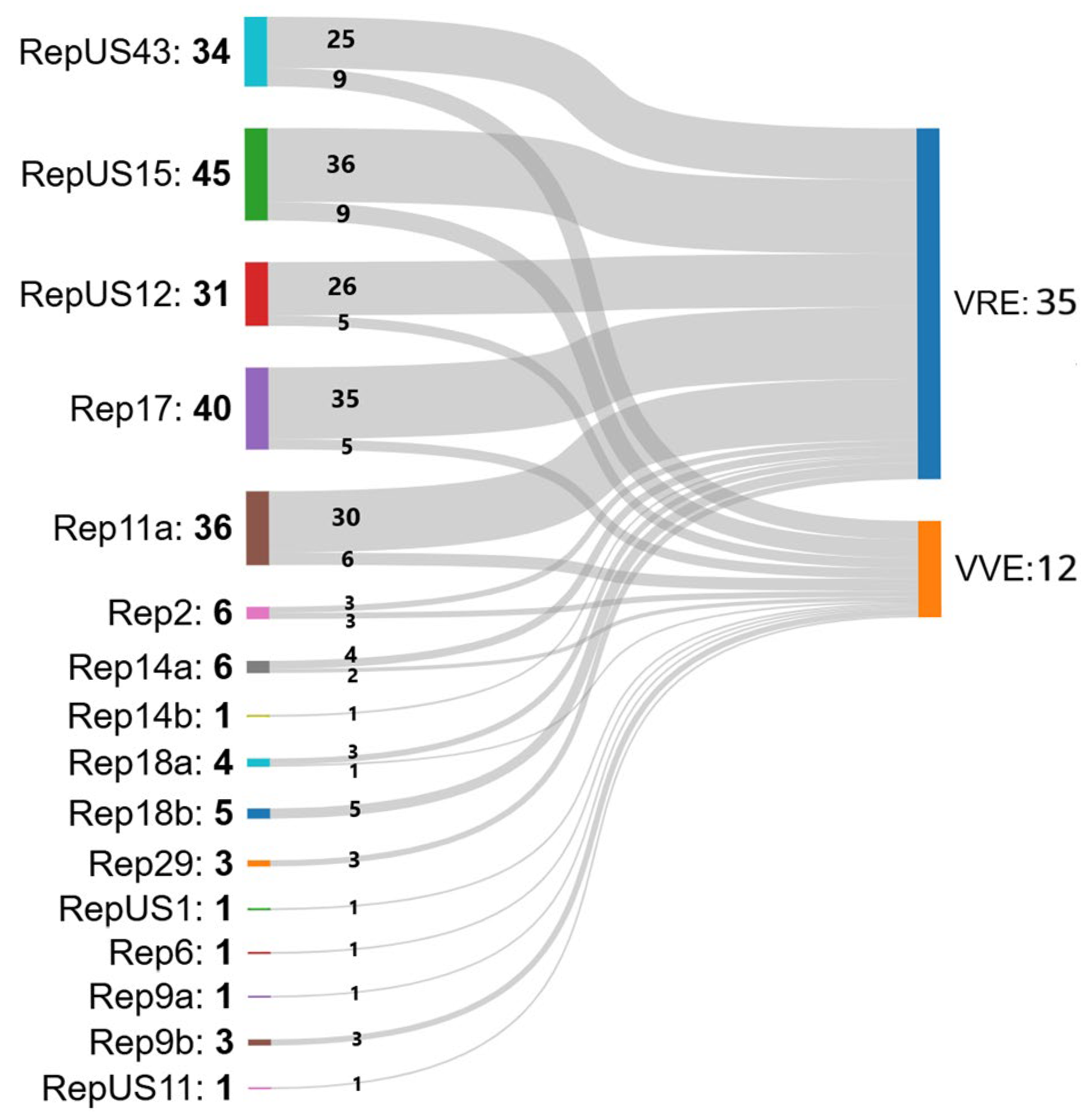
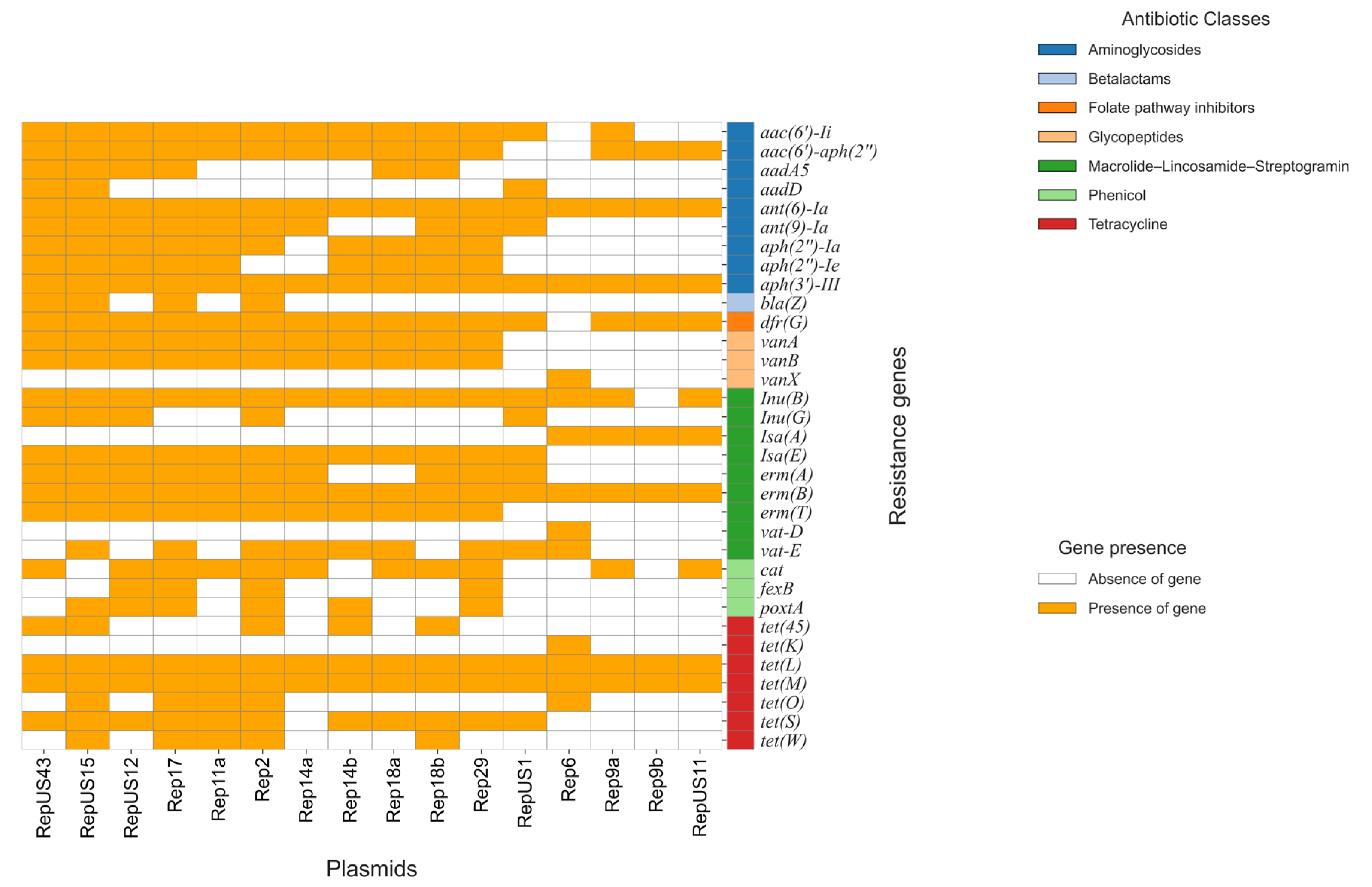
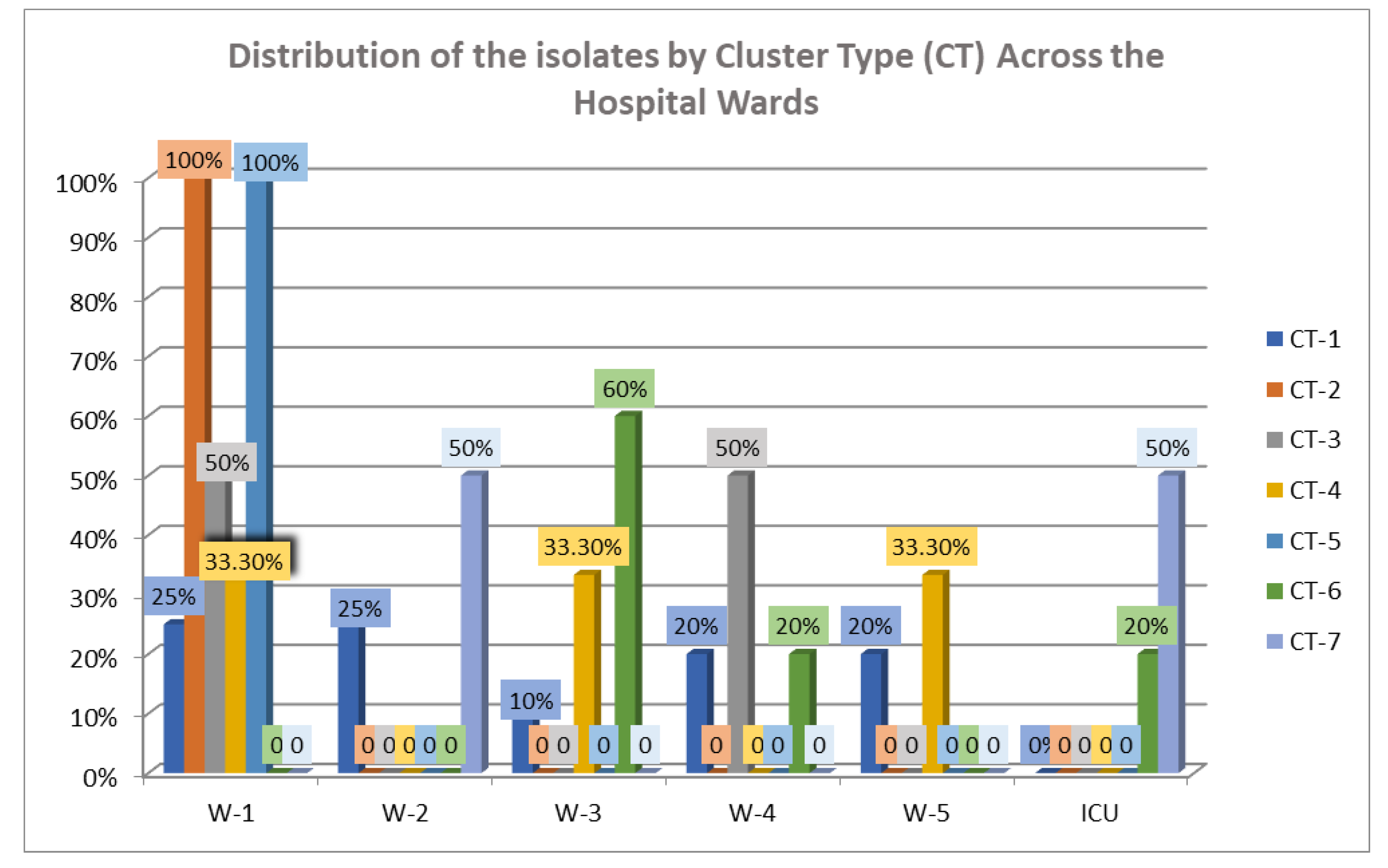
2.2. Whole-Genome Sequencing (WGS) Analysis and Multilocus (ML) Phylogenetic Tree
2.3. cgMLST Analysis
3. Discussion
4. Materials and Methods
4.1. Hospital Setting and PPS
4.2. Rectal Sampling and Phenotypic Characterization of Isolates
4.3. Whole-Genome Sequencing, Multilocus (ML) Phylogenetic Tree and Core-Genome Multilocus Sequence Typing (cgMLST) Analysis
4.3.1. Assembly and Annotations
4.3.2. Pangenomes
4.3.3. AMR Gene Analysis
4.3.4. Pathogenicity and Virulence
4.3.5. Phylogenetic Analysis
4.4. Statistical Evaluation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| VRE | Vancomycin-resistant enterococci |
| VVE | Vancomycin-variable enterococci |
| MA | Molecular assays |
| WGS | Whole-genome sequencing |
| PPS | Point prevalence survey |
| VREfaecium | Vancomycin-resistant Enterococcus faecium |
| ANI | Average nucleotide identity |
| cgMLST | Core-genome multilocus sequence typing |
| ML | Multilocus |
| E. faecium | Enterococcus faecium |
| E. faecalis | Enterococcus faecalis |
| CI | Confidence interval |
| IS | Insertion sequence |
| VVEfaecalis | Vancomycin-variable E. faecalis |
| VVEfaecium | Vancomycin-variable E. faecium |
| CT | Cluster type |
| ST | Sequence type |
| W | Ward |
| W-1 | Hepatology Unit |
| W-2 | Viral Immunodeficiency Unit |
| W-3 | Immune Systemic Infections Unit |
| W-4 | Respiratory System Infectious Diseases Unit |
| W-5 | High Intensity Care Infectious Disease Unit |
| ICU | Intensive Care Unit |
| MICs | Minimum inhibitory concentrations |
| AMP | Ampicillin |
| IMP | Imipenem |
| LZD | Linezolid |
| DAP | Daptomycin |
| TEC | Teicoplanin |
| TG | Tigecycline |
| VAN | Vancomycin |
| GEN-hl | Gentamycin high-level |
| S | Susceptible |
| R | Resistant |
| IE | Insufficient evidence |
| EUCAST | European Committee on Antimicrobial Susceptibility Testing |
| RGI | Resistance gene identifier |
| CARD | Comprehensive Antibiotic Resistance Database |
| CGE | Center for Genomic Epidemiology |
| MGE | Mobile genetic elements |
| MST | Minimum spanning tree |
| AMR | Antimicrobial resistance |
References
- Fiore, E.; Van Tyne, D.; Gilmore, M.S. Pathogenicity of Enterococci. Microbiol. Spectr. 2019, 7. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.N.; Sheriff, E.K.; Duerkop, B.A.; Chatterjee, A. Let Me Upgrade You: Impact of Mobile Genetic Elements on Enterococcal Adaptation and Evolution. J. Bacteriol. 2021, 203, e0017721. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Bertagnolio, S.; Dobreva, Z.; Centner, C.M.; Olaru, I.D.; Donà, D.; Burzo, S.; Huttner, B.D.; Chaillon, A.; Gebreselassie, N.; Wi, T.; et al. WHO global research priorities for antimicrobial resistance in human health. Lancet Microbe 2024, 5, 100902. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Global Report on Infection Prevention and Control. 2024. Available online: https://www.who.int/publications/i/item/9789240103986 (accessed on 6 June 2025).
- Gagnon, S.; Lévesque, S.; Lefebvre, B.; Bourgault, A.M.; Labbé, A.C.; Roger, M. vanA-containing Enterococcus faecium susceptible to vancomycin and teicoplanin because of major nucleotide deletions in Tn1546. J. Antimicrob. Chemother. 2011, 66, 2758–2762. [Google Scholar] [CrossRef] [PubMed]
- Raddaoui, A.; Chebbi, Y.; Frigui, S.; Latorre, J.; Ammeri, R.W.; Abdejlil, N.B.; Torres, C.; Abbassi, M.S.; Achour, W. Genetic characterization of vancomycin-resistant Enterococcus faecium isolates from neutropenic patients in Tunisia: Spread of the pandemic CC17 clone associated with high genetic diversity in Tn1546-like structures. J. Appl. Microbiol. 2024, 135, lxae225. [Google Scholar] [CrossRef] [PubMed]
- Navarra, A.; Cicalini, S.; D’Arezzo, S.; Pica, F.; Selleri, M.; Nisii, C.; Venditti, C.; Cannas, A.; Mazzarelli, A.; Vulcano, A.; et al. Vancomycin-Resistant Enterococci: Screening Efficacy and the Risk of Bloodstream Infections in a Specialized Healthcare Setting. Antibiotics 2025, 14, 304. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Antimicrobial Resistance Surveillance in Europe 2023–2021 Data. Available online: https://www.ecdc.europa.eu/en/publications-data/antimicrobial-resistance-surveillance-europe-2023-2021-data (accessed on 6 June 2025).
- Lee, S.Y.; Nam, J.H.; Kim, J.W.; Kim, S.H.; Yoo, J.S. Prevalence of Vancomycin-Variable Enterococci from the Bloodstream in the Korea Global Antibiotic Resistance Surveillance System, 2017–2022. Antibiotics 2024, 13, 1210. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lu, C.J.; Hung, W.C.; Lan, Z.H.; Lu, P.L.; Lin, C.Y.; Chen, Y.H.; Chen, T.C.; Huang, C.H.; Chang, Y.T.; Lee, C.Y.; et al. Characteristics and Prevalence of Vancomycin-variable Enterococcus faecium bacteremia in southern Taiwan. J. Microbiol. Immunol. Infect. 2024, 57, 926–936. [Google Scholar] [CrossRef] [PubMed]
- Kohler, P.; Eshaghi, A.; Kim, H.C.; Plevneshi, A.; Green, K.; Willey, B.M.; McGeer, A.; Patel, S.N. Toronto Invasive Bacterial Diseases Network (TIBDN). Prevalence of vancomycin-variable Enterococcus faecium (VVE) among vanA-positive sterile site isolates and patient factors associated with VVE bacteremia. PLoS ONE 2018, 13, e0193926. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sivertsen, A.; Pedersen, T.; Larssen, K.W.; Bergh, K.; Rønning, T.G.; Radtke, A.; Hegstad, K. A Silenced vanA Gene Cluster on a Transferable Plasmid Caused an Outbreak of Vancomycin-Variable Enterococci. Antimicrob. Agents Chemother. 2016, 60, 4119–4127. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hansen, T.A.; Pedersen, M.S.; Nielsen, L.G.; Ma, C.M.G.; Søes, L.M.; Worning, P.; Østergaard, C.; Westh, H.; Pinholt, M.; Schønning, K. Emergence of a vancomycin-variable Enterococcus faecium ST1421 strain containing a deletion in vanX. J. Antimicrob. Chemother. 2018, 73, 2936–2940. [Google Scholar] [CrossRef] [PubMed]
- Viswanath, L.S.; Sugumar, M.; Peela, S.C.M.; Walia, K.; Sistla, S. Detection of vancomycin variable enterococci (VVE) among clinical isolates of Enterococcus faecium collected across India-first report from the subcontinent. Indian J. Med. Microbiol. 2022, 40, 285–288. [Google Scholar] [CrossRef] [PubMed]
- Samad, M.A.; Sagor, M.S.; Hossain, M.S.; Karim, M.R.; Mahmud, M.A.; Sarker, M.S.; Shownaw, F.A.; Mia, Z.; Card, R.M.; Agunos, A.; et al. High prevalence of vancomycin non-susceptible and multi-drug resistant enterococci in farmed animals and fresh retail meats in Bangladesh. Vet. Res. Commun. 2022, 46, 811–822. [Google Scholar] [CrossRef] [PubMed]
- Wagner, T.M.; Janice, J.; Schulz, M.; Ballard, S.A.; da Silva, A.G.; Coombs, G.W.; Daley, D.A.; Pang, S.; Mowlaboccus, S.; Stinear, T.; et al. Reversible vancomycin susceptibility within emerging ST1421 Enterococcus faecium strains is associated with rearranged vanA-gene clusters and increased vanA plasmid copy number. Int. J. Antimicrob. Agents 2023, 62, 106849. [Google Scholar] [CrossRef] [PubMed]
- Sinel, C.; Cacaci, M.; Meignen, P.; Guérin, F.; Davies, B.W.; Sanguinetti, M.; Giard, J.C.; Cattoir, V. Subinhibitory Concentrations of Ciprofloxacin Enhance Antimicrobial Resistance and Pathogenicity of Enterococcus faecium. Antimicrob. Agents Chemother. 2017, 61, e02763-16. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Tran, T.T.; Panesso, D.; Gao, H.; Roh, J.H.; Munita, J.M.; Reyes, J.; Diaz, L.; Lobos, E.A.; Shamoo, Y.; Mishra, N.N.; et al. Whole-genome analysis of a daptomycin-susceptible Enterococcus faecium strain and its daptomycin-resistant variant arising during therapy. Antimicrob. Agents Chemother. 2013, 57, 261–268. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Coccitto, S.N.; Cinthi, M.; Simoni, S.; Pocognoli, A.; Zeni, G.; Mazzariol, A.; Morroni, G.; Mingoia, M.; Giovanetti, E.; Brenciani, A.; et al. Genetic analysis of vancomycin-variable Enterococcus faecium clinical isolates in Italy. Eur. J. Clin. Microbiol. Infect. Dis. 2024, 43, 673–682. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Turnidge, J.; Kahlmeter, G.; Cantón, R.; MacGowan, A.; Giske, C.G.; European Committee on Antimicrobial Susceptibility Testing. Daptomycin in the treatment of enterococcal bloodstream infections and endocarditis: A EUCAST position paper. Clin. Microbiol. Infect. 2020, 26, 1039–1043. [Google Scholar] [CrossRef] [PubMed]
- The European Committee on Antimicrobial Susceptibility Testing (EUCAST): 2020 version 10.0 Clinical Breakpoints and Dosing of Antibiotics. Available online: https://www.eucast.org/clinical_breakpoints/ (accessed on 2 January 2020).
- Meziane-Cherif, D.; Stogios, P.J.; Evdokimova, E.; Egorova, O.; Savchenko, A.; Courvalin, P. Structural and Functional Adaptation of Vancomycin Resistance VanT Serine Racemases. mBio 2015, 6, e00806. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wright, G.D.; Molinas, C.; Arthur, M.; Courvalin, P.; Walsh, C.T. Characterization of vanY, a DD-carboxypeptidase from vancomycin-resistant Enterococcus faecium BM4147. Antimicrob. Agents Chemother. 1992, 36, 1514–1518. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Roer, L.; Kaya, H.; Tedim, A.P.; Novais, C.; Coque, T.M.; Aarestrup, F.M.; Peixe, L.; Hasman, H.; Hammerum, A.M.; Freitas, A.R.; et al. VirulenceFinder for Enterococcus faecium and Enterococcus lactis: An enhanced database for detection of putative virulence markers by using whole-genome sequencing data. Microbiol. Spectr. 2024, 12, e0372423. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Tamai, S.; Suzuki, Y. Diversity of Fecal Indicator Enterococci among Different Hosts: Importance to Water Contamination Source Tracking. Microorganisms 2023, 11, 2981. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Surveillance of Antimicrobial Resistance in Europe, 2023 Data—Executive Summary. Available online: https://www.ecdc.europa.eu/en/publications-data/surveillance-antimicrobial-resistance-europe-2023-data-executive-summary (accessed on 7 June 2025).
- Mareković, I.; Markanović, M.; Lešin, J.; Ćorić, M. Vancomycin-Resistant Enterococci: Current Understandings of Resistance in Relation to Transmission and Preventive Strategies. Pathogens 2024, 13, 966. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Meschiari, M.; Kaleci, S.; Monte, M.D.; Dessilani, A.; Santoro, A.; Scialpi, F.; Franceschini, E.; Orlando, G.; Cervo, A.; Monica, M.; et al. Vancomycin resistant enterococcus risk factors for hospital colonization in hematological patients: A matched case-control study. Antimicrob. Resist. Infect. Control 2023, 12, 126. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Thaker, M.N.; Kalan, L.; Waglechner, N.; Eshaghi, A.; Patel, S.N.; Poutanen, S.; Willey, B.; Coburn, B.; McGeer, A.; Low, D.E.; et al. Vancomycin-variable enterococci can give rise to constitutive resistance during antibiotic therapy. Antimicrob. Agents Chemother. 2015, 59, 1405–1410. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Pourshafie, M.R.; Talebi, M.; Saifi, M.; Katouli, M.; Eshraghi, S.; Kühn, I.; Möllby, R. Clonal heterogeneity of clinical isolates of vancomycin-resistant Enterococcus faecium with unique vanS. Trop. Med. Int. Health 2008, 13, 722–727. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jensen, L.B.; Ahrens, P.; Dons, L.; Jones, R.N.; Hammerum, A.M.; Aarestrup, F.M. Molecular analysis of Tn1546 in Enterococcus faecium isolated from animals and humans. J. Clin. Microbiol. 1998, 36, 437–442. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Monteiro da Silva, B.N.; Faria, A.R.; Souza, S.D.S.R.; Colodette, S.S.; Morais, J.M.; Teixeira, L.M.; Merquior, V.L.C. Expression of VanA-type vancomycin resistance in a clinical isolate of Enterococcus faecium showing insertion of IS19 in the vanS gene. Int. J. Antimicrob. Agents 2020, 55, 105897. [Google Scholar] [CrossRef] [PubMed]
- Palazzo, I.C.; Pitondo-Silva, A.; Levy, C.E.; da Costa Darini, A.L. Changes in vancomycin-resistant Enterococcus faecium causing outbreaks in Brazil. J. Hosp. Infect. 2011, 79, 70–74. [Google Scholar] [CrossRef] [PubMed]
- Georges, M.; Odoyo, E.; Matano, D.; Tiria, F.; Kyany’a, C.; Mbwika, D.; Mutai, W.C.; Musila, L. Determination of Enterococcus faecalis and Enterococcus faecium Antimicrobial Resistance and Virulence Factors and Their Association with Clinical and Demographic Factors in Kenya. J Pathog. 2022, 2022, 3129439. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Werner, G.; Fleige, C.; Geringer, U.; van Schaik, W.; Klare, I.; Witte, W. IS element IS16 as a molecular screening tool to identify hospital-associated strains of Enterococcus faecium. BMC Infect. Dis. 2011, 11, 80. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Fioriti, S.; Simoni, S.; Caucci, S.; Morroni, G.; Ponzio, E.; Coccitto, S.N.; Brescini, L.; Cirioni, O.; Menzo, S.; Biavasco, F.; et al. Trend of clinical vancomycin-resistant enterococci isolated in a regional Italian hospital from 2001 to 2018. Braz. J. Microbiol. 2020, 51, 1607–1613. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kavanagh, N.L.; Kinnevey, P.M.; Egan, S.A.; McManus, B.A.; O’Connell, B.; Brennan, G.I.; Coleman, D.C. Protracted transmission and persistence of ST80 vancomycin-resistant Enterococcus faecium clonal complex types CT2933, CT2932 and CT1916 in a large Irish hospital: A 39-month whole-genome sequencing study. J. Hosp. Infect. 2024, 151, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.; Luo, L.; Zhou, H.; Xiao, Y.; Zeng, J.; Zhang, L.; Pu, J.; Zeng, J.; Zhang, N.; Jiang, Y.; et al. Emergence and ongoing outbreak of ST80 vancomycin-resistant Enterococcus faecium in Guangdong province, China from 2021 to 2023: A multicenter, time-series and genomic epidemiological study. Emerg. Microbes Infect. 2024, 13, 2361030. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- van Hal, S.J.; Beukers, A.G.; Timms, V.J.; Ellem, J.A.; Taylor, P.; Maley, M.W.; Newton, P.J.; Ferguson, J.K.; Lee, A.; Chen, S.C.; et al. Relentless spread and adaptation of non-typeable vanA vancomycin-resistant Enterococcus faecium: A genome-wide investigation. J. Antimicrob. Chemother. 2018, 73, 1487–1491. [Google Scholar] [CrossRef] [PubMed]
- The European Committee on Antimicrobial Susceptibility Testing—EUCAST. Clinical Breakpoints—Breakpoints and Guidance. Available online: https://www.eucast.org/ (accessed on 20 December 2024).
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data; ScienceOpen Inc.: Lexington, MA, USA, 2010. [Google Scholar]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mikheenko, A.; Prjibelski, A.; Saveliev, V.; Antipov, D.; Gurevich, A. Versatile genome assembly evaluation with QUAST-LG. Bioinformatics 2018, 34, i142–i150. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Manni, M.; Berkeley, M.R.; Seppey, M.; Simão, F.A.; Zdobnov, E.M. BUSCO Update: Novel and Streamlined Workflows along with Broader and Deeper Phylogenetic Coverage for Scoring of Eukaryotic, Prokaryotic, and Viral Genomes. Mol. Biol. Evol. 2021, 38, 4647–4654. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Pritchard, L.; Glover, R.H.; Humphris, S.; Elphinstone, J.G.; Toth, I.K. Genomics and taxonomy in diagnostics for food security: Soft-rotting enterobacterial plant pathogens. Anal. Methods 2016, 8, 12–24. [Google Scholar] [CrossRef]
- Alcock, B.P.; Huynh, W.; Chalil, R.; Smith, K.W.; Raphenya, A.R.; Wlodarski, M.A.; Edalatmand, A.; Petkau, A.; Syed, S.A.; Tsang, K.K.; et al. CARD 2023: Expanded curation, support for machine learning, and resistome prediction at the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2023, 51, D690–D699. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
| ID Strain | Organism 2 | Type 3 | Date | Antibiotic MIC Values (mg/L) 1 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| AMP | IMP 4 | LZD | DAP 5 | TEC | TG | VAN | GEN-hl | ||||
| TRCIO_01 | E. faecium | VRE | 6 May 2024 | >16, R | 2, S | >4 | >8, R | 0.25, S | >8, R | >500, R | |
| TRCIO_02 | E. faecium | VRE | 16 April, 2024 | >16, R | 2, S | >4 | >8, R | 0.25, S | >8, R | >500, R | |
| TRCIO_03 | E. faecium | VVE | 6 May 2024 | >16, R | 2, S | 4 | ≤0.5, S | 0.25, S | ≤0.5, S | >500, R | |
| TRCIO_04 | E. faecium | VRE | 17 April 2024 | >16, R | 2, S | >4 | >8, R | 0.25, S | >8, R | >500, R | |
| TRCIO_05 | E. faecium | VRE | 18 April 2024 | >16, R | 2, S | >4 | >8, R | 0.25, S | >8, R | >500, R | |
| TRCIO_06 | E. faecium | VRE | 3 May 2024 | >16, R | 2, S | 4 | >8, R | 0.125, S | >8, R | >500, R | |
| TRCIO_07 | E. faecium | VVE | 6 May 2024 | >16, R | 2, S | >4 | ≤0.5, S | 0.25, S | ≤0.5, S | >500, R | |
| TRCIO_08 | E. faecium | VRE | 6 May 2024 | >16, R | 2, S | >4 | >8, R | 0.25, S | >8, R | >500, R | |
| TRCIO_09 | E. faecium | VRE | 3 April 2024 | >16, R | 4, S | 4 | >8, R | ≤0.125, S | >8, R | >500, R | |
| TRCIO_10 | E. faecium | VRE | 18 April 2024 | >16, R | 2, S | >4 | >8, R | 0.25, S | >8, R | >500, R | |
| TRCIO_11 | E. faecium | VRE | 6 May 2024 | >16, R | 2, S | >4 | >8, R | 0.25, S | >8, R | >500, R | |
| TRCIO_12 | E. faecium | VRE | 6 May 2024 | >16, R | 4, S | 4 | >8, R | 0.125, S | >8, R | >500, R | |
| TRCIO_13 | E. faecium | VRE | 6 May 2024 | >16, R | 2, S | >4 | >8, R | 0.25, S | >8, R | >500, R | |
| TRCIO_14 | E. faecium | VRE | 7 April 2024 | >16, R | 2, S | 4 | >8, R | 0.25, S | >8, R | >500, R | |
| TRCIO_15 | E. faecium | VRE | 25 January 2024 | >16, R | 2, S | 4 | >8, R | ≤0.125, S | 8, R | >500, R | |
| TRCIO_16 | E. faecium | VVE | 6 May 2024 | >16, R | 1, S | 4 | ≤0.5, S | ≤0.125, S | ≤0.5, S | >500, R | |
| TRCIO_17 | E. faecalis | VVE | 6 May 2024 | 2, S | 2, S | 1, S | 4 | ≤0.5, S | ≤0.125, S | 1, S | ≤500, S |
| TRCIO_18 | E. faecium | VRE | 6 May 2024 | >16, R | 4, S | >4 | >8, R | 0.25, S | >8, R | >500, R | |
| TRCIO_19 | E. faecium | VRE | 6 May 2024 | >16, R | 2, S | 4 | >8, R | 0.125, S | >8, R | >500, R | |
| TRCIO_20 | E. faecium | VRE | 6 May 2024 | >16, R | 2, S | >4 | >8, R | 0.25, S | >8, R | >500, R | |
| TRCIO_21 | E. faecium | VRE | 20 February 2024 | >16, R | 2, S | 4 | ≤0.5, S | 0.25, S | >8, R | >500, R | |
| TRCIO_22 | E. faecium | VRE | 1 May 2024 | >16, R | 2, S | 4 | >8, R | 0.25, S | >8, R | ≤500, S | |
| TRCIO_23 | E. faecium | VRE | 11 April 2024 | >16, R | 2, S | 4 | >8, R | ≤0.125, S | >8, R | >500, R | |
| TRCIO_24 | E. faecium | VRE | 26 April 2024 | >16, R | 2, S | 2 | >8, R | ≤0.125, S | >8, R | ≤500, S | |
| TRCIO_25 | E. faecium | VRE | 14 January 2024 | 16, R | 2, S | 4 | >8, R | ≤0.125, S | >8, R | >500, R | |
| TRCIO_26 | E. faecium | VRE | 4 May 2024 | >16, R | 2, S | 4 | >8, R | <0.125, S | >8, R | >500, R | |
| TRCIO_27 | E. faecium | VRE | 6 May 2024 | >16, R | 2, S | >4 | >8, R | 0.25, S | >8, R | >500, R | |
| TRCIO_28 | E. faecium | VRE | 27 April 2024 | >16, R | >4, R | >4 | >8, R | ≤0.125, S | >8, R | >500, R | |
| TRCIO_29 | E. faecium | VVE | 30 April 2024 | >16, R | 2, S | >4 | ≤0.5, S | ≤0.25, S | ≤0.5, S | >500, R | |
| TRCIO_30 | E. faecium | VVE | 6 May 2024 | 4, S | 1, S | >4 | ≤0.5, S | ≤0.125, S | 2, S | ≤500, S | |
| TRCIO_31 | E. faecium | VRE | 6 May 2024 | >16, R | 2, S | 2 | >8, R | 0.125, S | >8, R | ≤500, S | |
| TRCIO_32 | E. faecium | VVE | 6 May 2024 | >16, R | 2, S | >4 | ≤0.5, S | 0.125, S | 1, S | >500, R | |
| TRCIO_33 | E. faecalis | VVE | 6 May 2024 | 2, S | 4, S | 2, S | 2 | ≤0.5, S | ≤0.125, S | 2, S | >500, R |
| TRCIO_34 | E. faecium | VRE | 6 May 2024 | >16, R | 4, S | >4 | >8, R | 0.125, S | >8, R | >500, R | |
| TRCIO_35 | E. faecium | VRE | 6 May 2024 | >16, R | 2, S | 4 | >8, R | 0.25, S | >8, R | >500, R | |
| TRCIO_36 | E. faecium | VRE | 6 May 2024 | >16, R | 2, S | 4 | >8, R | 0.25, S | >8, R | >500, R | |
| TRCIO_37 | E. faecium | VVE | 6 May 2024 | >16, R | 2, S | 4 | ≤0.5, S | 0.125, S | 2, S | >500, R | |
| TRCIO_38 | E. faecium | VVE | 6 May 2024 | >16, R | 2, S | >4 | ≤0.5, S | ≤0.125, S | ≤0.5, S | >500, R | |
| TRCIO_39 | E. faecalis | VVE | 3 May 2024 | 2, S | 2, S | 1, S | 2 | ≤0.5, S | ≤0.125, S | 1, S | ≤500, S |
| TRCIO_40 | E. faecium | VRE | 6 May 2024 | >16, R | 2, S | >4 | >8, R | 0.25, S | >8, R | >500, R | |
| TRCIO_41 | E. faecium | VRE | 9 April 2024 | >16, R | 2, S | 2 | >8, R | 0.125, S | >8, R | ≤500, S | |
| TRCIO_42 | E. faecium | VRE | 6 May 2024 | >16, R | 2, S | 4 | >8, R | ≤0.125, S | >8, R | >500, R | |
| TRCIO_43 | E. faecium | VRE | 25 December 2023 | >16, R | 4, S | 4 | >8, R | ≤0.125, S | >8, R | >500, R | |
| TRCIO_44 | E. faecium | VVE | 6 May 2024 | >16, R | 4, S | 4 | ≤0.5, S | 0.125, S | ≤0.5, S | >500, R | |
| TRCIO_45 | E. faecium | VRE | 24 April 2024 | >16, R | 2, S | >4 | >8, R | 0.25, S | >8, R | >500, R | |
| TRCIO_46 | E. faecium | VRE | 6 May 2024 | >16, R | 2, S | >4 | >8, R | 0.25, S | >8, R | >500, R | |
| TRCIO_47 | E. faecium | VRE | 6 May 2024 | >16, R | 2, S | >4 | >8, R | 0.125, S | >8, R | >500, R | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rotondo, C.; Antonelli, V.; Rossi, A.; D’Arezzo, S.; Selleri, M.; Properzi, M.; Turco, S.; Chillemi, G.; Dimartino, V.; Venditti, C.; et al. Surveillance and Characterization of Vancomycin-Resistant and Vancomycin-Variable Enterococci in a Hospital Setting. Antibiotics 2025, 14, 795. https://doi.org/10.3390/antibiotics14080795
Rotondo C, Antonelli V, Rossi A, D’Arezzo S, Selleri M, Properzi M, Turco S, Chillemi G, Dimartino V, Venditti C, et al. Surveillance and Characterization of Vancomycin-Resistant and Vancomycin-Variable Enterococci in a Hospital Setting. Antibiotics. 2025; 14(8):795. https://doi.org/10.3390/antibiotics14080795
Chicago/Turabian StyleRotondo, Claudia, Valentina Antonelli, Alberto Rossi, Silvia D’Arezzo, Marina Selleri, Michele Properzi, Silvia Turco, Giovanni Chillemi, Valentina Dimartino, Carolina Venditti, and et al. 2025. "Surveillance and Characterization of Vancomycin-Resistant and Vancomycin-Variable Enterococci in a Hospital Setting" Antibiotics 14, no. 8: 795. https://doi.org/10.3390/antibiotics14080795
APA StyleRotondo, C., Antonelli, V., Rossi, A., D’Arezzo, S., Selleri, M., Properzi, M., Turco, S., Chillemi, G., Dimartino, V., Venditti, C., Guerci, S., Gallì, P., Nisii, C., Arcangeli, A., Caraffa, E., Cicalini, S., & Fontana, C. (2025). Surveillance and Characterization of Vancomycin-Resistant and Vancomycin-Variable Enterococci in a Hospital Setting. Antibiotics, 14(8), 795. https://doi.org/10.3390/antibiotics14080795






